Abstract
Open reading frame 57 (ORF57) of gamma-2 herpesviruses is a key regulator of viral gene expression. It has been reported to enhance the expression of viral genes by transcriptional, posttranscriptional, or translational activation mechanisms. Previously we have shown that the expression of gH and gL of rhesus monkey rhadinovirus (RRV), a close relative of the human Kaposi's sarcoma-associated herpesvirus (KSHV), could be dramatically rescued by codon optimization as well as by ORF57 coexpression (J. P. Bilello, J. S. Morgan, and R. C. Desrosiers, J. Virol. 82:7231–7237, 2008). We show here that ORF57 coexpression and codon optimization had similar effects, except that the rescue of expression by codon optimization was temporally delayed relative to that of ORF57 coexpression. The transfection of gL mRNA directly into cells with or without ORF57 coexpression and with or without codon optimization recapitulated the effects of these modes of induction on transfected DNA. These findings suggested an important role for the enhancement of mRNA stability and/or the translation of mRNA for these very different modes of induced expression. This conclusion was confirmed by several different measures of gH and gL mRNA stability and accumulation with or without ORF57 coexpression and with or without codon optimization. Our results indicate that RRV gH and gL expression is severely limited by the stability of the mRNA and that ORF57 coexpression and codon optimization independently induce gH and gL expression principally by allowing accumulation and translation of these mRNAs.
INTRODUCTION
Rhesus monkey rhadinovirus (RRV) is a gamma-2 herpesvirus that is closely related to human Kaposi's sarcoma-associated herpesvirus (KSHV). The genomic organization of RRV is quite similar to that of KSHV; most of its genes are in corresponding locations and have the same polarity between the two viruses (1, 11, 36). ORF57 orthologues are expressed early in the lytic phase of herpesvirus replication (24, 28) and are conserved in all herpesviruses, reflecting an indispensable role in viral replication (15, 25). Although ORF57 orthologues have different nomenclatures among different viruses (MTA or ORF57 for KSHV, SM for Epstein-Barr virus, ICP27 for herpes simplex virus, etc.), they all function to enhance the expression of delayed-early and late genes of the virus (27, 30, 40). The deletion or interruption of ORF57 orthologues is deleterious for infectious virion production, demonstrating their critical role for virus replication (15, 25). However, despite similar activity in inducing viral gene expression, a wide variety of different mechanisms have been reported for the different ORF57 orthologues, ranging from transcription to posttranscription to translation.
KSHV ORF57 activates the de novo expression of viral genes in a promoter-specific manner, including genes for ori-Lyt (L), kaposin, ORF57, and thymidine kinase (TK) (33). The transcriptional activation of these promoters could be accomplished by ORF57 protein alone, or it could be further enhanced by the cooperation with the ORF50 gene product (also known as RTA) (20, 28, 33). However, as the name MTA (mRNA transcript accumulation) in KSHV implies, a posttranscriptional mechanism of activation by ORF57 orthologues has been studied and documented most extensively (4, 14, 21, 29). It has been reported that increased expression appears to be achieved in many cases via the facilitation of the nuclear export of target mRNAs (4, 6, 8, 29, 41). Many herpesviral mRNAs, most of which are intronless and therefore do not undergo splicing, seem to be poor substrates for nuclear export by the regular cellular transport machineries. The facilitation of nuclear export may be achieved at least in part by the subversion of cellular RNA export machineries for the efficient expression of viral genes (8, 17, 21, 41). ORF57 binds to RNA export factor (REF; also known as ALY), which is one component of the transcription export (TREX) complex. Subsequently, it recruits the complete TREX complex and Tip-associated protein (TAP; also known as NFX1) to the vicinity of its target mRNA (4, 7, 8, 29). As a result, many intronless herpesviral mRNAs can form ribonucleoprotein (RNP) particles and be efficiently exported to the cytoplasm.
ORF57 also has been reported to enhance the accumulation of target mRNA levels. KSHV ORF57 has been shown to enhance, by as-yet unknown mechanisms, mRNA levels of ORF59 and nut-1/polyadenylated nuclear (PAN) RNA (20, 26). However, it has been difficult to demonstrate experimentally that this enhancement of mRNA accumulation is independent of nuclear export. Still, the fact that nut-1/PAN RNA, which always localizes to the nucleus and is not exported to the cytoplasm (9, 39), is enhanced by ORF57 (15, 20, 25) supports the idea that enhanced RNA accumulation in this case is independent of nuclear export (35). Furthermore, the nuclear and cytoplasmic fractionation of KSHV ORF59 RNA showed no sign of enhancement of nuclear RNA export (32).
Apart from transcriptional enhancement, another unique function of ORF57 orthologues is the enhancement of translation. In a tethered function assay of Xenopus oocytes, herpes simplex virus (HSV) ICP27 has been shown to activate translation (22). Also, KSHV ORF57 activated the translation of intronless mRNAs through direct interaction with PYM, subsequently recruiting 48S preinitiation complex onto newly exported mRNAs (5). Thus, the detailed action of ORF57 orthologues seems to be quite different depending not only on the viral origin but also on the individual target gene.
In a previous study from our group, it was shown that RRV glycoproteins gH and gL are difficult to express from a standard expression cassette and that this blockage can be overcome either by codon optimization or by the coexpression of RRV ORF57 (3). Here, we studied the detailed aspects of gH and gL induction by RRV ORF57 and by codon optimization. By directly transfecting RNA, we have been able to bypass the step of nuclear RNA egress. By so doing, we have been able to demonstrate direct effects of ORF57 and codon optimization on allowing high levels of gL mRNA translation. Our additional studies demonstrate that RRV ORF57 greatly enhances steady-state levels of RRV RNAs, corresponding with high levels of gH and gL protein expression. By comparing the degradation kinetics of codon-optimized gH RNA in the absence/presence of ORF57, we further show that RRV ORF57 greatly enhances the target RNA level by slowing down its degradation. Our results demonstrate the role of RRV ORF57 in target RNA stabilization and subsequent translation independently of RNA egress from the nucleus.
MATERIALS AND METHODS
Plasmids, cell culture, and transfection.
The cDNA sequences encoding glycoprotein gH and gL of RRV strain 26-95 and codon-optimized (c.o.) versions of the sequences were cloned in a pCDNA6-v5-hisA vector (Invitrogen). The cDNA sequences encoding ORF57 of KSHV, HSV, and RRV (2, 12, 27) were cloned in pEF1-v5-hisA (Invitrogen). The prespliced form of the ORF57 sequence was amplified directly from the viral genome of the RRV 26-95 strain and cloned into pEF1-v5-hisA. The pEFIRES-p vector (16) was used to generate a C-terminal FLAG-tagged version of RRV ORF57.
For the insertion of an intron in the middle of the coding sequence of gH, gL, c.o. gH, and c.o. gL, the first intron (130 bp) of human beta-globin (NCBI accession number NG_000007) was amplified from HEK293T cells using Intron-F (GTTGGTATCAAGGTTACAAGACA) and Intron-R (CTAAGGGTGGGAAAATAGACCAA) primers. To preserve the original splicing donor and acceptor sites (AG↓gt and ag↓G, respectively) of the beta-globin gene, the amplified intron DNA fragment was inserted at base pair 694/695 of gH and c.o. gH and at base pair 294/295 of gL and c.o. gL by overlapping PCR mutagenesis.
Plasmid pNL1.5ERTEm26CTE, the parental plasmid that contains the RTEm26/CTE nuclear export sequence (37), was a kind gift of Barbara Felber (NIH). The RTEm26/CTE sequence was amplified as a blunt-end fragment using Pfx DNA polymerase (Invitrogen) and RTE-F (CCGTGGGGTGCGAGGCTAAGC) and CTE-R (CAAATCCCTCGGAAGCTGCGC) primers. The RTEm26CTE fragment was inserted into the 3′-untranslated region (UTR) of pEF1-v5-hisA vector using the PmeI restriction enzyme to yield the pEF1-v5-hisA-RTEm26/CTE vector. The cDNA encoding gH, gL, c.o. gH, or c.o. gL was cloned into the pEF1-v5-hisA-RTEm26/CTE vector using EcoRI and NotI restriction enzymes and then subcloned into a pCDNA6-v5-hisA vector using EcoRI and AvrII restriction enzymes to assess the role of the RTEm26/CTE cis-acting sequence element.
HEK293T cells were cultured and maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS; Invitrogen), glutamine, and penicillin-streptomycin. Cells, seeded onto six-well plates 1 day prior to transfection, were transfected with plasmid DNA using a CalPhos mammalian transfection kit (Clontech).
Immunoblotting.
Cells were harvested, resuspended, and lysed with lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.5% Triton X-100) supplemented with protease inhibitor cocktail (Sigma). After centrifugation (12,000 × g for 5 min), the supernatants were transferred to new tubes and their protein levels were measured using a bicinchoninic acid protein assay kit (Pierce). The total protein for each sample was normalized to the lowest concentration among samples. An equal volume of 2× SDS sample buffer was added to each lysate, and then the samples were boiled for 5 min and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins from the gels then were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories) and subjected to immunoblotting. Briefly, the membranes were blocked with phosphate-buffered saline (PBS) containing 5% skim milk for 30 min at room temperature and incubated with the appropriate primary antibody for 2 h, followed by a 1-h incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. For loading controls, beta-actin antibody (Abcam) was used. Specific signals were detected by an enhanced chemiluminescence system using a SuperSignal West Pico chemiluminescent substrate (Pierce).
In vitro transcription and RNA transfection.
The DNA fragments containing gL, c.o. gL, prespliced ORF57, and ORF57 coding sequences were amplified from pCDNA6-gL-v5-hisA, pCDNA6-c.o. gL-v5-hisA, pEF1-pre-ORF57-v5-hisA, and pEF1-ORF57-v5-hisA plasmids, respectively, using Pfx DNA polymerase and T7-F (TAATACGACTCACTATAGGG) and Vitro-R (T40CAGATGGCTGGCAACTAGAA) primers. After PCR amplification, each DNA fragment had, in addition to each coding sequence, a T7 priming site at the 5′ terminus and 40 stretches of T at the 3′ terminus. Each DNA fragment was purified after agarose gel electrophoresis (Qiagen gel purification kit) and used as a template in an in vitro transcription reaction. The mRNAs encoding gL, c.o. gL, prespliced ORF57, and ORF57 were synthesized from the aforementioned template using a MessageMAX T7 ARCA-capped message transcription kit (Epicentre Biotechnologies) according to the manufacturer's instructions. RNAs were quantified by measuring UV absorbance (NanoDrop; Thermo Scientific) and checked by denaturing glyoxal agarose gel electrophoresis (Ambion). HEK293T cells seeded onto a six-well plate were incubated for 1 day and transfected with mRNA using a TransIT-mRNA transfection kit (Mirus).
Northern blotting.
Total cellular RNA was isolated from transfected cells using an RNeasy plus minikit (Qiagen). A Northern blot was performed according to the procedure described in the NorthernMax-Gly kit manual (Ambion). Briefly, ∼3 μg of total RNA isolated as described above was mixed with an equal volume of glyoxal loading dye and incubated for 30 min at 50°C. The RNA was separated on a 0.8% glyoxal denaturing gel (110 V, 100 min), transferred onto a BrightStar-Plus nylon membrane (Ambion), and cross-linked with UV light. The membrane was prehybridized for 1 h at 42°C in Ultrahyb solution (Ambion) and incubated with 8 nM biotin-labeled antisense v5 probe (biotin-CGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACC-biotin) in Ultrahyb solution overnight at 42°C. The next morning, the membrane was washed three times, each time for 10 min, with NorthernMax high stringent wash buffer (Ambion) at 42°C. The bands that hybridized to the probe were detected using a chemiluminescent nucleic acid detection kit (Pierce) according to the manufacturer's procedure.
Fractionation of nuclear and cytoplasmic RNA.
Cells grown on a 10-cm dish were rinsed with PBS and detached using PBS containing EDTA (1 mM). Roughly 1/6 of the cells were subjected to immunoblotting, and the remaining 5/6 of cells were subjected to fractionation. For fractionation, cells first were pelleted by low-speed centrifugation (6,000 × g, 1 min), and then 1.0 ml of ice-cold cell fractionation buffer (PARIS kit; Ambion) and 3 μl of RNase OUT (Invitrogen) were added to the cell pellets. After incubation on ice with gentle mixing (10 min), the nuclear pellets and cytoplasmic supernatants were separated by centrifugation for 3 min at 500 × g. For the cytoplasmic fraction, 0.3 ml of supernatant was mixed with an equal volume of 2× lysis/binding buffer (PARIS kit), and 0.3 ml of 100% ethanol was added to the mixture. For the nuclear fraction, the nuclear pellet described above was lysed with 0.6 ml of RLT buffer (RNeasy Plus miniprep kit) and passed through a QIAshredder and gDNA elimination column (Qiagen). The total RNA from these nuclear and cytoplasmic fractions was purified further using an RNeasy plus miniprep kit according to the manufacturer's instructions.
Actinomycin treatment and measurement of RNA stability.
HEK293T cells were transfected with pCDNA6-c.o. gH-v5-hisA plasmid together with either pEF1-v5-hisA empty plasmid or pEF1-ORF57-v5-hisA plasmid. At 36 h posttransfection, cells were treated with 2 μM actinomycin (Sigma) to block de novo mRNA synthesis for 0 to 8 h before harvesting. Total RNA was isolated using an RNeasy Plus miniprep kit, and the bands corresponding to c.o. gH RNA were visualized by Northern blotting.
RESULTS
RRV ORF57 enhances the expression of c.o. gH, gH, c.o. gL, and gL proteins.
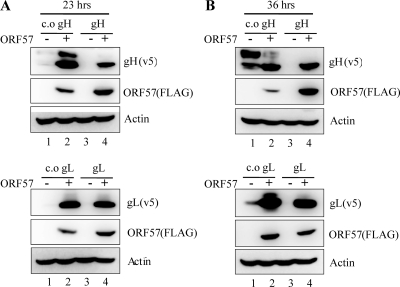
Bilello et al. showed that gH and gL protein expression from a cassette driven by human EF1 promoter was below the limit of detection (3). Similarly, we could not detect any gH or gL expression from an expression cassette driven by a cytomegalovirus (CMV) promoter at 23 (Fig. 1 A, lane 3), 36 (Fig. 1B, lane 3), or 48 h (data not shown) posttransfection. However, codon-optimized versions of gH and gL (c.o. gH and c.o. gL, respectively) showed readily detectable levels of expression by 36 h (Fig. 1B, lane 1) posttransfection even in the absence of RRV ORF57, confirming that codon optimization alone could increase the poor expression of gH and gL. However, the expression of gH and gL as well as c.o. gH and c.o. gL was greatly induced when RRV ORF57 also was expressed (Fig. 1A and B, lanes 2 and 4).
Fig. 1.
Induction of target gene expression by RRV ORF57. Plasmids encoding RRV c.o. gH, gH, c.o. gL, and gL were cotransfected with either the empty or the ORF57 expression plasmid, and the levels of protein expressed were measured at 23 (A) or 36 h (B) posttransfection by immunoblotting. The top band, but not the bottom band, of the gH doublet appears to be an O-glycosylated form of gH on the basis of Jacalin reactivity (38).
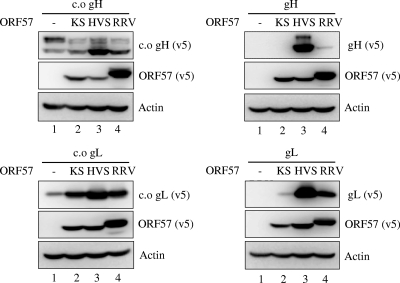
In addition to RRV ORF57, we also tested whether ORF57 orthologues from other gamma-2 herpesviruses, KSHV and herpesvirus saimiri (HVS), induced the expression of RRV gH, gL, c.o. gH, and c.o. gL. As shown in Fig. 2, ORF57 from KSHV and HVS also significantly induced the expression of c.o. gL and gL. However, ORF57 from KSHV, unlike ORF57 from HVS and RRV, did not induce the expression of c.o. gH and gH. Thus, there seems to be subtle differences among ORF57 orthologues of gamma-2 herpesviruses with respect to their target specificities. Interestingly, HVS ORF57 was even more potent than RRV ORF57 in inducing RRV gH, gL, c.o. gH, and c.o. gL expression.
Fig. 2.
Induction of RRV c.o. gH, gH, c.o. gL, and gL expression by KSHV, HVS, and RRV ORF57. Plasmids encoding RRV c.o. gH, gH, c.o. gL, and gL were cotransfected with the empty, KSHV ORF57, HVS ORF57, or RRV ORF57 expression vector, and the level of protein expression from each was measured at 36 h posttransfection by immunoblotting.
Effect of intron and RTEm26/CTE cis-acting sequence on the expression of gH and gL.
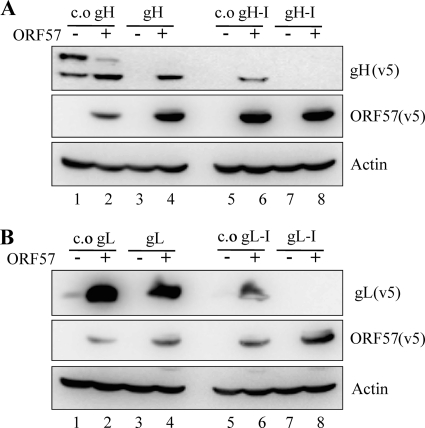
The gH and gL genes of RRV, like most other RRV genes, do not have introns that are spliced out during mRNA processing. It has been shown that TREX and the exon-junction complex (EJC) binds onto mRNA in a splicing-dependent manner to facilitate the efficient nuclear export of spliced mRNAs in eukaryotic cells (19, 23, 31). We asked whether the lack of appropriate ribonucleoprotein (RNP), due to a lack of splicing, was responsible for the poor expression of gH and gL mRNAs. We introduced the intron of the human beta-globin gene into the coding regions of gH, gL, c.o. gH, and c.o. gL to mimic eukaryotic splicing. The sites of intron insertion were carefully selected to closely mimic the splicing donor and acceptor sites of the human beta-globin gene (see Materials and Methods). The poor expression of gH and gL protein was not rescued by intron insertion into these genes (Fig. 3 A and B, lanes 7). Moreover, the protein expression from c.o. gH and c.o. gL was not readily detectable when the intron of the beta-globin gene was inserted into the coding region of these genes (Fig. 3A and B, lanes 1 versus lanes 5). The intron that was inserted into the c.o. gH and c.o. gL genes was at least partially functional, since protein expression was detected from these in the presence of ORF57 (Fig. 3A and B, lanes 5 versus lanes 6). Overall, the introduction of a heterologous intron into the gH and gL genes had an adverse effect on protein expression.
Fig. 3.
Insertion of an artificial intron does not rescue expression of gH and gL. Intron1 of the human beta-globin gene was inserted into the coding regions of c.o. gH, gH, c.o. gL, and gL, and protein expression from the resulting constructs (c.o. gH-I and gH-I [A], c.o. gL-I and gL-I [B]) were measured together with the expression from the parental constructs in HEK293T cells at 36 h posttransfection by immunoblotting.
The introduction of a cis-acting sequence was another approach we have taken to attempt to rescue the poor expression of intronless gH and gL mRNAs. RTEm26/CTE has been shown to enhance by several hundredfold the expression of subgenomic gag and env genes from human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) (37). Thus, the RTEm26/CTE sequence was introduced into the 3′-UTR region of the gH and gL expression cassettes, and protein expression levels were compared to those of each parental vector (Fig. 4). The introduction of RTEm26/CTE sequence into gH mRNA failed to rescue gH protein expression (Fig. 4A, lane 3), while it did rescue gL protein expression (Fig. 4B, lane 3). The insertion of the RTEm26/CTE sequence into the 3′-UTR of the gL gene caused the significant accumulation of gL RNA in both the cytoplasm and the nucleus (Fig. 4D, lanes 5 and 6). Upon the coexpression of RRV ORF57, the levels of gL RNA in both compartments were greatly increased (Fig. 4D, lanes 11 and 12). Greatly increased RNA levels of gL and codon-optimized gL also were induced by RRV ORF57 (Fig. 4D, lanes 7 to 10). On the other hand, the levels of gH RNA remained below the limit of detection by the insertion of the same RTEm26/CTE sequence (Fig. 4C, lanes 5 and 6). However, they were greatly increased by the coexpression of RRV ORF57 (Fig. 4C, lanes 11 and 12). Similarly to the RNAs of gL, RNAs from gH and the codon-optimized version of gH were greatly increased by RRV ORF57 (Fig. 4C, lanes 7 to 10). Overall, the gH and gL RNA levels both in the cytoplasm and in the nucleus faithfully recapitulated the cognate protein expression presented in Fig. 4A and B. The main effect of RRV ORF57 was to enhance the accumulated RNA levels of target genes both in the cytoplasm and in the nucleus.
Fig. 4.
Tethering of RTEm26/CTE rescues gL but not gH expression. The RTCm26/CTE sequence was inserted at the 3′-untranslated region of the gH and gL genes. The resulting plasmids (gH-R/C and gL-R/C) as well as parental plasmids and the codon-optimized constructs were transfected into HEK293T cells together with either the empty or the RRV ORF57 expression vector. The expression of c.o. gH (lanes 1 and 4), gH (lanes 2 and 5), and gH-R/C (lanes 3 and 6) (A) as well as c.o. gL (lanes 1 and 4), gL (lanes 2 and 5), and gL-R/C (lanes 3 and 6) (B) were measured at 36 h posttransfection by immunoblotting. (C and D) HEK293T cells were transfected with different combinations of plasmid DNAs as shown, and cells were harvested at 36 h posttransfection. The isolated RNAs from cytoplasm (C; 2 μg/lane) and nucleus (N; 1 μg/lane) were analyzed by Northern blotting. (C) Levels of c.o. gH (lanes 1, 2, 7, and 8), gH (lanes 3, 4, 9, and 10), and gH-R/C (lanes 5, 6, 11, and 12) RNA were analyzed. (D) Levels of c.o. gL (lanes 1, 2, 7, and 8), gL (lanes 3, 4, 9, and 10), and gL-R/C (lanes 5, 6, 11, and 12) RNA were analyzed. The 28S and 18S as well as the 45S and 32S rRNA bands from ethidium bromide staining are shown as loading and fractionation controls.
Translational activation by RRV ORF57.
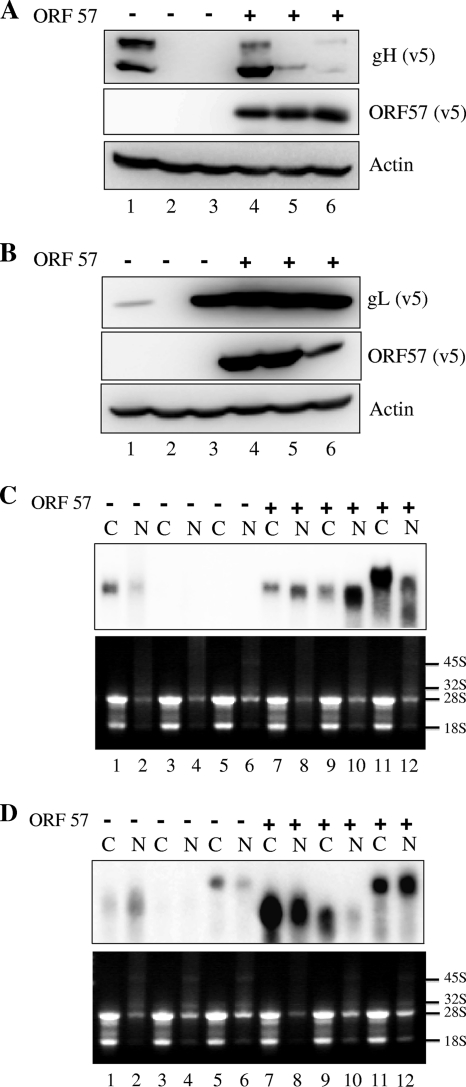
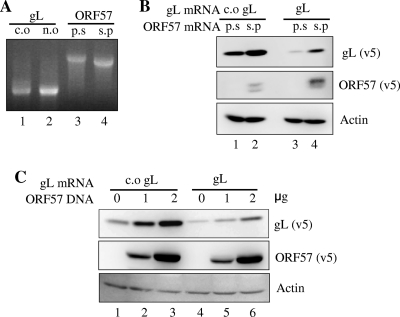
To assess the effect of RRV ORF57 on translation, mRNAs of RRV gL, c.o. gL, prespliced ORF57, and spliced ORF57 were synthesized in vitro (Fig. 5 A) and transfected into HEK-293T cells. The prespliced ORF57 RNA serves as a negative control since it does not express ORF57 in the absence of splicing inside the cell. Both c.o. gL and gL expression were enhanced in the presence of ORF57 protein (Fig. 5B, lanes 1 and 3 versus lanes 2 and 4), indicating increased levels of translation by RRV ORF57 acting directly on the transfected mRNA. Given that each mRNA was synthesized in vitro and transfected directly into cells in this assay, it is not likely that any events related to nuclear export processes were involved. The increased levels of translation could result from the direct activation of translation by ORF57 (5, 13, 22) or from the increased stabilization of the RNA by ORF57 (see below). Increasing levels of ORF57 protein expression were achieved from increasing inputs of ORF57 plasmid DNA. Under these conditions, both c.o. gL and gL expression showed a concomitant increase, corroborating the observations of Fig. 5B. It is noteworthy that the transfection of gL mRNA yielded detectable levels of gL protein even in the absence of ORF57 (Fig. 5B, lane 3, and C, lane 4), while the transfection of gL plasmid DNA did not (Fig. 1 to 4). These data suggest that gL mRNA can be translated as long as it exists above a certain threshold level in the cytoplasm.
Fig. 5.
Activation of translation by RRV ORF57. (A) RNA encoding c.o. gL, gL, prespliced ORF57, and spliced ORF57 were synthesized in vitro and analyzed in a denaturing glyoxal agarose gel. (B) Codon-optimized (c.o.) gL or nonoptimized (n.o) gL RNA was transfected into HEK293T cells together with either prespliced (p.s) or spliced (s.p) ORF57 RNA, and the expression of protein from each RNA was measured at 36 h posttransfection by immunoblotting. (C) An increasing amount of ORF57 plasmid DNA was transfected instead of ORF57 RNA, and the expression of cognate proteins was measured at 36 h posttransfection by immunoblotting.
Accumulation of target RNAs: posttranscriptional control by RRV ORF57.
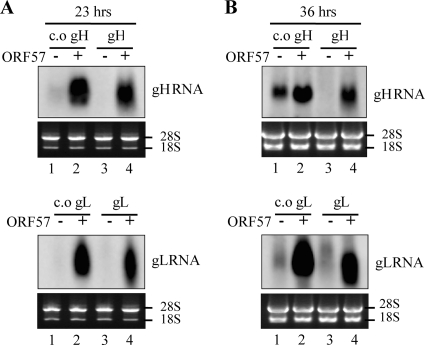
The ability to detect gL protein, at least to some extent, when gL mRNA was transfected directly (Fig. 5) suggested to us that there is a problem with RNA accumulation when it is made from transfected plasmid DNA. We therefore examined the steady-state levels of RNA made from the transfection of the plasmid DNA. Several observations are worth noting from the data shown in Fig. 6. First, no detectable levels of gH and gL RNAs were observed in both earlier (23 h) (Fig. 6A, lane 3) and later (36 h) (Fig. 6B, lane 3) time points. Given that they were driven by a strong CMV promoter, this is surprising but consistent with our inability to detect protein expression following the transfection of these expression cassettes. Second, we observed a very large increase in the amount of each RNA in the presence of the ORF57 protein (Fig. 6A and B, lanes 1 versus lanes 2 and lanes 3 versus lanes 4). Third, no detectable levels of c.o. gH and c.o. gL RNA were observed at 23 h posttransfection in the absence of ORF57; however, they were detected at 36 h posttransfection at smaller amounts. Overall, the RNA levels faithfully recapitulated the protein levels presented in Fig. 1, indicating that the steady-state levels of RNA were critical to the levels of protein expression.
Fig. 6.
Accumulation of target RNAs by RRV ORF57. Plasmids encoding RRV c.o. gH, gH, c.o. gL, and gL were cotransfected with either the empty or the ORF57 expression vector. Total RNA was isolated at 23 h (A) or 36 h (B) posttransfection, and an equal amount of total RNA (4 μg) was loaded into each lane. The levels of each target RNA were measured by Northern blotting. The 28S and 18S rRNA bands from ethidium bromide staining are shown as loading controls.
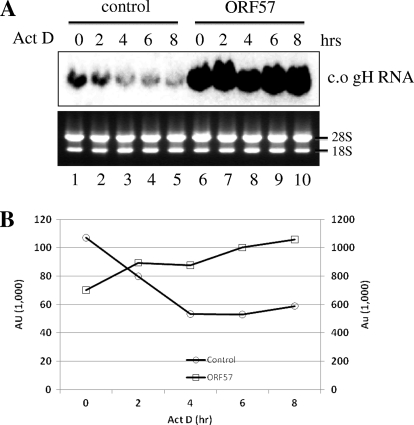
The large increases of RNA levels in the presence of ORF57 protein are not likely to result from direct transcriptional activation by ORF57. RRV ORF57 did not activate the transcription of the renilla luciferase gene driven from the same CMV promoter (data not shown). Furthermore, it has been shown in previous reports that KSHV ORF57 did not activate transcription from a CMV promoter (14, 20). We therefore hypothesized that gH and gL mRNAs are naturally unstable and that ORF57 allows gH, gL, c.o. gH, and c.o. gL mRNAs to accumulate to much higher levels than they would otherwise by inhibiting their degradation. To investigate this, we compared the degradation rate of c.o. gH RNA in the absence or presence of RRV ORF57. The c.o. gH was chosen as a model because it allowed some levels of accumulation even in the absence of ORF57 at 36 h posttransfection (Fig. 6B, lane 1). HEK293T cells were transfected with c.o. gH plasmid together with either the empty vector or the ORF57 plasmid and subsequently were treated with actinomycin D to block de novo RNA synthesis. As shown in Fig. 7, the degradation of c.o. gH RNA was much faster in the absence than the presence of ORF57. The c.o. gH RNA seemed to be quite stable in the presence of ORF57 during the 8-h assay period without any noticeable degradation (Fig. 7, lanes 6 to 10).
Fig. 7.
Posttranscriptional stabilization of c.o. gH RNA by RRV ORF 57. Plasmid encoding c.o. gH was transfected into HEK293T cells together with either the empty or the RRV ORF57 expression vector. At 36 h posttransfection, cells were treated with 2 μM actinomycin D for the indicated times. Total RNA was isolated, and an equal amount of RNA (4 μg) was loaded into each lane. (A) The c.o. gH RNA was detected by Northern blotting. The 28S and 18S rRNA bands from ethidium bromide staining are shown as loading controls. (B) The intensity of each band (AU) was analyzed using the Image Gauge program (Fuji Film) and was graphically represented. (Note that the y axis scale for c.o. gH RNA is 10 times greater in the presence of ORF57 than in the absence of ORF57.)
Distribution of c.o. gH RNA in nucleus and cytoplasm.
We measured RNA amounts in the cytoplasm and nucleus after fractionation following the transfection of plasmid DNA expression cassettes. In the absence of ORF57, cytoplasmic and nuclear levels of c.o. gH RNA were barely detectable at 26 h posttransfection (Fig. 8, lanes 1 and 2) and still very low at 36 h posttransfection (Fig. 8, lanes 5 and 6). However, they were readily detectable at both 26 and 36 h posttransfection in the presence of ORF57 (Fig. 8, lanes 3, 4, 7, and 8). Since c.o. gH RNA levels were much higher in the presence than in the absence of ORF57 (about 4-fold at 26 h and 7-fold at 36 h), it was difficult to accurately evaluate the effect of ORF57 on nuclear export. However, the c.o. gH RNA in the cytoplasm was increased about 2.4-fold from 26 to 36 h posttransfection (compare lane 7 to lane 3) in the presence of ORF57. The increase of c.o. gH RNA in the cytoplasm during the same time period was about 1.7-fold in the absence of ORF57 (compare lane 5 to lane 1). While there was little increase between 26 and 36 h in the absolute amount of c.o. gH RNA in the absence of ORF57, whether cytoplasmic or nuclear, there was a large increase in the amount of c.o. gH RNA in both compartments in the presence of ORF57. These results again suggest a defect in the accumulation of target RNA in the absence of ORF57.
Fig. 8.
Cellular localization of gH RNA. A plasmid encoding c.o. gH was transfected into HEK293T cells together with either the empty or the RRV ORF57 expression vector. Cells were harvested at 26 or 36 h posttransfection and fractionated to separate the cytoplasmic (C) and nuclear (N) RNA. (A) Cytoplasmic RNA (2.2 μg) and nuclear RNA (1.5 μg) were loaded into each corresponding lane, and c.o. gH RNA was detected by Northern blotting. The 28S and 18S as well as the 45S and 32S rRNA bands from ethidium bromide staining are shown as loading and fractionation controls. (B) The intensity of each band (in arbitrary units [AU]) was analyzed using Image Gauge (Fuji Film) and was graphically represented.
DISCUSSION
Gamma-2 herpesviruses have elaborate systems to control gene expression during the virus life cycle. The timing and intensity of viral gene expression is orchestrated by comprehensive, interrelated regulatory mechanisms. ORF57 is expressed in the immediate-early phase of lytic replication (20, 24, 28) and is a key orchestrator of the viral lytic cycle by inducing delayed early and late gene expression (15, 20, 25, 26, 32). The extremely low levels of expression of envelope glycoproteins until the final stage of lytic replication may serve in part to limit any cytotoxicity of virus-encoded glycoproteins and may also aid virus replication by limiting the exposure of these antigens to the host immune response. RRV gH and gL mRNAs, from which envelope glycoproteins are made in the late phase of lytic replication, seem from our work here to be quite unstable in nature. These transcripts were not above the level of detection even when their synthesis was driven from a strong CMV promoter.
In trying to pinpoint the site or sites of action of any ORF57 orthologue, it is important to realize the close linkage of some cellular processes and the difficulty in distinguishing one site of action from a closely linked process. Examples include translation and mRNA stability, the nuclear egress of mRNA and mRNA stability, and the transcription and nuclear egress of mRNA. In looking at the literature on ORF57 action, it is often difficult to distinguish what is multifunctional activity from what is a single activity manifest in different, linked, functional processes (6, 17, 20, 26, 32). Similar difficulties can be seen in the data reported here. gL protein synthesis was significantly higher when gL mRNA was transfected in the context of ORF57 expression (Fig. 5). Is this an effect of ORF57 directly on the translation of mRNA or on the stability of mRNA? Following the transfection of DNA expression cassettes, gH and gL RNAs accumulated to much greater extents in the presence of ORF57 (Fig. 6 and 8). Is this an effect on RNA egress from the nucleus or on RNA stability? Several lines of evidence from our experiments argue for a major effect of RRV ORF57 on the stabilization of RRV gH and gL mRNAs. By examining both mRNA and protein levels over time following DNA transfection, we have demonstrated a clear defect in gH and gL mRNA accumulation in the absence of ORF57, while RRV ORF57 allowed high levels of the accumulation of gH and gL mRNAs (Fig. 6). It is noteworthy that we did not observe any preferential accumulation of mRNA in any specific cell compartment. Instead, we observed higher levels of gH and gL total RNA, which seemed to be contributed from both nucleus and cytoplasm proportionally in the presence of ORF57. The preferential accumulation of target RNA in the cytoplasm has been used to support a role for some ORF57s in the egress of RNA from the nucleus (4, 6, 29, 41). In our study, the overall increase of total RNA levels of the target gene was contributed by nuclear and cytoplasmic compartments (Fig. 8). This supports RNA stabilization in both the nucleus and cytoplasm as a major site of action of RRV ORF57.
It generally has been considered that the nuclear export of cellular mRNA needs an active transport process (reviewed in references 10 and 18). Therefore, the major increase of target gene RNA in both the cytoplasm and nucleus in our study (Fig. 8) seems to be best explained by the posttranscriptional stabilization of target RNA in the nucleus and subsequent nuclear egress of that RNA. The notion of nuclear stabilization agrees well with the cellular localization of ORF57, which resides principally in the nucleus (14, 20, 34). Also, the stabilization of target RNA in the nucleus by ORF57 has been well documented for KSHV PAN RNA (35). Since KSHV PAN RNA always resides in the nucleus and is not exported to the cytoplasm, the stabilization process in this case appears to be independent of nuclear export. The enhanced levels of gH and gL RNAs were not due to increased RNA synthesis. We confirmed that RRV ORF57 does not affect de novo transcription from a CMV promoter, which also has been observed for other ORF57s in previous reports (14, 20).
The introduction of the RTEm26/CTE sequence into the 3′-UTR of the gL gene rescued gL protein expression while it did not rescue gH protein expression (Fig. 4). Since the introduction of the RTEm26/CTE sequence into the gL transcript raised the amounts of gL RNA from undetectable to readily detectable levels in both nuclear and cytoplasmic compartments, it seems likely that the RTEm26/CTE element stabilized the accumulation of this RNA. In a previous study of retroviral gag RNA and protein synthesis, the RTEm26/CTE element was found to affect all steps of posttranscriptional regulation, including nuclear export, stabilization, and translation (37).
KSHV ORF57 has been reported to cause a noticeable shift of gL RNA distribution to the cytoplasm, indicating its major role in the process of target RNA egress (4). In our results described here, RRV ORF57 did not cause such a significant change in the distribution of gL RNA between the nucleus and cytoplasm. Instead, we observed great induction of total gL RNA in the presence of RRV ORF57. Thus, the actions of ORF57s from KSHV and RRV on their gL RNAs seem to be different. Further work will be needed to discern whether this reflects a true difference in the mode of action of those ORF57s or whether these are different manifestations of the same mode of action.
ACKNOWLEDGMENTS
We thank Patrick Neilan, Anusha Anukanth, and Elizabeth Stansell for reading the manuscript; Alexander Hahn, Thomas Postler, and all the members of R. C. Desrosiers' laboratory for their helpful discussions; and Deborah Letourneau for her kind assistance. We also thank John P. Bilello for reagents and valuable insights throughout this study.
This work was supported by NIH grants AI 072004 and AI 063928 to R.C.D. and by the primate center base grant RR00168.
Footnotes
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Alexander L., et al. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bello L. J., et al. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80(Pt 12):3207–3215 [DOI] [PubMed] [Google Scholar]
- 3. Bilello J. P., Morgan J. S., Desrosiers R. C. 2008. Extreme dependence of gH and gL expression on ORF57 and association with highly unusual codon usage in rhesus monkey rhadinovirus. J. Virol. 82:7231–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyne J. R., Colgan K. J., Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyne J. R., Jackson B. R., Taylor A., Macnab S. A., Whitehouse A. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 29:1851–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyne J. R., Whitehouse A. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen I. H., Li L., Silva L., Sandri-Goldin R. M. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen I. H., Sciabica K. S., Sandri-Goldin R. M. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conrad N. K., Steitz J. A. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 24:1831–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cullen B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desrosiers R. C., et al. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duan W., Wang S., Liu S., Wood C. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403–413 [DOI] [PubMed] [Google Scholar]
- 13. Fontaine-Rodriguez E. C., Knipe D. M. 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 82:3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta A. K., Ruvolo V., Patterson C., Swaminathan S. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han Z., Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hobbs S., Jitrapakdee S., Wallace J. C. 1998. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1alpha promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252:368–372 [DOI] [PubMed] [Google Scholar]
- 17. Johnson L. A., Li L., Sandri-Goldin R. M. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83:6335–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim V. N., Dreyfuss G. 2001. Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events. Mol. Cells 12:1–10 [PubMed] [Google Scholar]
- 19. Kim V. N., et al. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20:2062–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirshner J. R., Lukac D. M., Chang J., Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koffa M. D., et al. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larralde O., et al. 2006. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J. Virol. 80:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Hir H., Izaurralde E., Maquat L. E., Moore M. J. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lukac D. M., Kirshner J. R., Ganem D. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Majerciak V., Pripuzova N., McCoy J. P., Gao S. J., Zheng Z. M. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 81:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Majerciak V., Yamanegi K., Nie S. H., Zheng Z. M. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 27. Majerciak V., Yamanegi K., Zheng Z. M. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 80:11968–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malik P., Blackbourn D. J., Cheng M. F., Hayward G. S., Clements J. B. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155–2166 [DOI] [PubMed] [Google Scholar]
- 29. Malik P., Blackbourn D. J., Clements J. B. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001–33011 [DOI] [PubMed] [Google Scholar]
- 30. Malik P., Schirmer E. C. 2006. The Kaposi's sarcoma-associated herpesvirus ORF57 protein: a pleurotropic regulator of gene expression. Biochem. Soc. Trans. 34:705–710 [DOI] [PubMed] [Google Scholar]
- 31. Masuda S., et al. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nekorchuk M., Han Z., Hsieh T. T., Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palmeri D., Spadavecchia S., Carroll K. D., Lukac D. M. 2007. Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 81:13299–13314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phelan A., Clements J. B. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78(Pt 12):3327–3331 [DOI] [PubMed] [Google Scholar]
- 35. Sahin B. B., Patel D., Conrad N. K. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 6:e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Searles R. P., Bergquam E. P., Axthelm M. K., Wong S. W. 1999. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smulevitch S., et al. 2006. RTE and CTE mRNA export elements synergistically increase expression of unstable, Rev-dependent HIV and SIV mRNAs. Retrovirology 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stansell E., Canis K., Haslam S. M., Dell A., Desrosiers R. C. 2011. Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate. J. Virol. 85:582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun R., Lin S. F., Gradoville L., Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 93:11883–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swaminathan S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell Biochem. 95:698–711 [DOI] [PubMed] [Google Scholar]
- 41. Williams B. J., et al. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387:295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]