Abstract
Virus replication and pulmonary disease pathogenesis in ferrets following intranasal infection with a pandemic influenza virus strain (A/California/4/09 [CA09]), a human seasonal influenza H1N1 virus isolate (A/New Caledonia/20/99 [Ncal99]), a classical swine influenza H1N1 virus isolate (A/Swine/Iowa/15/30 [Sw30]), or an avian H1N1 virus isolate (A/Mallard/MN/A108-2355/08 [Mal08]) were compared. Nasal wash virus titers were similar for Ncal99 and Sw30, with peak virus titers of 105.1 50% tissue culture infectious doses (TCID50)/ml and 105.5 TCID50/ml occurring at day 3 postinfection (p.i.), respectively. The mean peak titer for CA09 also occurred at day 3 p.i. but was higher (107 TCID50/ml). In contrast, the peak virus titers (103.6 to 104.3 TCID50/ml) for Mal08 were delayed, occurring between days 5 and 7 p.i. Disease pathogenesis was characterized by microscopic lesions in the nasal turbinates and lungs of all ferrets; however, Sw30 infection was associated with severe bronchointerstitial pneumonia. The results demonstrate that although CA09 is highly transmissible in the human population and replicates well in the ferret model, it causes modest disease compared to other H1N1 viruses, particularly Sw30 infection.
INTRODUCTION
In April 2009, a novel H1N1 influenza A virus was determined to be the cause of outbreaks of respiratory illness in Mexico (4). Within weeks of discovery, this virus was transmitted across communities in North America and was subsequently identified in many areas of the world by May 2009 (1, 5). On 11 June 2009, the World Health Organization (WHO) declared a worldwide pandemic, indicating uncontained community-level transmission of the novel pandemic influenza A virus (pH1N1) in multiple areas of the world (1). Worldwide transmission of pH1N1 continued in both the Northern and Southern Hemispheres from June, and the end of the pandemic was declared in October 2010 (9). This was the first pandemic influenza virus of the H1N1 subtype since the 1918 pandemic, which caused unprecedented worldwide mortality (20). The majority of the fatalities associated with 2009 pH1N1 occurred among individuals with underlying medical conditions such as cardiac and respiratory disease, immune suppression, and pregnancy, with obesity being a key factor associated with fatal outcomes (34). In severe cases, tracheitis, bronchiolitis, and diffuse alveolar damage have been noted with pulmonary edema and increased neutrophilic infiltrates, similar to the pathology observed after infection with other influenza virus strains, including the 1918 H1N1 isolate (11).
Several studies have examined novel swine-origin pH1N1 infection in ferrets because they have proven to be a good model for studying various aspects of human disease associated with influenza virus (18, 24, 26). However, there is some disparity among the published reports regarding the pathogenicity and transmission of pH1N1. For example, a North American isolate, A/California/4/09 (CA09; H1N1), caused no overt clinical signs or marked changes in weight and temperature after intranasal inoculation of ferrets (18, 24, 26). In contrast, ferrets infected with A/Netherlands/602/09, a Eurasian pH1N1 isolate that differs by eight amino acids from CA09, exhibited lethargy, sneezing, ruffled fur, inappetance, nasal discharge, and weight loss (26). In all studies, the levels of virus shedding were higher in trachea and the lungs of ferrets intranasally inoculated with the pH1N1 strain than with the seasonal H1N1 isolates examined for comparison, indicating enhanced replication of pH1N1. However, pulmonary pathology associated with pH1N1 infection was generally found to be intermediate between that of highly pathogenic H5N1 and seasonal H1N1 influenza virus (35).
The human 2009 pH1N1 strain is believed to have originated from a North American swine virus and contains a combination of genes not previously reported in humans or swine (10). These genes are from Eurasian swine virus (neuraminidase [NA] and M), classical swine H1N1 influenza virus (hemagglutinin [HA], nucleoprotein [NP], and NS), and the triple reassortant swine (TRS) virus (PB2, PB1, and PA) (10) lineages. Several of the genes appear to have been originally seeded from avian influenza viruses (PB2, PA, NA, and M), whereas the PB1 gene appears to have been seeded from the human H3N2 lineage. Except for the recent pandemic virus, study of H1N1 isolates has been mainly in the context of evaluating vaccines and antivirals, and the pathogenesis or host response after infection in ferrets has not been extensively examined. There is a lack of data available on the comparison of pH1N1 disease pathogenesis with viruses other than seasonal H1N1 isolates. In this study, we evaluated and compared disease pathology caused by pH1N1 in the context of related H1N1 influenza viruses. While the pandemic H1N1 isolate replicated more efficiently in ferrets, it was less pathogenic than the classical swine H1N1 isolate in ferrets, suggesting that replication efficiency of influenza virus may not strictly correlate with pathogenicity.
MATERIALS AND METHODS
H1N1 Viruses.
Influenza A virus A/California/4/2009 (CA09) was provided by the Center for Disease Control and Prevention (Atlanta, GA). Influenza A virus A/Swine/Iowa/15/30 (Sw30) was kindly supplied by Richard Webby (St. Jude Children's Research Hospital, Memphis, TN), and A/Mallard/MN/AI08-2355/08 (Mal08) was kindly provided by David Stallknecht (Southeast Cooperative Wildlife Disease Study, Athens, GA). Sw30, Mal08, and A/New Caledonia/20/99 (NCal99) were grown in the allantoic cavity of 10-day old embryonated chicken eggs for 48 h at 37°C. CA09 was grown in Madin-Darby canine kidney (MDCK) cells for 72 h at 35°C. Stock viruses were aliquoted and stored at −80°C until use. The 50% tissue culture infectious doses (TCID50) of the stock viruses were determined by the Reed and Muench method.
Ferret studies.
Outbred male ferrets (Triple F Farms, Sayre, PA), 3 to 4 months old, seronegative by hemagglutination inhibition (HAI) assay to A/Brisbane/59/07 (H1N1), A/Uruguay/716/07 (H3N2), and the influenza virus they were inoculated with, were used for these studies. Prior to infection, a subcutaneous implantable temperature transponder (Bio Medic Data Systems, Seaford, DE) was placed in each ferret for identification and temperature readings. A minimum of three ferrets lightly anesthetized with isoflurane were inoculated intranasally per group with 0.2 ml (0.1 ml per nostril) of either a low (102 TCID50) or high (105 TCID50) dose of virus. Weights and temperatures were monitored daily for 7 to 10 days after inoculation. All animal studies were performed according to guidelines approved by the University of Georgia Investigational Animal Care and Use Committee.
Determination of virus titers.
Nasal washes were obtained from ferrets on days 1, 3, 5, 7, and 9 postinoculation (p.i.). Ferrets were anesthetized with ketamine (30 mg/kg), and nasal washes were performed using 2.0 ml of phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin (BSA), penicillin (4,000 U/ml) (Calbiochem, Gibbstown, NJ), streptomycin (800 μg/ml) (Sigma, St. Louis, MO), polymyxin B (400 U/ml) (MP Biochmemicals, LLC, Solon, OH), and gentamicin (100 μg/ml) (Gibco, Carlsbad, CA). Nasal washes were collected into 1.5-ml tubes, and the TCID50 was determined for each sample as previously described (12). Briefly, 10-fold serial dilutions of samples were made from 10−1 to 10−6 in 1× minimal essential medium (MEM) with TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (1 μg/ml; Worthington Biochemical Corporation, Lakewood, NJ). Dilutions of each sample were added to MDCK cells (4 wells for each dilution; 200 μl/well), and the cells were incubated for 48 h at 37°C or 35°C for samples from ferrets inoculated with CA09. The contents of each well were tested for hemagglutination by incubating 50 μl of tissue culture supernatant with 50 μl of 0.5% chicken red blood cells (CRBCs) for 30 min at room temperature. The TCID50 was calculated by the Reed and Muench method (30).
Serological analysis.
Serum samples collected from each ferret prior to infection and at day 14 p.i. were treated with receptor-destroying enzyme (RDE) and tested for serum antibodies to the virus that the ferrets were infected with in a hemagglutination inhibition assay with 0.5% CRBCs or turkey red blood cells (TRBCs) as previously described (38). Viruses were diluted to contain four agglutinating units in sterile PBS.
Virus neutralization (VN) assays were performed in MDCK cells plated in microtiter plates, as previously described (17). The TCID50 was determined for each virus and was calculated by the Reed and Muench method (30). Serum samples collected at 14 days p.i. (dpi) were treated with RDE and heat inactivated as mentioned above. Twofold serial dilutions of serum samples, beginning at 1:20, were made in 1× MEM containing 4% BSA and TPCK-trypsin (1 μg/ml). The homologous virus was standardized to contain 200 TCID50, an equal volume of virus was added to each serum dilution, and the mixture was incubated for 60 min at 37°C. Confluent monolayers of MDCK cells in 96-well plates were washed three times with sterile PBS, and 200 μl of virus-antiserum mixture was added to each well (four wells per mixture). After incubation at 37°C for 72 h, 50 μl of supernatant from each well was tested for HA activity by using 0.5% CRBCs. Geometric mean titers were calculated for each serum sample.
Histopathological examination of tissues.
A total of six ferrets were inoculated with a low dose (102 TCID50/ml) of H1N1 virus. At days 3 and 7 p.i., tissue samples were harvested from three animals from each group infected with a low dose (102 TCID50/ml) of H1N1 virus. Tissue samples from the following organs were collected in 10% buffered formalin for histological examination: trachea, lung, heart, thymus, liver, spleen, stomach, small intestine, large intestine, mesenteric lymph node, pancreas, kidney, adrenal gland, bladder, nasal turbinates, and brain. Formalin-fixed tissues were embedded in paraffin wax, sectioned (4 to 5 μm thick), and placed on glass slides. Tissues were examined histologically using routine hematoxylin and eosin staining to detect lesions consistent with viral infection.
Immunohistochemical staining for the detection of influenza virus viral antigen in nasal and lung sections was performed as previously described (15). Briefly, slides were deparaffinized using HistoChoice (Amersco, Solon, OH) and rehydrated in graduated ethanol baths. Sections were treated in 10 mM citric acid (pH 6) as described previously (7). Slides were then treated with 3% H2O2 for 20 min, blocked using an avidin-biotin block and serum-free protein block (Dako USA, Carpinteria, CA) for 30 min, and incubated with influenza virus NP-specific monoclonal antibody (Biodesign International, Saco, ME) for 60 min at 37°C. The slides were washed with PBS (three times for 3 min each time at room temperature), and incubated with biotinylated goat anti-mouse antibody for 60 min at room temperature and then with ABC substrate according to the manufacturer's specifications (ABC Elite kit; Vectorlabs, Burlingame, CA). DAB (diaminobenzidine) was used as chromogen, and slides were lightly counterstained with Lillie-Mayer hematoxylin for 2 min and mounted using Permount mounting medium (Fisher Scientific, Fair Lawn, NJ).
Real-time RT-PCR.
Tissue homogenates and whole blood were used for PCR analysis. RNA was extracted using TRIzol LS Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's recommended protocol until the phase separation step. RNA was further purified using an RNeasy kit (Qiagen, Inc., Valencia, CA). One-step real-time reverse transcription-PCR (RT-PCR) for the detection of the H1N1 M gene was performed following the protocol established by the WHO Collaborating Centre for influenza at the Centers for Disease Control and Prevention (Atlanta, GA) using a One-Step RT-PCR kit (Qiagen, Inc.) (22). Controls included RNA extracted from virus stocks. Samples were analyzed by real-time PCR with a Stratagene Mx3000P quantitative PCR (QPCR) system (Agilent Technologies, Inc., Santa Clara, CA) for 45 cycles. When all controls meet the requirements outlined in the protocol, a sample was considered presumptive-positive for influenza A virus if the reaction growth curve crossed the threshold line within 35 cycles.
NA enzyme inhibition assay.
Neuraminidase (NA) activity was measured by a modified fluorescence-based NA enzyme inhibition assay using the fluorigenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid sodium salt hydrate (MUNANA; Sigma-Aldrich, St. Louis, MO) (13, 29). Wild-type and resistant A/MS/3/01 (H1N1) viruses were provided by the Neuraminidase Inhibitor Susceptibility Network (Australia) and were used as controls in this assay. Briefly, viruses were standardized to equivalent NA enzyme activity in the linear range of the curve and incubated with the NA inhibitor (NAI) oseltamivir carboxylate (oseltamivir) [(3R,4R,5S)-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylic acid] at concentrations of 0.00005 to 10 μM. Oseltamivir obtained from F. Hoffmann-La Roche was dissolved in distilled water, and aliquots were stored at −20°C until use. After incubation with oseltamivir MUNANA at a final concentration of 100 μM in 325 mM 2-(N-morpholino)ethanesulfonic acid ([MES] pH 6.5; Sigma), buffer containing 10 mM CaCl2 was added. The reaction was terminated after 1 h at 37°C by addition of 150 μl of freshly prepared stop solution (25% ethanol and 12.5% glycine; Fisher Scientific, Rochester, NY) in distilled water. The fluorescence of the released 4-methylumbelliferone was measured in a Safire2 multifunctional monochromator-based microplate reader (Tecan, San Jose, CA) using excitation and emission wavelengths of 355 and 460 nm, respectively. The drug concentration that inhibited 50% of the NA enzymatic activity (EC50) was determined from the dose-response curve using Microsoft Office Excel 2007 (Microsoft Corp., Redmond, WA) and GraphPad Prism, version 5.01 (GraphPad Software, San Diego, CA). EC50s are reported as the means of two independent determinations.
RESULTS
Clinical signs in ferrets caused by H1N1 viruses of different host origin.
We compared the clinical disease caused by pH1N1 A/California/4/09 (CA09) to that caused by a human seasonal H1N1 isolate, A/New Caledonia/20/99 (NCal99), a vaccine component for the seasonal influenza virus vaccine from 2000 to 2007, and a classical swine H1N1 isolate, A/Swine/Iowa/15/30 (Sw30), because three gene segments (HA, NP, and NS) of the CA09 isolate are derived from classical swine H1N1 virus (10). A North American avian H1N1 (Mal08) isolate was included for comparison because several of the gene segments of the pandemic H1N1 were derived from the North American avian gene pool (10). Ferrets inoculated with CA09, NCal99, and Mal08 showed no overt clinical signs, including respiratory symptoms, weight loss, or change in body temperature during the observation period. Ferrets inoculated with both a low and high dose of Sw30 remained alert throughout the study; however, on days 4 to 6 p.i., ferrets in both groups exhibited reduced activity. A rise in body temperature was measured on day 4 p.i. in one ferret inoculated with 102 TCID50/0.2 ml of Sw30, and the body temperature (40.7°C) remained elevated for 2 days. This result showed that infection with Sw30 caused overt clinical disease signs in ferrets.
Replication of H1N1 viruses from different host origins in ferrets.
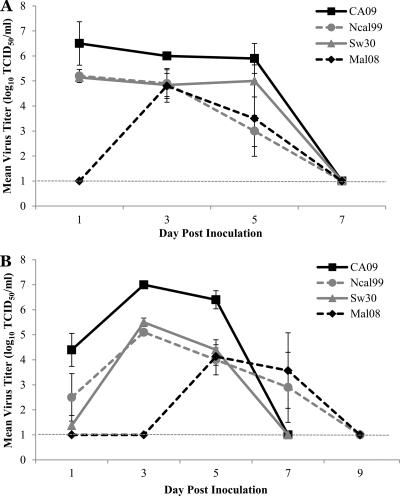
To determine the virus dose required to induce disease in ferrets infected with H1N1 influenza viruses from different origins, ferrets were intranasally inoculated with 0.2 ml (0.1 ml per nostril) of either a low (102 TCID50) or high (105 TCID50) dose of virus, and nasal washes were collected beginning at day 1 p.i. until virus was no longer detected. With the high-dose inoculum, virus titers of >105 TCID50 were isolated at day 1 p.i. for all virus groups except Mal08, and these represent mean peak virus titers (Fig. 1A). The mean peak virus titer, 104.8 TCID50/ml, for the Mal08 infection group was reached at day 3 p.i. Ferrets inoculated with a low dose of virus (102 TCID50) had mean peak virus titers on day 3 p.i. for all viruses examined except the Mal08, which reached mean peak titers at day 5 p.i. (104.1 TCID50/ml) (Fig. 1B). The mean peak virus titers for NCal99 and Sw30 were comparable at 105.1 TCID50/ml and 105.5 TCID50/ml while the mean peak titer for CA09 was approximately 2 logs higher (107 TCID50/ml). Both the high- and low-dose virus titers of CA09 further support the efficient replication of this isolate. Mean virus titers in nasal washes decreased by day 5 p.i., and virus was undetectable at day 7 p.i. for the CA09 and Sw30 infection groups. Virus was isolated from one of three ferrets inoculated with NCal99 at day 7 p.i. (103.4 TCID50/ml) and one of three ferrets in the Mal08 infection group (103.6 TCID50/ml). By day 9 p.i., virus was undetectable in nasal washes from all ferrets inoculated with any H1N1 virus.
Fig. 1.
Replication kinetics in the upper respiratory tract of H1N1 isolates in ferrets. Groups of three ferrets were intranasally inoculated with 0.2 ml (0.l ml per nostril) of a high dose (105 TCID50) (A) or low dose (102 TCID50) (B) of each H1N1 isolate tested. Nasal washes were collected on days 1, 3, 5, 7, and 9 after inoculation. Data are presented as mean virus titer ± standard deviation (log10 TCID50/ml) on the indicated day. The limit of detection was 101 TCID50/ml.
It has been previously reported that swine, equine, and avian influenza A viruses can be isolated not only from the respiratory tract but also from the intestinal tract of ferrets, whereas most human influenza viruses are recovered only from the respiratory tract (21). To determine if the H1N1 influenza virus isolates studied here replicated in the gastrointestinal tract, rectal swabs were collected from ferrets on days 3 and 5 p.i. for virus isolation. Swab samples were considered to be positive for virus if at least one of the three embryonated chicken eggs was positive by the hemagglutination test. Virus was not detected at any time point in rectal swabs of ferrets inoculated with either dose of CA09 (Table 1). However, virus was detected at day 3 p.i. in the rectal swab of 1 of 3 ferrets inoculated with 102 TCID50/0.2 ml NCal99, and virus was undetectable at day 5 p.i. for both groups of NCal99-infected ferrets. Rectal swabs were positive for virus on day 3 p.i. from 2 of 3 ferrets inoculated with 102 TCID50/0.2 ml of Sw30, and rectal swabs for all three ferrets in this group were positive for virus on day 5 p.i. In addition, virus was detected in 1 of 3 ferrets inoculated with 105 TCID50/0.2 ml of Sw30, and the rectal swab for the same ferret was also positive at day 5 p.i. Virus was detected from rectal swabs collected from 3 of 6 ferrets inoculated with a low dose of Mal08 on day 3 p.i. and from 2 of 3 ferrets on day 5 p.i. Rectal swabs were not collected from ferrets inoculated with a high dose of the Mal08 virus given the preponderance of virus via low-dose inoculation.
Table 1.
Virus isolation results after inoculation of ferrets with H1N1 isolates
| Virus | Inoculum dose (TCID50/ml) | Virus detection (no. of positive animals/total no. tested [titers]) by sample typea |
|||||
|---|---|---|---|---|---|---|---|
| Rectal swab at: |
Other at day 3 |
||||||
| Day 3 | Day 5 | Small intestine | Olfactory bulb | Lung | Nasal washes | ||
| A/California/4/09 | 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 6/6 (6.7, 6.4, 6.4, >6, >6, 5.4) |
| A/New Caledonia/20/99 | 102 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 6/6 (6.0, 5.7, 5.4, 5.4, 5.0, 5.0) |
| A/Swine/Iowa/15/30 | 102 | 5/6 | 3/3 | 0/3 | 0/3 | 2/3b (<1c, 3.7, 4) | 6/6 (6.4, 6.2, 5.4, 5.4, 5.2, 4.9) |
| A/Mallard/MN/A108-2355/08 | 102 | 3/6 | 2/3 | 0/3 | 0/3 | 0/3 | 2/6 (3.2, 2.7, <1, <1, <1, <1) |
Titers are expressed as log10 TCID50/ml.
Confirmed by real-time PCR.
Limit of detection.
To determine if virus replicated in internal organs, three ferrets from each virus group were euthanized, and the olfactory bulb, a section of small intestine, and lung tissue from each ferret was collected at day 3 p.i. Virus was isolated from nasal washes collected prior to necropsy from all ferrets on day 3 p.i. with the exception of the ferrets in the Mal08 infection group due to the delayed replication kinetics of the virus; however, rectal swabs collected from ferrets in the Mal08 infection group were found to be positive for virus on day 3 p.i. (Table 1). Tissue samples were homogenized, and virus titers were determined by endpoint titration in MDCK cells. Virus was below the limit of detection (<101 TCID50/ml) in the olfactory bulb and intestinal samples of ferrets in all H1N1 virus groups, and virus was undetectable from lung samples of ferrets inoculated with the NCal99, Mal08, or CA09 H1N1 isolate. In contrast, virus was isolated from the lungs of two of the three ferrets inoculated with Sw30 (103.7 and 104 TCID50/ml). Isolation of virus in the lungs of the Sw30 ferrets was confirmed using real-time RT-PCR for the detection of the influenza virus M gene. RT-PCR threshold cycle (CT) values were greater than 35 for all other organ samples as well as whole blood obtained from all ferrets, confirming the absence of virus in these samples.
Pathology associated with H1N1 infections in ferrets.
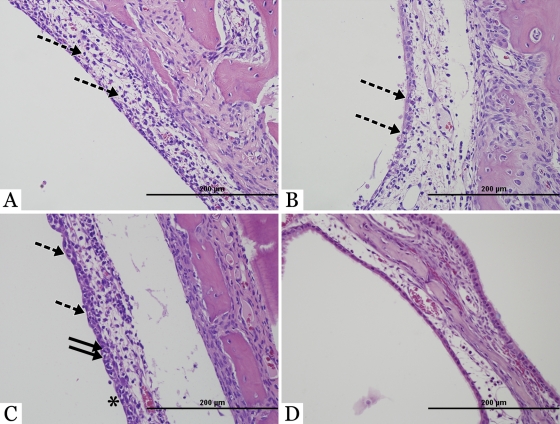
To determine if infection by different H1N1 viruses resulted in variation in pathology, three ferrets from each virus group were euthanized, and organs were harvested at days 3 and 7 p.i. Gross lesions were not apparent in any tissues at the time of necropsy. Microscopic lesions were observed only in the liver, nasal turbinates, and lungs following infection by any H1N1 virus (Table 2 and Fig. 2 and 3). Chronic portal hepatitis was observed in the liver of all ferrets. These changes were interpreted to be unrelated to the experimental viral infection; rather, they were likely the result of a previous ascending enteric infection or intestinal parasitism because no inflammation or parasites were found in the intestines during necropsy. Specific histologic lesions related to the experimental viral infection were rhinitis and bronchointerstitial pneumonia of various degrees of severity. Histologically, rhinitis manifested as expansion of the mucosa with edema and infiltration with inflammatory cells (lymphocytes, plasma cells, and lesser numbers of neutrophils). The surface epithelial cells were often attenuated or necrotic. Additionally, neutrophils were observed migrating through the surface epithelial cells toward the nasal cavity. In many instances, inflammatory exudate composed of many degenerate neutrophils, cellular and nuclear debris, and mucus was present in the nasal cavity, sometimes adhered to the mucosal surface. The severity of rhinitis was classified as mild, moderate, marked, or severe, based upon the degree of inflammation and accumulation of inflammatory exudate on the nasal epithelial surface. Rhinitis was observed in the nasal turbinates of all ferrets at day 3 p.i., with the exception of the nasal turbinates from ferrets in the Mal08 infection group which were essentially normal (Fig. 2). At 7 dpi, one ferret in the Mal08 infection group had mild rhinitis while another had moderate rhinitis (data not shown). Infection with CA09 virus resulted in moderate rhinitis in one ferret and mild rhinitis in another ferret at 3 dpi (Fig. 2A). Rhinitis persisted at day 7 p.i. and was of moderate severity in ferrets inoculated with CA09 with scattered degenerating epithelial cells (data not shown). NCal99 infection in all three ferrets resulted in mild rhinitis at 3 dpi with slightly edematous mucosa and neutrophils transmigrating the epithelium (Fig. 2B). Histologically, these nasal lesions were similar to the ones described above except that they were less severe. By day 7 p.i., rhinitis persisted and was of moderate severity, with further lesions observed which included scattered degenerating epithelial cells in larger numbers, particularly in the nasal turbinates of ferrets inoculated with NCal99 (data not shown). Moderate to severe rhinitis was observed in the nasal turbinates of ferrets inoculated with Sw30 (Fig. 2C), with attenuation and degeneration of the surface epithelium, edematous expansion of the nasal mucosa, and neutrophils transmigrating though the epithelium. In all groups the histologic characteristics of rhinitis at day 7 p.i. were not significantly different from those at day 3 p.i. Immunohistochemistry for influenza virus viral antigen was performed on the nasal sections from ferrets at day 3 p.i. Nasal sections were uniformly negative in all groups except the Sw30 infection group, which revealed viral antigen in the nuclei of surface epithelial cells, scattered stromal epithelial cells in the submucosa, and intraepithelial neutrophils of the ferret nasal passages (data not shown).
Table 2.
Histopathology results after inoculation of ferrets with H1N1 isolates
| Organ | Severity of lesions by virus and day p.i. (clinical symptom)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| A/California/4/09 |
A/New Caledonia/20/99 |
A/Swine/Iowa/15/30 |
A/Mallard/MN/A108-2355/08b |
|||||
| 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | |
| Brain | − | − | − | − | − | − | − | − |
| Trachea | − | − | − | − | − | − | − | − |
| Nasal turbinates | + (rhinitis) | ++ (rhinitis) | + (rhinitis) | ++ (rhinitis) | ++ (rhinitis) | ++ (rhinitis) | − | + (rhinitis) |
| Lung | − | ++ (bronchopneumonia) | + (bronchopneumonia) | − | + (bronchopneumonia) | ++ (bronchopneumonia) | + (bronchopneumonia) | + (bronchopneumonia) |
| Heart | − | − | − | − | − | − | − | − |
| Thymus | − | − | − | − | − | − | − | − |
| Stomach | − | NS | NS | − | − | − | − | − |
| Small intestine | − | − | − | − | − | − | −* | −* |
| Large intestine | − | − | − | − | − | − | − | − |
| Mesenteric lymph node | − | − | − | − | − | − | − | − |
| Pancreas | − | − | − | − | − | − | − | − |
| Liver | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) | + (portal hepatitis) |
| Spleen | + (lymphoid hyperplasia) | + (lymphoid hyperplasia) | + (lymphoid hyperplasia) | + (lymphoid hyperplasia) | + (lymphoid hyperplasia) | + (lymphoid hyperplasia) | − | − |
| Kidney | − | − | − | − | − | − | − | − |
| Adrenal gland | − | − | − | NS | − | − | NS | − |
| Bladder | − | − | − | − | − | − | − | − |
−, minimal; +, mild; ++, moderate; +++, severe. NS, no sample.
*, one of three animals had mild enteritis, and virus was detected in rectal swabs at 3 dpi.
Fig. 2.
Histopathology in the upper respiratory tract of ferrets. Hematoxylin- and eosin-stained images of nasal turbinates from ferrets at day 3 after intranasal inoculation with a low dose (102 TCID50) of A/California/4/09 (A), A/New Caledonia/20/99 (B), A/Swine/Iowa/15/30 (C), or A/Mallard/MN/A108-2355/08 (D). (A and B) Mild rhinitis with edematous mucosa and multiple neutrophils (dashed arrows) was observed after inoculation with human H1N1 isolates. (C) A classical swine isolate caused moderate rhinitis, with multiple neutrophils migrating through the epithelium (dashed arrows), degenerating epithelial cells (solid arrows), and inflammatory cells on the mucosal surface (asterisk). However, after inoculation with an avian H1N1 isolate (D) the nasal turbinates were histologically normal.
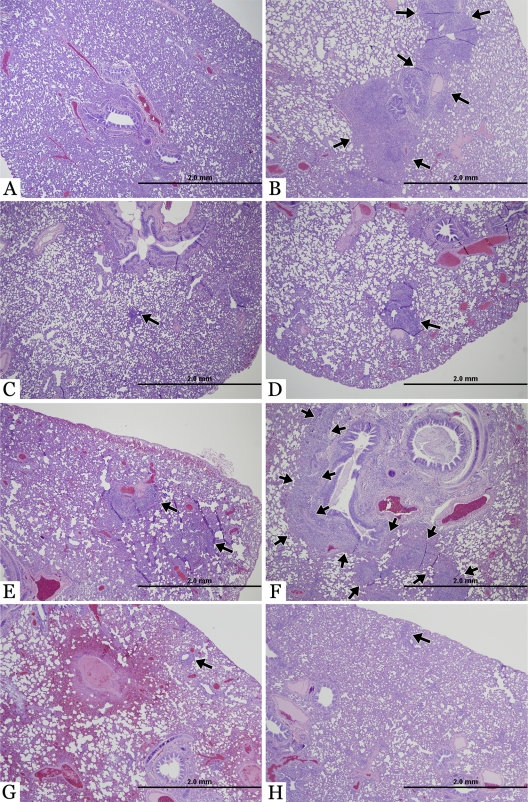
Fig. 3.
Lung pathology in ferrets inoculated with H1N1 isolates. Hematoxylin- and eosin-stained images of microscopic lesions in the lungs of ferrets inoculated with a low dose (102 TCID50) of virus. (A) Histology of a normal lung at 3 dpi from a ferret infected with A/California/4/09. (B) Low-magnification photomicrograph of moderate focal bronchopneumonia in a ferret infected with A/California/4/09 at 7 dpi. The locally extensive pneumonic process is highlighted with arrows. (C) Minimal focal bronchopneumonia in a ferret infected with A/New Caledonia/20/99 at 3 dpi. The focal pneumonic process is highlighted with an arrow. (D) Mild focal bronchopneumonia in a ferret infected with A/New Caledonia/20/99 at 7 dpi. (E) Mild focal bronchopneumonia (arrows) in a ferret infected with A/Swine/Iowa/15/30 at 3 dpi. (F) Moderate to marked locally extensive bronchopneumonia (highlighted with arrows) in a ferret infected with A/Swine/Iowa/15/30 at 7 dpi. With infection with A/Mallard/MN/A108-2355/08 there is mild focal bronchointerstitial pneumonia in the lungs (arrow) at both 3 dpi (G) and 7 dpi (H).
The most significant lesions among all organs examined were observed in the lungs (Fig. 3); however, the severity of the lesions differed among groups and by the number of days postinfection. The anatomic pattern of pneumonia was bronchointerstitial, demonstrating a combination of bronchiolar necrosis and alveolar damage, with the latter being localized to the alveoli adjacent to affected bronchioles. Bronchiolar necrosis affected solely the lining epithelial cells, usually in a multifocal fashion. Epithelium of the other, nonaffected areas of the bronchioles alternated between attenuation (cuboidal to flattened bronchiolar epithelium) and hyperplasia (piled up bronchiolar epithelium). Alveolar damage was histologically characterized by small, patchy areas of necrosis, thickening of the alveolar walls, occasional fibrin strands within alveoli, and neutrophilic to lymphoplasmacytic inflammatory cells in various numbers within alveolar walls and alveoli. The pneumonic process was further described according to its severity (mild, moderate, marked, or severe), based upon the degree of inflammation and its extent throughout the parenchyma. At day 3 p.i., lesions were not observed in the lungs of ferrets inoculated with CA09 (Fig. 3A) although the lungs of ferrets inoculated with CA09 revealed more severe bronchointerstitial pneumonia on day 7 p.i. than was observed in ferrets from this group at day 3 p.i., with local inflammation centered on bronchioles (Fig. 3B). At 3 dpi, a mild focal bronchointerstitial pneumonia was observed in the cranial, caudal, and accessory lobe of the lungs of ferrets inoculated with NCal99 (Fig. 3C). By 7 dpi, mild bronchointerstitial pneumonia was observed in the caudal lobe of only one ferret inoculated with NCal99. Mild bronchointerstitial pneumonia was also detected at day 3 p.i. in the caudal and accessory lobe of the lungs of ferrets inoculated with Sw30 (Fig. 3E). The most severe bronchointerstitial pneumonia was observed in ferrets inoculated with Sw30 (Fig. 3F), with extensive marked inflammation centered on bronchioles. In contrast to the other viruses, infection with Mal08 resulted in mild bronchointerstitial pneumonia at both days 3 and 7 p.i. (Fig. 3G and H). Immunohistochemistry for influenza virus viral antigen was performed on the lung sections from ferrets at day 3 p.i. and were uniformly negative in all groups.
Serologic response to H1N1 viruses isolated from different hosts.
To determine the serum antibody response of ferrets to the various H1N1 isolates, blood was collected from all ferrets before infection and at 14 days p.i. to test for seroconversion. All ferrets were shown to be seronegative by the hemagglutination inhibition (HAI) assay for currently circulating human H3 and H1 influenza virus strains, as well as the homologous virus with which they were inoculated (data not shown). All ferrets seroconverted after inoculation with H1N1 influenza viruses as measured by an HAI assay, with the exception of the Mal08 ferrets (Table 3). Mean serum antibody titers for ferrets inoculated with CA09 were higher than the mean antibody titers of ferrets inoculated with either NCal99 or Sw30. Ferrets inoculated with a low dose of CA09 had a mean serum antibody titer of 1:1,067 while the mean antibody titer of ferrets inoculated with the same dose of either NCal99 or Sw30 was 1:427. Serum antibody responses for ferrets in the Mal08 infection group were undetectable by the HAI assay, but the mean serum antibody titers were found to be 1:1,271 (range, 1:381 to 1:2,153) for the low-dose group and 1:1,943 (range, 1:1,522 to 1:2,153) for the high-dose group by virus microneutralization assay.
Table 3.
Serum antibody titers in ferrets at 14 days postinoculation
| Virus titer | Mean HAI titer at 14 dpi (range) by virus |
|||
|---|---|---|---|---|
| A/California/4/09 | A/New Caldonia/20/99 | A/Swine/Iowa/15/30 | A/Mallard/MN/A108-2355/08 | |
| 102 | 1,067 (640–≥1280) | 427 (320–640) | 427 (320–640) | <20 (<10–20)a |
| 105 | 533 (320–≥1280) | 640 | 427 (320–640) | 27 (20–40)b |
Mean VN titer (range) of 1,271 (381–2,153).
Mean VN titer (range) of 1,943 (1,522–2,153).
It has been reported that infection with 1918 influenza virus and/or closely related human H1N1 viruses elicited neutralizing antibodies to pH1N1 isolate, CA09 (18). Experimental data showed that persons 60 years and older had some level of cross-reactive antibodies to the novel swine origin H1N1 strain (14). At the amino acid level, homology between the HA of CA09 and Sw30 is 85%; however, serum from ferrets infected with Sw30 cross-reacted with CA09 (1:160). Interestingly, serum from ferrets infected with CA09 did not cross-react to Sw30 (<1:20). Also the homology between the HAs of Sw30 and NCal99 (84%) is similar to the homology between Sw30 and CA09 (85%), and serum antibodies against NCal99 did not cross-react with Sw30. Homology between NCal99 and CA09 is lower (79%), and serum antibodies did not cross-react. This suggests that Sw30 shares an antigenic epitope with CA09.
Sensitivity of H1N1 virus isolates to a neuraminidase inhibitor.
Mutations in the conserved residues of the NA that are also associated with decreased susceptibility to NAI have been shown to decrease the sialidase activity and/or stability of the NA, thus compromising viral fitness (39). In some cases, these viruses have also been shown to have reduced virulence and rates of replication and transmissibility in mice and ferrets (3, 33). To determine if the NA plays a role in the differences observed in the replication kinetics of the H1N1 isolates in ferrets, the susceptibility of the viruses to oseltamivir was tested (39). To determine the level of susceptibility of the H1N1 isolates to the neuraminidase inhibitor oseltamivir, the efficacy of the compound was tested against each virus. The H1N1 isolates were all sensitive to the neuraminidase inhibitor oseltamivir, with the CA09 isolate being the most sensitive (EC50 of 1.7) and the Sw30 the least sensitive (EC50 of 15.4) (Table 4). Sequence analysis of the Mal08 NA showed the presence of 274Y, which is known to be involved in decreased susceptibility to oseltamivir; however, the EC50 against the compound was 14.2.
Table 4.
Virus sensitivity to oseltamivir in the NAI assay
| Influenza H1N1 virus | NA mutation | Oseltamivir sensitivity (EC50 [nM])a |
|---|---|---|
| A/MS/3/01 | 5.4 | |
| A/MS/3/01 | 274Y | 599 |
| A/California/4/09 | 1.7 | |
| A/New Caldonia/20/99 | 2.4 | |
| A/Swine/Iowa/15/30 | 15.4 | |
| A/Mallard/MN/A108-2355/08 | 274Y | 14.2 |
Values were determined by a fluorescence-based assay and are the average of two independent replicates.
DISCUSSION
While the 2009 H1N1 pandemic virus transmitted efficiently from human-to-human and spread to over 214 countries worldwide, in general the virus caused mild illness with very little mortality. Previous reports assessing the pathogenicity of the prototypic pandemic H1N1 strain A/California/4/09 (CA09) in mammalian models suggested that it was more pathogenic than seasonal H1N1 influenza viruses, with a wider distribution of virus replication and lesions in the upper and lower respiratory tract of ferrets (18, 26). Here, we characterized the pathogenicity of the A/California/4/09 (H1N1) virus in ferrets in the context of related precursor H1N1 viruses. Consistent with other studies, CA09 replicated to higher titers in the upper respiratory tract of ferrets than the seasonal, avian, and classical swine H1N1 isolates tested. In addition, the pandemic strain was more immunogenic in ferrets, inducing high levels of serum antibodies after low-dose intranasal inoculation. Contrary to other reports, it caused a milder clinical disease than what was previously reported, and lesions were not apparent in the lungs of ferrets inoculated with the pandemic strain until day 7 after inoculation with a low dose of virus. The most severe pulmonary lesions were observed in ferrets inoculated with a classical swine H1N1 isolate, a precursor virus contributing several gene segments to the H1N1 pandemic strain.
With the exception of the 1918 H1N1 pandemic strain, replication of H1N1 influenza virus and the pathology associated with this subtype of virus in ferrets had not been extensively studied outside the context of vaccine testing. Until recently, few studies have characterized the pathology caused by either seasonal H1N1 influenza viruses or swine influenza virus in ferrets. Previous reports assessing the pandemic H1N1 isolate in mammals showed that the virus could replicate to high titers in both the upper and lower respiratory tract compared to seasonal influenza virus (18, 24, 26). One study where virus was isolated from lungs on day 3 after intranasal inoculation with CA09 showed that infected ferrets had more severe bronchopneumonia with viral antigen in the peribronchial glands and fewer alveolar cells than produced by infection with a recent seasonal human H1N1 isolate (18). Lung lesions at days 3 and 6 after inoculation included alveolar spaces filled with edema, inflammatory cells, and cell debris. However, virulence and mortality are dose and strain dependent, with mortality observed only in ferrets inoculated with higher doses of the pandemic strain. In our studies intranasal inoculation of a low dose of the pandemic H1N1 caused similar or less severe pulmonary lesions than inoculation with other H1N1 isolates examined, with Sw30 demonstrating the most severe pulmonary lesions. This may be more consistent with the clinical course in the human population as most cases were uncomplicated, mild, and had a clinical course similar to that observed for seasonal influenza virus infection. The details of pathology induced by mild influenza virus infection in humans remain unknown because most infections are self-limiting and resolve. Therefore, it is difficult to know whether disease in animal models accurately mirrors the disease in humans or at least the disease aspect under investigation. A low dose may more accurately reflect what would be encountered by humans.
The 2009 pandemic strain originated from triple reassortant swine (TRS) viruses with a gene constellation comprised of segments from Eurasian swine (NA and M) and classical swine H1N1 (HA, NP, and NS) influenza viruses; thus, antigenically the 2009 H1N1 pandemic virus is most similar to classical swine H1N1 isolates (10). Swine influenza virus infections in humans have been reported throughout the world, with the first isolation of a swine influenza virus from a human in 1974 although influenza virus was first isolated from a pig in 1930 (27, 31, 32). Despite this, there have been few studies documenting the pathology associated with swine influenza virus isolates in ferrets, the animal model for humans. Recently, it has been shown that an early classical swine H1N1 influenza virus isolated in 1931 caused severe clinical disease and lung pathology, with necrotizing bronchiolitis and alveolitis similar to those observed with the 1918 pandemic virus (25). This was in stark contrast to a contemporary seasonal human H1N1 isolate which resulted in mild clinical disease. Another study showed that recent Eurasian swine isolates (H1N1 and H3N2) replicated in ferrets, caused transient fever, and were readily transmitted from pigs to contact pigs and ferrets (6). TRS H1N1 influenza viruses isolated from humans in 2007 and 2008 were similar to the 2009 pandemic H1N1 in terms of their pathogenicity in ferrets (2). Intranasal inoculation with the TRS isolates caused mild clinical disease, replicated in the respiratory and intestinal tissues, and elicited lung pathology more pronounced than seasonal H1N1 infection and generally similar to that observed after pH1N1 infection in ferrets, although with slightly less pathology in the lung parenchyma. Here, we showed that the classical swine H1N1 isolate replicated in both the upper and lower respiratory tract of ferrets and caused mild clinical disease symptoms, including a slight elevation in temperature with reduced activity. In addition, this isolate replicated in the gastrointestinal tract and caused the most severe lesions in the lungs of ferrets even after inoculation with a low dose of virus. This suggests that the pathogenicity of influenza viruses isolated from pigs should be more carefully monitored and characterized in an attempt to assess the pandemic potential of such viruses.
The neuraminidase (NA) of influenza virus is responsible for release of newly assembled virus particles from the cell surface by cleaving α-ketosidic linkages between terminal sialic acid and adjacent sugar residues (37). Studies have shown that mutations that are part of the NA enzyme catalytic site or that surround it may impair virus fitness, and these mutations generally decrease susceptibility to neuraminidase inhibitors. In addition, human influenza virus isolates which harbor NA mutations that confer resistance to NA inhibitors have been generally attenuated in animal models (16, 19). A recent study showed that a 2009 pandemic H1N1 isolate and its NA mutant counterpart, resistant to oseltamivir, caused a similar disease course in ferrets without apparent attenuation of clinical signs (8). In addition, the NA inhibitor-resistant mutant retained its ability to efficiently transmit by direct contact. Interestingly, the mutant isolate had a significant growth delay during the first 12 h postinfection in two cell lines. These authors suggested that the growth delay could be due to delayed release of virus progeny or impaired viral penetration into the host respiratory tract due to reduced NA enzyme efficiency. This could be the case with the avian H1N1 isolate tested in our study since it carries the resistance marker and there was a delay in replication in the ferrets. On the other hand, the CA09 isolate used in our studies was highly sensitive to oseltamivir and replicated more efficiently in the ferrets than the other isolates tested. Although we cannot directly compare the isolates studied due to the high rate of divergence between the NA segments, perhaps the NA of the CA09 virus has more efficient activity enabling more efficient spread of the virus to susceptible host cells.
While most cases of pandemic H1N1 influenza virus infection were mild and self-limited, more deaths occurred in a younger age group than normally occur with seasonal influenza virus. A majority of the severe cases of pandemic H1N1 were among immunocompromised patients. Of fatal cases for which information was available, it was estimated that 91% had underlying medical conditions such as cardiac and respiratory diseases, immunosuppression, pregnancy, and obesity (11). Immune status of the host is a factor in pathogenicity, and it has been suggested that the severity of disease seen in this subset of the population could be related to a less effective innate and adaptive immune response. While the innate immune response plays a primary role in clearance of infection in naïve animals, antibodies produced against the HA are neutralizing and, thus, are the primary immune mediator for protection of the host against reinfection. In ferrets, high levels of serum antibodies were produced after inoculation with a low dose of the pandemic H1N1 virus, which perhaps accounts for the modest severity of disease that was observed. The ferrets used in these studies were immune competent. It would be interesting to assess the pathogenicity of the pandemic H1N1 isolate in immunocompromised ferrets to determine if the disease severity mimics more closely that seen in immunocompromised humans.
The ferret has become the established animal model for studying influenza disease pathogenesis because it is generally accepted that influenza virus infection in ferrets closely mimics that in humans with respect to clinical signs, pathogenesis, and antibody responses, and ferrets are naturally susceptible to infection with human influenza A viruses (23). Ferrets also share similarities to humans in terms of lung physiology, airway morphology, and cell types present in the respiratory tract, including the distribution of α-2,6-linked sialic acids, the receptor for human influenza viruses (28, 36). Shortly after a novel H1N1 swine-origin influenza virus entered the human population, the pathogenesis of the virus was assessed in ferrets by several research groups. While the isolates tested replicated to high titers in the respiratory tract of ferrets, there were some discrepancies among these reports. In addition, while our studies corroborate the high level of replication of this virus in the upper respiratory tract of ferrets, there was a difference in the replication capacity of the virus in the lower respiratory tract, most likely due to the lower dose of virus in the inoculum and the minimal inoculum volume (0.2 ml) used here. In order to better understand the pathogenicity of influenza viruses, research using the ferret as an animal model should be standardized so as to produce results that most accurately reflect the disease in humans and to allow for comparison of results from different experiments. Future studies should consider the method of virus inoculation as influenza virus can be transmitted not only by direct contact but also by aerosols, and each method of inoculation could have different outcomes.
ACKNOWLEDGMENTS
We thank Spencer Poore, Scott Johnson, and Rhonda Wheeler for their excellent technical assistance.
This work was supported by the Georgia Research Alliance and in part by NIH grant HHSN266200700006C.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Anonymous. 2009. New influenza A (H1N1) virus: global epidemiological situation, June 2009. Wkly. Epidemiol. Rec. 84:249–257 [PubMed] [Google Scholar]
- 2. Belser J. A., et al. 2011. Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J. Virol. 85:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carr J., et al. 2002. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 54:79–88 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control Prevention 2009. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467–470 [PubMed] [Google Scholar]
- 5. Centers for Disease Control Prevention 2009. Update: novel influenza A (H1N1) virus infections—worldwide, May 6, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:453–458 [PubMed] [Google Scholar]
- 6. De Vleeschauwer A., Van Poucke S., Braeckmans D., Van Doorsselaere J., Van Reeth K. 2009. Efficient transmission of swine-adapted but not wholly avian influenza viruses among pigs and from pigs to ferrets. J. Infect. Dis. 200:1884–1892 [DOI] [PubMed] [Google Scholar]
- 7. Dilbeck P. M., McElwain T. F. 1994. Immunohistochemical detection of Coxiella burnetti in formalin-fixed placenta. J. Vet. Diagn. Invest. 6:125–127 [DOI] [PubMed] [Google Scholar]
- 8. Duan S., et al. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fraser C., et al. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garten R. J., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill J. R., et al. 2010. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch. Pathol. Lab. Med. 134:235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Govorkova E. A., Kaverin N. V., Gubareva L. V., Meignier B., Webster R. G. 1995. Replication of influenza A viruses in a green monkey kidney continuous cell line (Vero). J. Infect. Dis. 172:250–253 [DOI] [PubMed] [Google Scholar]
- 13. Gubareva L. V., Webster R. G., Hayden F. G. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antiviral Res. 53:47–61 [DOI] [PubMed] [Google Scholar]
- 14. Hancock K., et al. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945–1952 [DOI] [PubMed] [Google Scholar]
- 15. He F., Du Q., Ho Y., Kwang J. 2009. Immunohistochemical detection of Influenza virus infection in formalin-fixed tissues with anti-H5 monoclonal antibody recognizing FFWTILKP. J. Virol. Methods 155:25–33 [DOI] [PubMed] [Google Scholar]
- 16. Herlocher M. L., et al. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190:1627–1630 [DOI] [PubMed] [Google Scholar]
- 17. Humberd J., Guan Y., Webster R. G. 2006. Comparison of the replication of influenza A viruses in Chinese ring-necked pheasants and chukar partridges. J. Virol. 80:2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Itoh Y., et al. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ives J. A., et al. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307–317 [DOI] [PubMed] [Google Scholar]
- 20. Johnson N. P., Mueller J. 2002. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105–115 [DOI] [PubMed] [Google Scholar]
- 21. Kawaoka Y., Bordwell E., Webster R. G. 1987. Intestinal replication of influenza A viruses in two mammalian species. Brief report. Arch. Virol. 93:303–308 [DOI] [PubMed] [Google Scholar]
- 22. Lam W. Y., et al. 2010. Development and comparison of molecular assays for the rapid detection of the pandemic influenza A (H1N1) 2009 virus. J. Med. Virol. 82:675–683 [DOI] [PubMed] [Google Scholar]
- 23. Maher J. A., DeStefano J. 2004. The ferret: an animal model to study influenza virus. Lab. Anim. (NY) 33:50–53 [DOI] [PubMed] [Google Scholar]
- 24. Maines T. R., et al. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Memoli M. J., et al. 2009. An early “classical” swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology 393:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munster V. J., et al. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myers K. P., Olsen C. W., Gray G. C. 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 44:1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plopper C. G., Hill L. H., Mariassy A. T. 1980. Ultrastructure of the nonciliated bronchiolar epithelial (Clara) cell of mammalian lung. III. A study of man with comparison of 15 mammalian species. Exp. Lung Res. 1:171–180 [DOI] [PubMed] [Google Scholar]
- 29. Potier M., Mameli L., Belisle M., Dallaire L., Melancon S. B. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 30. Reed L. J., Muench H. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 31. Shope R. E. 1931. Swine influenza: I. Experimental transmission and pathology J. Exp. Med. 54:349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith T. F., et al. 1976. Isolation of swine influenza virus from autopsy lung tissue of man. N. Engl. J. Med. 294:708–710 [DOI] [PubMed] [Google Scholar]
- 33. Tai C. Y., et al. 1998. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob. Agents Chemother. 42:3234–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vaillant L., La Ruche G., Tarantola A., Barboza P. 2009. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill. 14:pii19309. [DOI] [PubMed] [Google Scholar]
- 35. van den Brand J. M., et al. 2010. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J. Infect. Dis. 201:993–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Riel D., et al. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wagner R., Matrosovich M., Klenk H. D. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12:159–166 [DOI] [PubMed] [Google Scholar]
- 38. Webster R. G., Laver W. G., Kilbourne E. D. 1968. Reactions of antibodies with surface antigens of influenza virus. J. Gen. Virol. 3:315–326 [DOI] [PubMed] [Google Scholar]
- 39. Yen H. L., et al. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]