Abstract
Discovery of genetically distinct hantaviruses in multiple species of shrews (order Soricomorpha, family Soricidae) and moles (family Talpidae) contests the conventional view that rodents (order Rodentia, families Muridae and Cricetidae) are the principal reservoir hosts and suggests that the evolutionary history of hantaviruses is far more complex than previously hypothesized. We now report on Rockport virus (RKPV), a hantavirus identified in archival tissues of the eastern mole (Scalopus aquaticus) collected in Rockport, TX, in 1986. Pairwise comparison of the full-length S, M, and L genomic segments indicated moderately low sequence similarity between RKPV and other soricomorph-borne hantaviruses. Phylogenetic analyses, using maximum-likelihood and Bayesian methods, showed that RKPV shared a most recent common ancestor with cricetid-rodent-borne hantaviruses. Distributed widely across the eastern United States, the fossorial eastern mole is sympatric and syntopic with cricetid rodents known to harbor hantaviruses, raising the possibility of host-switching events in the distant past. Our findings warrant more-detailed investigations on the dynamics of spillover and cross-species transmission of present-day hantaviruses within communities of rodents and moles.
INTRODUCTION
Hantaviruses (genus Hantavirus), like viruses of other Bunyaviridae genera (Orthobunyavirus, Phlebovirus, Nairovirus, and Tospovirus), possess a negative-sense, single-stranded, tripartite RNA genome consisting of large (L), medium (M), and small (S) segments which encode an RNA-dependent RNA polymerase (RdRP), two envelope glycoproteins (Gn, Gc), and a nucleocapsid protein (NP), respectively (29, 37, 43, 44). However, hantaviruses are unique in that they have no known insect or arthropod host and instead are harbored by rodents (order Rodentia, families Muridae and Cricetidae) (36, 56). Hantaviruses hosted by rodents in the subfamilies Murinae and Arvicolinae cause hemorrhagic fever with renal syndrome in Eurasia (30, 55, 56), while those carried by rodents in the subfamilies Neotominae and Sigmodontinae cause hantavirus cardiopulmonary syndrome in the Americas (10, 35).
The segregation of hantaviruses into clades that parallel the molecular phylogeny of rodents in the Murinae, Arvicolinae, Neotominae, and Sigmodontinae subfamilies has suggested that hantaviruses have coevolved with their reservoir rodent hosts (20, 22, 37). Recently, this premise has been strenuously challenged on the basis of the disjunction between the evolutionary rates of the host and virus species (39, 40). That is, rather than codivergence, host switching and local species-specific adaptation have been proposed to account for the similarities between the host and virus phylogenies. Since some sympatric and syntopic rodent species occasionally serve as reservoirs for the same hantavirus, host switching or cross-species transmission have clearly occurred during the evolution of hantaviruses (33). Topografov virus in the Siberian lemming (Lemmus sibiricus) is an often-cited example (53). On the other hand, full-genome analysis of Thottapalayam virus (TPMV), a hantavirus isolated from the Asian house shrew (Suncus murinus) more than 40 years ago (6, 58), shows an early evolutionary divergence from rodent-associated hantaviruses (46, 54).
Moreover, the recent discovery of genetically diverse hantaviruses in shrews of multiple species (order Soricomorpha, family Soricidae), including Tanganya virus in the Therese's shrew (Crocidura theresae) (27), Imjin virus in the Ussuri white-toothed shrew (Crocidura lasiura) (49), Camp Ripley virus in the northern short-tailed shrew (Blarina brevicauda) (4), Cao Bang virus in the Chinese mole shrew (Anourosorex squamipes) (48), Seewis virus in the Eurasian common shrew (Sorex araneus) (47), Ash River virus in the masked shrew (Sorex cinereus) (2), Jemez Springs virus in the dusky shrew (Sorex monticolus) (2), and Kenkeme virus in the flat-skulled shrew (Sorex roboratus) (25), as well as in moles (family Talpidae), including Asama virus (ASAV) in the Japanese shrew mole (Urotrichus talpoides) (3), Oxbow virus (OXBV) in the American shrew mole (Neurotrichus gibbsii) (23), and Nova virus (NVAV) in the European common mole (Talpa europaea) (24), suggests that the evolutionary history of hantaviruses is more complex than previously conjectured.
In particular, the highly divergent hantavirus in the European common mole (24) predicts the existence of additional talpid-borne hantaviruses. High on the list of candidate talpid hosts has been the eastern mole (Scalopus aquaticus) (subfamily Scalopinae), which is widely distributed across the eastern United States (57). Here, we report on the molecular phylogeny of Rockport virus (RKPV), a newfound hantavirus in the eastern mole. The unexpected finding of an ancestry shared by RKPV and cricetid-rodent-borne hantaviruses is consistent with cross-species virus transmission in the distant past but leaves unanswered questions about which mammalian lineage served as the original host of primordial hantaviruses.
MATERIALS AND METHODS
Tissues.
Frozen livers from 60 eastern moles, archived in the Museum of Southwestern Biology at the University of New Mexico in Albuquerque, were analyzed. Moles were collected between 1984 and 1992 from the eastern United States (Fig. 1A), including Florida, Kansas, South Carolina, Tennessee, and Texas (Table 1).
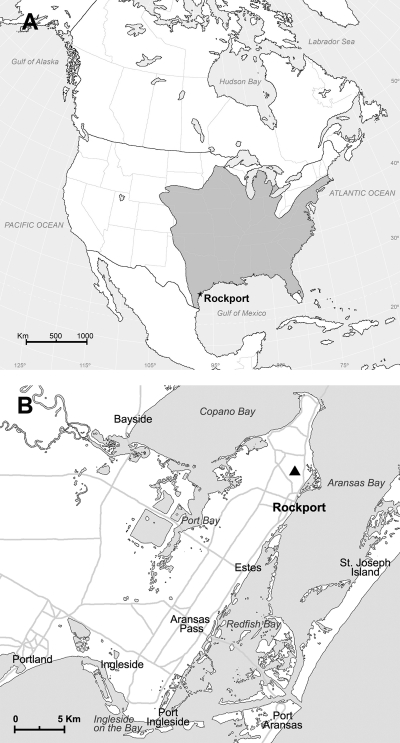
Fig. 1.
Map showing the geographic distribution of the eastern mole (Scalopus aquaticus) in the United States (A) and the location of Aransas National Wildlife Refuge in Rockport, TX, where hantavirus-infected eastern moles were captured in October 1986 (B).
Table 1.
Characteristics of eastern moles (Scalopus aquaticus) from various capture sites tested for hantavirus RNA
| State | County | Capture site | Capture date | No. tested | No. positive |
|---|---|---|---|---|---|
| Florida | Pinellas | Seminole Castleberry | July 1988 | 4 | 0 |
| Kansas | Rooks | Stockton | September 1992 | 1 | 0 |
| South Carolina | Aiken | Savannah River Plant | July 1984 | 2 | 0 |
| October 1987 | 5 | 0 | |||
| December 1987 | 8 | 0 | |||
| January 1988 | 7 | 0 | |||
| February 1988 | 1 | 0 | |||
| June 1989 | 2 | 0 | |||
| Barnwell | Savannah River Plant | June 1984 | 2 | 0 | |
| July 1984 | 1 | 0 | |||
| February 1988 | 9 | 0 | |||
| May 1989 | 10 | 0 | |||
| Tennessee | Shelby | Memphis (Raleigh) | July 1988 | 1 | 0 |
| Texas | Aransas | Aransas National Wildlife Refuge | August 1986 | 5 | 4 |
| Motley | Pease River | March 1981 | 2 | 0 |
RNA extraction and reverse transcription (RT)-PCR analysis.
Total RNA was extracted from tissues, using the PureLink Micro-to-Midi total RNA purification kit (Invitrogen, San Diego, CA), and then reverse transcribed, using the SuperScript III first-strand synthesis system (Invitrogen) and oligonucleotide primer (OSM55, 5′-TAGTAGTAGACTCC-3′), designed from the conserved 3′ ends of the S, M, and L segments of hantaviruses. For amplification of hantavirus genes, a two-step PCR was performed in 20-μl reaction mixtures containing 250 μM deoxynucleoside triphosphate (dNTP), 2 mM MgCl2, 1 U of AmpliTaq polymerase (Roche, Basel, Switzerland), and a 0.25 μM concentration of each primer. Initial denaturation at 94°C for 5 min was followed by two cycles each of denaturation at 94°C for 40 s, 2°C step-down annealing from 48°C to 38°C for 40 s, and elongation at 72°C for 1 min and then 32 cycles of denaturation at 94°C for 40 s, annealing at 42°C for 40 s, and elongation at 72°C for 1 min, in a GeneAmp PCR 9700 thermal cycler (Perkin-Elmer, Waltham, MA). Amplicons were separated by electrophoresis on 1.5% agarose gels and purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). DNA was sequenced directly using an ABI Prism 377XL genetic analyzer (Applied Biosystems, Foster City, CA).
Genetic analysis.
Complete S, M, and L genomic nucleotide and amino acid sequences of RKPV were aligned with representative rodent- and soricomorph-borne hantavirus sequences, using the ClustalW method (TranslatorX server and BioEdit 7.0.5) (1, 15, 52). Nucleotide sequences were also analyzed using multiple recombination detection methods within the RDP3 Beta34 software. The NP secondary structure was predicted from the entire amino acid sequence of the RKPV S segment using five methods at the NPS@ structure server (9): DSC (26), HNN (26), MLRC (14), PHD (42), and PREDATOR (12). COILS (31) was also used to scan the NP for coiled-coil regions. To determine the glycosylation and transmembrane sites for RKPV Gn and Gc, NetNlyc 1.0 and Predictprotein (13) and TMHMM version 2.0 (28) were used, respectively.
Phylogenetic analysis.
To determine the phylogenetic relationship of RKPV with well-characterized hantaviruses, phylogenetic trees, based on the entire coding regions of the S, M, and L segments, were generated using the maximum-likelihood (ML) method implemented in PAUP* (Phylogenetic Analysis Using Parsimony, 4.0b10) (51) and the RAxML BlackBox Web server (50), as well as a Bayesian approach (19) using MrBayes 3.1 (41). The optimal evolutionary model was estimated as the generalized time-reversible plus invariant-sites plus gamma-distributed (GTR+I+Γ) model of evolution, as selected by jModelTest version 0.1 (38). ML topologies were evaluated by bootstrap analysis of 1,000 neighbor-joining iterations (in PAUP*) or 1,000 ML iterations (in RAxML). Bayesian analysis consisted of 2 million Markov chain Monte Carlo (MCMC) generations sampled every 100 generations to ensure convergence across two runs of four chains each, with average standard deviations of split frequencies of less than 0.01 and effective sample sizes over 100, resulting in consensus trees supported by posterior-node probabilities. Phylogenetic trees were readdressed to construct a tanglegram of host and associated hantaviruses in TreeMap 2.0b (7, 8, 23).
mtDNA host phylogeny.
Genomic DNA was extracted from tissues using the QIAamp DNA minikit (Qiagen) to verify the taxonomic identities of the hantavirus-infected eastern moles and to study their phylogenetic relationships. The complete 1,140-nucleotide cytochrome b gene was amplified by PCR using well-tested primers (forward, 5′-CGAAGCTTGATATGAAAAACCATCGTTG-3′; and reverse, 5′-CTGGTTTACAAGACCAGAGTAAT-3′) (21). Host phylogenies based on mitochondrial DNA (mtDNA) cytochrome b sequences, along with published sequences for shrews and moles for this gene region, were generated, using the ML and Bayesian methods described previously (2–4, 23, 24). The tree was based on 3,000,000 MCMC generations, sampled every 100 generations, and burn-in after 10,000 trees.
Nucleotide sequence accession numbers.
GenBank accession numbers for the RKPV S segment were HM015218, HM015223, and HM015224; that for the RKPV M segment was HM015219; those for the RKPV L segment were HM015220, HM015221, and HM015222; and those for the Scalopus aquaticus cytochrome b gene were HM461914, HM461915, HM461916, and HM461917.
RESULTS
RT-PCR detection of hantavirus.
Of the 60 eastern moles studied, hantavirus RNA was detected in four of five Scalopus aquaticus moles captured in Aransas National Wild-life Refuge in Rockport (latitude 28.042°N, longitude 97.052°W), TX, in October 1986 (Fig. 1B). Despite using a series of oligonucleotide primers that proved useful for the amplification of RKPV and other soricomorph-borne hantaviruses, three or more separate attempts to detect hantavirus RNA in each of the remaining 55 eastern moles captured elsewhere in Texas and in Florida, Kansas, South Carolina, and Tennessee failed (Table 1).
Genetic analysis.
The complete genome of RKPV, designated strain MSB57412, was amplified from one of the four hantavirus-positive eastern moles. Full-length S and L segment sequences were also obtained from RKPV strains MSB57411 and MSB57413.
The full-length 1,830-nucleotide S genomic segment of RKPV strains MSB57411, MSB57412, and MSB57413 contained a single open reading frame (ORF), encoding a 428-amino-acid NP (nucleotide positions 33 to 1319), and 32- and 511-nucleotide 3′ and 5′ noncoding regions (NCR). The putative nonstructural protein (NSs) ORF was absent. By employment of prediction software available in the NPS@structure server, the RKPV NP secondary structure was shown to resemble those of other rodent-, soricid-, and talpid-borne hantaviruses, showing 48.7% α helices, 9.95% β sheets, and two major α-helical domains with the characteristic coiled-coil domain in the N-terminal region (residues 1 to 35 and 51 to 68) and a central β-pleated sheet at the presumed RNA-binding domain (residues 175 to 217).
Despite technical difficulties previously experienced in amplifying and sequencing other soricomorph-borne hantaviruses, the full-length RKPV M segment was obtained from one eastern mole, and from another, a partial sequence of 500 nucleotides was obtained. The complete M genomic segment of RKPV strain MSB57412 was 3,647 nucleotides, with a predicted glycoprotein of 1,136 amino acids (starting at nucleotide position 57) and a 179-nucleotide 5′ NCR. Like those of other rodent- and soricomorph-borne hantaviruses, the RKPV glycoprotein precursor had the highly conserved WAASA amino acid motif (amino acid positions 632 to 636) and four potential N-linked glycosylation sites (three in Gn at amino acid positions 135, 401, and 577 and one in Gc at position 929).
The full-length, 6,558-nucleotide L genomic segment of RKPV strains MSB57411, MSB57412, and MSB57413 encoded a 2,153-amino-acid RNA-dependent RNA polymerase (RdRP) (nucleotide positions 44 to 6505) and exhibited six major conserved motifs (designated premotif A and motifs A, B, C, D, and E), which have been reported for the RNA polymerase function in RNA viruses, including hantaviruses.
Percentages of sequence similarity at the nucleotide and amino acid levels were assessed between the S, M, and L genomic segments of RKPV strain MSB57412 and representative rodent- and soricomorph-borne hantaviruses. RKPV was highly divergent from other hantaviruses, with divergence ranging from 28.4 to 48.2% (nucleotide) and 20.8 to 57.9% (amino acid). RKPV sequences were even more divergent from crocidurine shrew-derived hantaviruses, such as TPMV strain VRC66412 and Imjin virus (MJNV) strain Cl05-11, differing overall by more than 36.7% (nucleotide) and 38.2% (amino acid). On the other hand, RKPV exhibited a higher degree of sequence homology with cricetid-rodent-borne hantaviruses at the nucleotide (S, 67.1 to 70.7%; M, 63.4 to 65.4%; L, 70.1 to 71.6%) and amino acid (S, 72.2 to 79.2%; M, 61.6 to 63.1%; L, 76.0 to 77.9%) levels. The degrees of sequence variation among RKPV strains MSB57411, MSB57412, and MSB57413 were 0.1 to 1.3% (nucleotide) and 0 to 0.2% (amino acid) for the S segment and 0.3 to 1.9% (nucleotide) and 0.5 to 0.6% (amino acid) for the L segment. An exhaustive search for recombination within the full-length S, M, and L segments of RKPV, using multiple recombination detection methods, revealed no convincing evidence of genetic recombination.
Phylogenetic analysis.
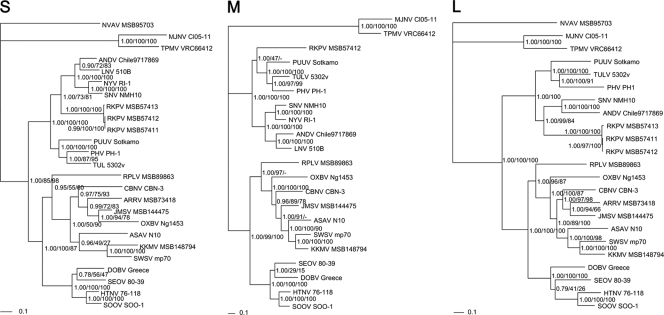
Phylogenetic trees, based on the coding regions of the full-length S, M, and L segments, revealed identical topologies by the ML and Bayesian methods (Fig. 2). Consistently and unexpectedly, the newfound mole-borne hantavirus clustered with Andes virus (ANDV) and Sin Nombre virus (SNV), two prototype hantaviruses harbored by sigmodontine and neotomine rodents, in both the S and the L genomic-segment-based phylogenetic trees, and with Puumala virus (PUUV), Tula virus (TULV), and Prospect Hill virus (PHV), well-characterized arvicolid rodent-associated hantaviruses, in the M genomic-segment phylogenetic tree (Fig. 2). The subfamilies Sigmodontinae, Neotominae, and Arvicolinae are all within the family Cricetidae. Phylogenetic trees, based on the deduced amino acid sequences of the S, M, and L segment-encoded proteins of RKPV and other representative hantaviruses, also revealed similar topologies, with RKPV sharing an ancestral node with hantaviruses harbored by cricetid rodents. Other shrew- and rodent-borne hantaviruses formed two well-defined groups according to their host subfamily (Soricinae and Crocidurinae for shrews; Murinae, Arvicolinae, and Neotominae/Sigmodontinae for rodents) in the hantavirus evolutionary tree.
Fig. 2.
Phylogenetic trees generated by the maximum-likelihood and Bayesian methods, using the best-fit GTR+I+Γ model of evolution as estimated from the data, based on the alignment of the entire coding regions of the 1,287-nucleotide S, 3,411-nucleotide M, and 6,462-nucleotide L genomic segments of RKPV. The phylogenetic positions of RKPV strains MSB57411, MSB57412, and MSB57413 are shown in relationship to those of representative murid-rodent-borne hantaviruses, including Hantaan virus (HTNV 76-118; GenBank accession numbers NC_005218, NC_005219, NC_005222), Soochong virus (SOOV SOO-1; AY675349, AY675353, DQ056292), Dobrava virus (DOBV Greece; NC_005233, NC_005234, NC_005235), and Seoul virus (SEOV 80-39; NC_005236, NC_005237, NC_005238); arvicolid-rodent-borne hantaviruses, including Tula virus (TULV M5302v; NC_005227, NC_005228, NC_005226), Puumala virus (PUUV Sotkamo; NC_005224, NC_005223, NC_005225), and Prospect Hill virus (PHV PH-1; Z49098, X55129, EF646763); and sigmodontine and neotomine-rodent-borne hantaviruses, including Andes virus (ANDV Chile 9717869; NC_003466, NC_003467, NC_003468), Sin Nombre virus (SNV NMH10; NC_00521, NC_005215, NC_005217), New York virus (NYV RI-1; U09488, NYU36801), and Laguna Negra virus (LNV 510B; AF005727, AF005728); soricine-shrew-borne hantaviruses, including Cao Bang virus (CBNV CBN-3; EF543524, EF543526, EF543525) from the Chinese mole shrew (Anourosorex squamipes), Camp Ripley virus (RPLV MSB89863; FJ790772, EF540774, EF540771) from the northern short-tailed shrew (Blarina brevicauda), Seewis virus (SWSV mp70; EF636024, EF636025, EF636026) from the Eurasian common shrew (Sorex araneus), Ash River virus (ARRV MSB73418; EF650086, EF619961) from the masked shrew (Sorex cinereus), Jemez Spring virus (JMSV MSB144475; FJ593499, FJ593500, FJ593501) from the dusky shrew (Sorex monticolus), and Kenkeme virus (KKMV MSB148794; GQ306148, GQ306149, GQ306150) from flat-skulled shrews (Sorex roboratus); crocidurine-shrew-borne hantavirus, including Imjin virus (MJNV Cl05-11; EF641804, EF641798, EF641806) from the Ussuri white-toothed shrew (Crocidura lasiura) and Thottapalayam virus (TPMV VRC66412; AY526097, EU001329, EU001330) from the Asian house shrew (Suncus murinus); talpid-borne hantavirus, including Asama virus (ASAV N10; EU929072, EU929075, EU929078) from the Japanese shrew mole (Urotrichus talpoides), Oxbow virus (OXBV Ng1453; FJ539166, FJ539167, FJ593497) from the American shrew mole (Neurotrichus gibbsii), and Nova virus (NVAV MSB95703; FJ539168, FJ593498) from the European common mole (Talpa europaea). The numbers at each node are posterior probabilities (left), and maximum-likelihood (middle) and neighbor-joining (right) bootstrap supports from 1,000 bootstrap replicates, respectively. The scale bar indicates the number of nucleotide substitutions per site. The GenBank accession numbers for the RKPV S segment were HM015218, HM015223, and HM015224, that for the RKPV M segment was HM015219, and those for the RKPV L segment were HM015220, HM015221, and HM015222.
Virus-host phylogeny analysis.
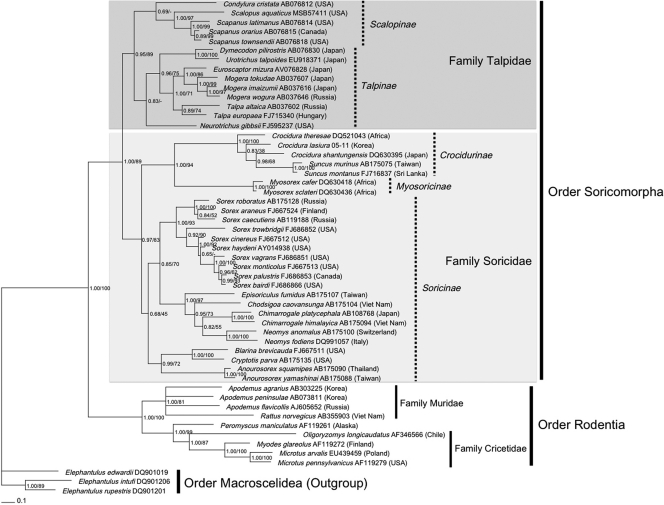
The 1,140-nucleotide cytochrome b gene from eastern moles, in which RKPV strains were detected, was sequenced to confirm the identity of Scalopus aquaticus. A phylogenetic tree based on the entire mtDNA gene revealed two well-supported lineages: one for Rodentia and the other for Soricomorpha (Fig. 3). The Soricomorpha lineage was divided into two families (Talpidae and Soricidae), with Scalopus aquaticus in the cluster comprised of the subfamily Scalopinae. Another subfamily, Talpinae, included Talpa europaea, the host of the divergent NVAV, as well as Neurotrichus gibbsii and Urotrichus talpoides, the hosts of OXBV and ASAV, respectively. Within the family Talpidae, each genus formed monophyletic clades in the subfamilies Uropsilinae, Scalopinae, and Talpinae. The most divergent and basal lineage within the family Talpidae comprised shrew-like moles (genus Uropsilus, subfamily Uropsilinae) and showed a phylogenetic history moderately different from that of shrew moles (tribes Neurotrichini and Urotrichini, subfamily Talpinae). Members of the genus Scalopus were most closely related to North American moles in the genus Scapanus. Phylogenetic analysis based on amino acid sequences of the cytochrome b gene did not provide additional insights into the evolution of hantaviruses and their reservoir hosts (data not shown).
Fig. 3.
Bayesian phylogenetic tree, based on the 1,140-nucleotide cytochrome b region of mitochondrial DNA of mammals within the order Soricomorpha (families Talpidae and Soricidae) and the order Rodentia (families Muridae and Cricetidae). Vertical bars indicate the subfamily within the Talpidae and Soricidae. The tree was rooted using Elephantulus (order Macroscelidea [GenBank accession numbers DQ901019, DQ901206, and DQ901201]) as the outgroup. Numbers at the nodes indicate posterior probability values (left of slash) and bootstrap values of maximum likelihood (right of slash) based on 1,000 bootstrap replicates, respectively.
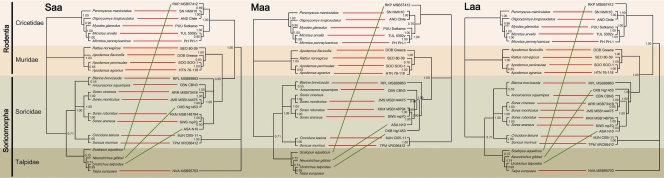
To compare the phylogenetic relationships of hantaviruses with their hosts, tanglegrams, constructed using TreeMap 2.0b (Fig. 4), generally indicated codivergence, with most hantavirus lineages segregating according to the subfamily of the reservoir hosts. RKPV, however, showed discordant matching with its host, much like two other mole-borne hantaviruses, OXBV and ASAV. Moreover, RKPV did not cluster with NVAV, an Old World talpid-borne hantavirus, but was more closely positioned with hantaviruses hosted by rodents in the family Cricetidae.
Fig. 4.
Tanglegrams, generated by TreeMap 2.0b, using consensus maximum-likelihood topologies based on the amino acid sequences of the nucleocapsid protein (labeled Saa), Gn and Gc glycoproteins (labeled Maa), and viral RNA-dependent RNA polymerase (labeled Laa) of RKPV MSB57412 and representative rodent-, shrew-, and mole-borne hantaviruses and cytochrome b mtDNA sequences of the respective reservoir host species. Numbers are posterior probabilities for each node. Virus and host names are provided in the legend to Fig. 2. The concordance of host and hantavirus cladograms was high (red line), except for hantaviruses in moles (ASAV, OXBV, and RKPV), which showed evidence of host switching (green lines).
DISCUSSION
Four genera of moles within the subfamily Scalopinae, namely, Scalopus (eastern mole), Condylura (star-nosed mole), Parascalops (hairy-tailed mole), and Scapanus (western North American mole), and one genus in the subfamily Talpinae, namely, Neurotrichus (American shrew mole), are found in the United States. Previously, we reported evidence for host switching during the evolution of a hantavirus hosted by the American shrew mole (23). We now report a previously unrecognized, distinctly “rodent-like” hantavirus in the eastern mole, the most widely distributed mole species in North America (57).
At least 16 subspecies of Scalopus aquaticus are currently recognized (17, 57), but the phylogeographic variation in this species has not been assessed. Due to inadequate sequence coverage for the eastern mole, it was impossible to establish the subspecies of the RKPV-infected eastern moles in this study. However, it is likely that they are of the subspecies texanus. To what extent other subspecies or geographic variants of Scalopus aquaticus harbor genetic variants of RKPV or entirely different hantaviruses requires further investigation. However, the detection of RKPV in eastern moles only from Rockport, TX, and the failure to detect RKPV in eastern moles from Florida, Kansas, South Carolina, and Tennessee raise interesting possibilities. RKPV in the eastern mole may simply represent spillover, with the eastern mole serving as a secondary host to an as-yet-unidentified present-day rodent reservoir host. Based on the basal position of RKPV in the phylogenetic trees, however, it is more likely that RKPV represents a bona fide mole-borne hantavirus resulting from cross-species transmission in the past, with subsequent host-specific divergence. The focal finding of RKPV in Texas provides the basis for detailed investigations on the transmission of present-day hantaviruses in phylogenetically diverse but distinct small-mammal communities.
Male eastern moles are generally solitary, although they may share burrows or tunnels with other moles in areas where their home ranges overlap (16, 17). However, their generally low population density and a subterranean existence, compared to the high population density and above-ground existence of sympatric rodent species, presumably offer limited opportunities for direct contact. Nevertheless, cross-species transmission of hantaviruses might occur through infectious secretions and excretions. Our previous studies of ASAV (3) and OXBV (23), two shrew mole-borne hantaviruses, indicate probable host switching with soricine shrews. Similarly, the polyphyletic relationship of RKPV and rodent-borne hantaviruses is suggestive of a host-switching event deep in the evolutionary history of these clades. Three of the four hantaviruses described from the family Talpidae (ASAV, OXBV, NVAV, and RKPV) have discordant coevolutionary relationships. The role of this unique host group in the evolution of hantaviruses, as a source or sink for host switching requires further investigation.
Consistently with recent molecular phylogenetic studies (3, 23, 24), our findings confirm that moles serve as hosts of hantaviruses. RKPV in the eastern mole is a genetically distinct hantavirus species by virtue of amino acid sequence differences of 20.8% and 36.9% for the NP and Gn/Gc glycoprotein, respectively, which satisfies the criteria set forth by the International Committee for Taxonomy of Viruses (11, 34). New criteria, based on an exhaustive analysis of hantavirus genomes, have been reported for the demarcation of hantavirus into species (amino acid distance of >10% for S or >12% for M) and into groups (amino acid distance of >24% for S or >32% for M) (32). Based on the these guidelines, RKPV and cricetid-rodent-borne hantaviruses belong to the same group, with S segment amino acid distances of 22.9% for SNV, 20.8% for ANDV, 27.8% for PUUV, 25.2% for PHV, and 22.7% for TULV. This grouping conformed to the results of our phylogenetic analysis.
Apart from the fact that RKPV represents the first example of a hantavirus harbored by a New World mole in the subfamily Scalopinae, the phylogenetic analyses further expand conventional thinking about the complex evolutionary history of hantaviruses. The emerging conceptual framework indicates multiple independent host-switching events through deep evolutionary time, or across deep divergences, followed by local host-specific adaptation and establishment of parallel enzootic cycles. Moreover, the collective data suggest that soricomorph-borne hantaviruses are somewhat more catholic in their host range than present-day rodent-borne hantaviruses, suggesting that ancestral shrews or moles may have served as the early hosts of primordial hantaviruses.
The published literature consists of only a few articles estimating the age of hantaviruses. For example, based on the rates of nucleotide substitutions per site per year for the SNV M and S segments, Black and colleagues concluded that SNV evolved within the past 37 to 106 years (5).
Using a mean rate of 4.245 × 10−4 substitutions per site per year, calculated for hantaviruses by Ramsden and coworkers (40), we estimated that RKPV, ANDV, and SNV shared a common ancestor 900 years before present (±233 years; 95% highest posterior density [HPD]) based on the S segment maximum clade credibility tree. On the other hand, by using a mean rate of 3.62 × 10−6 substitutions per site per year, derived from the work of Hughes and Friedman (20) and Sironen and coworkers (45), RKPV, ANDV, and SNV were shown to have last shared a common ancestor 106,449 years ago (±26,786 years; 95% HPD). Such age estimates, however, are biologically implausible, because they fail to explain how hantaviruses can be found in myriad species within two phylogenetically disparate orders of small mammals that have evolved in widely separated geographic regions across five continents over millions of years.
Although Ramsden and coworkers demonstrated that the divergence dates of hantaviruses were more recent than those of their hosts (40), divergence dates between viruses and hosts sometimes fail to coincide, mainly because RNA viruses evolve so rapidly that the signal is lost (overwhelmed by noise due to error-prone RdRP) long before time scales over which the host diverged are reached. Holmes (18) has argued that evolution in RNA viruses becomes incalculable with respect to rates and timing due to saturation of changes (homoplasy) after 50,000 years (e.g., for divergences of >50,000 years before present).
Because the sequence database of hantaviruses from shrews, moles, and other soricomorphs remains incomplete, it is premature to definitively conclude that recent host-switching events coupled with subsequent divergence are singularly responsible for the similarities between the phylogenies of hantaviruses and their mammalian reservoir hosts. The issue is not whether the evolution of hantaviruses is a direct consequence of either host switching or cophylogeny. Rather, both mechanisms apparently influenced the evolution of hantaviruses. That is, when viewed within the context of molecular phylogeny and zoogeography, the close association between distinct hantavirus clades and specific subfamilies of rodents, shrews, and moles is likely the result of alternating and periodic codivergence through deep evolutionary time. By more fully exploring the vast genetic diversity and phylogenetic divergence of present-day hantavirus species (including as-yet-unidentified soricid- and talpid-borne hantaviruses), the temporal and spatial scales for these events in this fascinating host/pathogen system will become more clear.
ACKNOWLEDGMENTS
We thank the field biologists who originally collected and carefully archived the natural history specimens.
This work was supported in part by U.S. Public Health Service grants from the National Institute of Allergy and Infectious Diseases (R01AI075057 and U54AI065359) and the National Center for Research Resources (P20RR018727 and G12RR003061), National Institutes of Health.
Footnotes
Published ahead of print on 1 June 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Abascal F., Zardoya R., Telford M. J. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arai S., et al. 2008. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am. J. Trop. Med. Hyg. 78:348–351 [PMC free article] [PubMed] [Google Scholar]
- 3. Arai S., et al. 2008. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides). Proc. Natl. Acad. Sci. U. S. A. 105:16296–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arai S., et al. 2007. Hantavirus in northern short-tailed shrew, United States. Emerg. Infect. Dis. 13:1420–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Black W. C., IV, Doty J. B., Hughes M. T., Beaty B. J., Calisher C. H. 2009. Temporal and geographic evidence for evolution of Sin Nombre virus using molecular analyses of viral RNA from Colorado, New Mexico and Montana. Virol. J. 6:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carey D. E., Reuben R., Panicker K. N., Shope R. E., Myers R. M. 1971. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. Indian J. Med. Res. 59:1758–1760 [PubMed] [Google Scholar]
- 7. Charleston M. A., Page R. D. M. 1998. TreeMap 2.0b. Macintosh program for co-phylogenetic analysis, 2.0b ed University of Oxford, Oxford, United Kingdom [Google Scholar]
- 8. Charleston M. A., Perkins S. L. 2006. Traversing the tangle: algorithms and applications for cophylogenetic studies. J. Biomed. Inform. 39:62–71 [DOI] [PubMed] [Google Scholar]
- 9. Combet C., Blanchet C., Geourjon C., Deléage G. 2000. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 25:147–150 [DOI] [PubMed] [Google Scholar]
- 10. Duchin J. S., et al. 1994. Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. N. Engl. J. Med. 330:949–955 [DOI] [PubMed] [Google Scholar]
- 11. Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on the Taxonomy of Viruses, 2nd ed, p. 704–707 Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 12. Frishman D., Argos P. 1996. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 9:133–142 [DOI] [PubMed] [Google Scholar]
- 13. Gavel Y., von Heijne G. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3:433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guermeur Y., Geourjon C., Gallinari P., Deléage G. 1999. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15:413–421 [DOI] [PubMed] [Google Scholar]
- 15. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxf.) 41:95–98 [Google Scholar]
- 16. Hanawalt F. A. 1922. Habits of the common mole: Scalopus aquaticus machrinus (Rafinesque). Ohio J. Sci. 22:164–169 [Google Scholar]
- 17. Harvey M. J. 1976. Home range, movements, and diel activity of the eastern mole, Scalopus aquaticus. Am. Midl. Nat. 95:436–445 [Google Scholar]
- 18. Holmes E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 77:3893–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 20. Hughes A. L., Friedman R. 2000. Evolutionary diversification of protein-coding genes of hantaviruses. Mol. Biol. Evol. 17:1558–1568 [DOI] [PubMed] [Google Scholar]
- 21. Irwin D. M., Kocher T. D., Wilson A. C. 1991. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32:128–144 [DOI] [PubMed] [Google Scholar]
- 22. Jackson A. P., Charleston M. A. 2004. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 21:45–57 [DOI] [PubMed] [Google Scholar]
- 23. Kang H. J., et al. 2009. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii). Virology 388:8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang H. J., et al. 2009. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS One 4:e6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang H. J., Arai S., Hope A. G., Cook J. A., Yanagihara R. 2010. Novel hantavirus in the flat-skulled shrew (Sorex roboratus). Vector Borne Zoonotic Dis. 10:593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King R. D., Sternberg M. J. 1996. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 5:2298–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klempa B., et al. 2007. Novel hantavirus sequences in shrew, Guinea. Emerg. Infect. Dis. 13:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 29. Kukkonen S. K., Vaheri A., Plyusnin A. 2005. L protein, the RNA-dependent RNA polymerase of hantaviruses. Arch. Virol. 150:533–556 [DOI] [PubMed] [Google Scholar]
- 30. Lee H. W., Lee P. W., Johnson K. M. 1978. Isolation of the etiologic agent of Korean hemorrhagic fever. J. Infect. Dis. 137:298–308 [DOI] [PubMed] [Google Scholar]
- 31. Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 32. Maes P., et al. 2009. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect. Genet. Evol. 9:813–820 [DOI] [PubMed] [Google Scholar]
- 33. Nemirov K., Henttonen H., Vaheri A., Plyusnin A. 2002. Phylogenetic evidence for host switching in the evolution of hantaviruses carried by Apodemus mice. Virus Res. 90:207–215 [DOI] [PubMed] [Google Scholar]
- 34. Nichol S. T., et al. 2005. Family Bunyaviridae, p. 695–716. In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses, 2nd ed Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 35. Nichol S. T., et al. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914–917 [DOI] [PubMed] [Google Scholar]
- 36. Plyusnin A., Morzunov S. P. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47–75 [DOI] [PubMed] [Google Scholar]
- 37. Plyusnin A., Vapalahti O., Vaheri A. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677–2687 [DOI] [PubMed] [Google Scholar]
- 38. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 39. Ramsden C., Holmes E. C., Charleston M. A. 2009. Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for co-divergence. Mol. Biol. Evol. 26:143–153 [DOI] [PubMed] [Google Scholar]
- 40. Ramsden C., et al. 2008. High rates of molecular evolution in hantaviruses. Mol. Biol. Evol. 25:1488–1492 [DOI] [PubMed] [Google Scholar]
- 41. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 42. Rost B., Sander C., Schneider R. 1994. PHD—an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10:53–60 [DOI] [PubMed] [Google Scholar]
- 43. Schmaljohn C. S., Dalrymple J. M. 1983. Analysis of Hantaan virus RNA: evidence for a new genus of Bunyaviridae. Virology 131:482–491 [DOI] [PubMed] [Google Scholar]
- 44. Schmaljohn C. S., Hasty S. E., Harrison S. A., Dalrymple J. M. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005–1012 [DOI] [PubMed] [Google Scholar]
- 45. Sironen T., Vaheri A., Plyusnin A. 2001. Molecular evolution of Puumala hantavirus. J. Virol. 75:11803–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song J.-W., Baek L. J., Schmaljohn C. S., Yanagihara R. 2007. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg. Infect. Dis. 13:980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song J.-W., et al. 2007. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol. J. 4:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song J.-W., et al. 2007. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg. Infect. Dis. 13:1784–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song J.-W., et al. 2009. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura). J. Virol. 83:6184–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 51. Swofford D. L. 2003. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 52. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vapalahti O., et al. 1999. Isolation and characterization of a hantavirus from Lemmus sibiricus: evidence for host switch during hantavirus evolution. J. Virol. 73:5586–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yadav P. D., Vincent M. J., Nichol S. T. 2007. Thottapalayam virus is genetically distant to the rodent-borne hantaviruses, consistent with its isolation from the Asian house shrew (Suncus murinus). Virol. J. 4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yanagihara R. 1990. Hantavirus infection in the United States: epizootiology and epidemiology. Rev. Infect. Dis. 12:449–457 [DOI] [PubMed] [Google Scholar]
- 56. Yanagihara R., Gajdusek D. C. 1988. Hemorrhagic fever with renal syndrome: a historical perspective and review of recent advances, p. 151–188. In Gear J. H. S. (ed.), CRC handbook of viral and rickettsial hemorrhagic fevers. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 57. Yates T. L., Schmidly D. J. 1978. Scalopus aquaticus. Mamm. Species 105:1–4 [Google Scholar]
- 58. Zeller H. G., et al. 1989. Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae. Arch. Virol. 108:211–227 [DOI] [PubMed] [Google Scholar]