Abstract
In 271 Enterobacter blood culture isolates from 12 hospitals, extended-spectrum beta-lactamase (ESBL) prevalence varied between 0% and 30% per hospital. High prevalence was associated with dissemination, indicating the potential relevance of infection control measures. Screening with cefepime or Vitek 2, followed by a cefepime/cefepime-clavulanate Etest, was an accurate strategy for ESBL detection in Enterobacter isolates (positive predictive value, 100%; negative predictive value, 99%).
TEXT
No guidelines have been issued for extended-spectrum beta-lactamase (ESBL) detection in members of the Enterobacteriaceae with inducible chromosomal AmpC beta-lactamases, such as Enterobacter spp., although these species may cause hospital outbreaks (7, 9, 10), are frequently multidrug resistant (MDR) (9, 10), and may constitute a reservoir for plasmid-mediated ESBLs (3, 12, 13) for other Enterobacteriaceae species. The absence of such recommendations likely has two reasons. First, phenotypic detection of ESBLs in members of the Enterobacteriaceae coexpressing an AmpC beta-lactamase is complex, because AmpC expression may mask the synergy required for ESBL detection between third-generation cephalosporins and clavulanic acid. This problem may be circumvented by demonstrating synergy between clavulanic acid and cefepime, a fourth-generation cephalosporin hydrolyzed by ESBLs but generally not by AmpC beta-lactamases (11, 13). Second, the outcome of an ESBL detection test in Enterobacteriaceae spp. with inducible AmpC is considered of limited therapeutic consequence, since most clinicians consider cephalosporins, including cefepime, inappropriate for treatment of infections caused by these species (6, 8). As a result, data are sparse on ESBL prevalence in Enterobacteriaceae spp. with inducible chromosomal AmpC beta-lactamases.
The aims of this study were (i) to develop a phenotypic ESBL detection strategy in Enterobacter species for the routine clinical laboratory and (ii) to determine whether infection control measures could potentially reduce the ESBL prevalence in Enterobacter species by assessing the association between prevalence and clonal relatedness of ESBL-positive isolates per hospital.
For this study, all Enterobacter blood culture isolates (one per patient) obtained in 2006 and 2007 from 12 Dutch laboratories were included. Identification was performed by the participating laboratories using either Vitek 2 (bioMérieux, France) or Phoenix (Becton Dickinson). A total of 271 blood culture isolates were included; 227 (84%) were Enterobacter cloacae and 44 (16%) were Enterobacter aerogenes. Based on detection of TEM, SHV, and CTX-M ESBL genes by sequencing, 37 (14%) were ESBL producers (36 E. cloacae isolates and 1 E. aerogenes isolate). The most prevalent ESBL genes were CTX-M-9 (54%) and SHV-12 (38%), in accordance with results from previous studies (1, 7, 9, 13). CTX-M-15, CTX-M-3, and CTX-M-39 were each detected once.
To develop a phenotypic ESBL detection strategy, the Vitek 2 advanced expert system (AES) (version 5.01; AST N048 cards) and MIC breakpoints of several indicator cephalosporins were evaluated, using detection of ESBL genes as the reference test. Susceptibility testing was performed by broth microdilution (BMD), except for cefpodoxime (Etest; bioMérieux, Marcy l'Etoile, France), using CLSI and EUCAST breakpoints. As a confirmatory phenotypic test, the PM/PML Etest (cefepime/cefepime plus clavulanic acid) was evaluated.
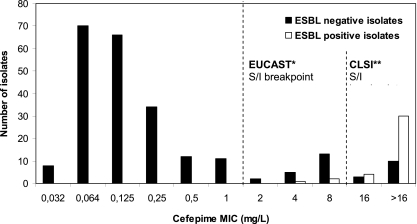
The test characteristics of the ESBL screening methods based on different cephalosporin MICs and the Vitek 2 AES are shown in Table 1. For all cephalosporins included, the MIC-based screening method provided a sensitivity and negative predictive value (NPV) of 100%. Using the Vitek 2 AES, the sensitivity was 92% (34/37) and the NPV was 99% (203/206). However, since a cefepime MIC of >1 mg/liter as determined by BMD (Fig. 1) or an ESBL warning of the Vitek 2 AES had the highest specificity (86 to 87% versus 37 to 68% for the other methods), these screening methods performed best. The relative high specificity will require a lower number of ESBL confirmation tests of the isolates (25% versus 42 to 68%) compared to the indicator cephalosporins recommended by CLSI for ESBL screening in Escherichia coli and Klebsiella spp. (Table 1).
Table 1.
Test characteristics of screening methods for ESBL production in 271 Enterobacter isolates (37 ESBL positive; 234 ESBL negative), using genotypic detection of ESBL as the reference methoda
| ESBL screening method | MIC (mg/liter) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Isolates meeting screening criteria (% of all isolates) (n = 271) |
|---|---|---|---|---|---|---|
| Broth microdilution | ||||||
| Ceftriaxoneb and/or ceftazidimeb | >1 | 100 | 63 | 30 | 100 | 124 (46) |
| Cefotaximeb and/or ceftazidime | >1 | 100 | 68 | 33 | 100 | 113 (42) |
| Cefpodoximeb | >4 | 100 | 37 | 20 | 100 | 185 (68) |
| Cefepime | >1 | 100 | 86d | 53d | 100 | 70 (26)d |
| Vitek 2 | ||||||
| AES “ESBL production” | 92c | 87d | 52d | 99c | 65 (24)d |
PPV, positive predictive value; NPV, negative predictive value; AES, advanced expert system.
Indicator cephalosporins and ESBL screening breakpoints recommended by CLSI for ESBL screening in E. coli, Klebsiella spp., and Proteus mirabilis. These breakpoints are equivalent to the EUCAST clinical susceptible/intermediate breakpoints, except for cefpodoxime (EUCAST susceptible breakpoint, ≤1 mg/liter).
P > 0.05 for comparison of sensitivity and NPV between Vitek 2 AES and the broth microdilution ESBL screening methods. Three ESBL-producing E. cloacae isolates (2 CTX-M-9, 1 SHV-12) were identified as ESBL-positive “high-level cephalosporinase” producers instead of ESBL producers.
P < 0.001 for comparison of specificity, PPV, and number of isolates meeting screening criteria between Vitek 2 AES or the MIC of cefepime (>1 mg/liter) and the other broth microdilution ESBL screening methods.
Fig. 1.
Distribution of cefepime MICs in Enterobacter blood culture isolates. *, 0% of the ESBL positive isolates and 86% of the ESBL negative isolates were susceptible to cefepime using the EUCAST breakpoint. **, 8% of the ESBL positive isolates and 94% of the ESBL negative isolates were susceptible to cefepime using the CLSI breakpoint. S/I, susceptible/intermediate.
It should be noted that a cefepime MIC of >1 mg/liter measured with BMD methodology was 100% sensitive, while a cefepime MIC of >1 mg/liter measured with Vitek 2 had a sensitivity of 54% (P < 0.001) (data not shown). This difference is explained by significantly lower cefepime MIC results from Vitek 2 compared to those from BMD (a MIC90 of 2 mg/liter versus a MIC90 of >16 mg/liter, respectively; P < 0.001), as reported by others (5).
The PM/PML Etest was evaluated as a confirmatory test on 124 isolates with a BMD MIC of ceftriaxone of >1 mg/liter and/or a MIC of ceftazidime of >1 mg/liter. The test characteristics were as follows: sensitivity, 86% (32/37); specificity, 95% (83/87); positive predictive value (PPV), 100% (32/32); and NPV, 99% (83/84). The test result was false negative in one CTX-M-9-positive isolate and off range in eight isolates, of which four contained an ESBL gene. These test characteristics are in line with a previous report using the less-expensive cefepime-clavulanate combination disks (13).
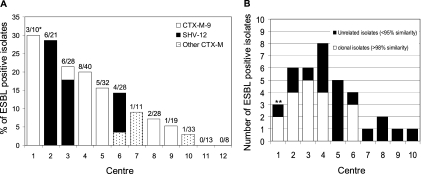
The ESBL prevalence varied between 0% and 30% per hospital (Fig. 2). A prevalence of >14% was observed in 6 hospitals. DiversiLab strain typing (4) demonstrated that a high prevalence was associated with a high level of clonality of E. cloacae in 5 hospitals (Fig. 2). All isolates within a single clonal lineage carried the same ESBL gene. Interestingly, in 5 of the 7 hospitals with different strain types, only one ESBL genotype was found, either exclusively SHV-12 or CTX-M-9, suggesting that plasmid transfer may have occurred. The ESBL prevalence in Enterobacter spp. was almost twice as high as the ESBL prevalence in invasive E. coli and Klebsiella pneumonia isolates from the same period in the Netherlands (4.7% and 6.9%, respectively [http://www.rivm.nl/earss/database/]). A likely explanation for this difference was the lack of a laboratory protocol for ESBL detection in Enterobacter spp., resulting in a lack of infection control measures and thus an increased likelihood of nosocomial spread.
Fig. 2.
ESBL prevalence (A) and clonal relatedness (B) in Enterobacter blood culture isolates (n = 271) per participating center. (A) Numbers on top of bars in panel A indicate the number of ESBL-positive Enterobacter isolates/total number of Enterobacter blood culture isolates per participating center (*). (B) Center 1, genotype 104 (n = 2); center 2, genotype 110 (n = 4); center 3, genotype 85 (n = 5); center 4, genotype 104 (n = 2) and genotype 54 (n = 2); center 6, genotype 85 (n = 3) (**). Clonal relatedness implicates >98% similarity in DiversiLab typing (9).
All isolates were susceptible to meropenem, imipenem, and tigecycline. Of the ESBL-producing isolates, 40% were MDR, i.e., simultaneously resistant to ciprofloxacin, cotrimoxazole, and tobramycin or gentamicin, versus 3% in the non-ESBL isolates. Of the 4 ESBL-positive strains involved in clonal dissemination (Fig. 2), 3 were MDR, including the 2 clones that were detected in different hospitals (genotypes 104 and 85). An increasing prevalence of MDR strains will augment the use of carbapenems, an undesirable development in the face of the worldwide emergence of carbapenemase-producing Enterobacteriaceae isolates (2).
In conclusion, the results of this study provide a practical and accurate strategy for phenotypic detection of ESBLs in Enterobacter spp. We recommend using this strategy in the routine clinical setting since infection control measures can potentially reduce the ESBL prevalence in Enterobacter spp., as a clear association was observed per hospital between a high prevalence and clonal relatedness.
Acknowledgments
This study was funded by an unrestricted grant from MSD (Whitehouse Station, NJ) and Wyeth Pharmaceuticals (Collegeville, PA).
For their help and for kindly providing the isolates, we thank Karola Waar (Izore Center for Infectious Diseases Friesland, Leeuwarden, the Netherlands), Gijs Ruijs (Laboratory of Clinical Microbiology and Infectious Diseases, Isala klinieken, Zwolle, the Netherlands), Fre Sebens (Department of Medical Microbiology and Infection Control, Deventer Ziekenhuis, Deventer, the Netherlands), and Willem Vogels (Martini Hospital Groningen, Groningen, the Netherlands).
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Biendo M., et al. 2008. Molecular typing and characterization of extended-spectrum TEM, SHV and CTX-M beta-lactamases in clinical isolates of Enterobacter cloacae. Res. Microbiol. 159:590–594 [DOI] [PubMed] [Google Scholar]
- 2. Bilavsky E., Schwaber M. J., Carmeli Y. 2010. How to stem the tide of carbapenemase-producing enterobacteriaceae?: proactive versus reactive strategies. Curr. Opin. Infect. Dis. 23:327–331 [DOI] [PubMed] [Google Scholar]
- 3. Crowley B., Ratcliffe G. 2003. Extended-spectrum beta-lactamases in Enterobacter cloacae: underestimated but clinically significant! J. Antimicrob. Chemother. 51:1316–1317 [DOI] [PubMed] [Google Scholar]
- 4. Fluit A. C., et al. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48:3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lat A., et al. 2011. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs against KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J. Clin. Microbiol. 49:1795–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Limaye A. P., Gautom R. K., Black D., Fritsche T. R. 1997. Rapid emergence of resistance to cefepime during treatment. Clin. Infect. Dis. 25:339–340 [DOI] [PubMed] [Google Scholar]
- 7. Manzur A., et al. 2007. Nosocomial outbreak due to extended-spectrum-beta-lactamase-producing Enterobacter cloacae in a cardiothoracic intensive care unit. J. Clin. Microbiol. 45:2365–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Medeiros A. A. 1997. Relapsing infection due to Enterobacter species: lessons of heterogeneity. Clin. Infect. Dis. 25:341–342 [DOI] [PubMed] [Google Scholar]
- 9. Paauw A., Fluit A. C., Verhoef J., Leverstein-van Hall M. A. 2006. Enterobacter cloacae outbreak and emergence of quinolone resistance gene in Dutch hospital. Emerg. Infect. Dis. 12:807–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Potron A., et al. 2009. Nosocomial spread of ESBL-positive Enterobacter cloacae co-expressing plasmid-mediated quinolone resistance Qnr determinants in one hospital in France. J. Antimicrob. Chemother. 64:653–654 [DOI] [PubMed] [Google Scholar]
- 11. Stürenburg E., Sobottka I., Noor D., Laufs R., Mack D. 2004. Evaluation of a new cefepime-clavulanate ESBL Etest to detect extended-spectrum beta-lactamases in an Enterobacteriaceae strain collection. J. Antimicrob. Chemother. 54:134–138 [DOI] [PubMed] [Google Scholar]
- 12. Szabó D., et al. 2005. SHV-type extended-spectrum beta-lactamase production is associated with reduced cefepime susceptibility in Enterobacter cloacae. J. Clin. Microbiol. 43:5058–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Towne T. G., Lewis J. S., Herrera M., Wickes B., Jorgensen J. H. 2010. Detection of SHV-type extended-spectrum beta-lactamase in Enterobacter isolates. J. Clin. Microbiol. 48:298–299 [DOI] [PMC free article] [PubMed] [Google Scholar]