Abstract
Canine parvovirus type 2 (CPV-2) is a severe enteric pathogen of dogs, causing high mortality in unvaccinated dogs. After emerging, CPV-2 spread rapidly worldwide. However, there is now some evidence to suggest that international transmission appears to be more restricted. In order to investigate the transmission and evolution of CPV-2 both nationally and in relation to the global situation, we have used a long-range PCR to amplify and sequence the full VP2 gene of 150 canine parvoviruses obtained from a large cross-sectional sample of dogs presenting with severe diarrhea to veterinarians in the United Kingdom, over a 2-year period. Among these 150 strains, 50 different DNA sequence types (S) were identified, and apart from one case, all appeared unique to the United Kingdom. Phylogenetic analysis provided clear evidence for spatial clustering at the international level and for the first time also at the national level, with the geographical range of some sequence types appearing to be highly restricted within the United Kingdom. Evolution of the VP2 gene in this data set was associated with a lack of positive selection. In addition, the majority of predicted amino acid sequences were identical to those found elsewhere in the world, suggesting that CPV VP2 has evolved a highly fit conformation. Based on typing systems using key amino acid mutations, 43% of viruses were CPV-2a, and 57% CPV-2b, with no type 2 or 2c found. However, phylogenetic analysis suggested complex antigenic evolution of this virus, with both type 2a and 2b viruses appearing polyphyletic. As such, typing based on specific amino acid mutations may not reflect the true epidemiology of this virus. The geographical restriction that we observed both within the United Kingdom and between the United Kingdom and other countries, together with the lack of CPV-2c in this population, strongly suggests the spread of CPV within its population may be heterogeneously subject to limiting factors. This cross-sectional study of national and global CPV phylogeographic segregation reveals a substantially more complex epidemic structure than previously described.
INTRODUCTION
Sequence analysis has revolutionized our knowledge of the spatial and temporal dynamics of infection, allowing a greater understanding of the evolution and molecular epidemiology of pathogens. This is particularly important for rapidly evolving pathogens, such as RNA viruses (e.g., feline calicivirus [9], foot and mouth disease virus [8], and influenza virus [33]) and also for certain single-stranded DNA viruses with high mutation rates, such as canine parvovirus (CPV) (37).
CPV type 2 (CPV-2) consists of a 5.5-kb single-stranded linear DNA genome (5) encoding nonstructural (NS1 and -2) and capsid (VP1, -2, and -3) proteins at the 5′ and 3′ ends, respectively (1). The three structural proteins are derived from the same open reading frame (ORF) by proteolytic cleavage and alternate RNA splicing, with the full, infectious capsid consisting predominately of VP2 (34).
The virus first emerged as a new causative agent of severe enteritis in dogs in 1978 (3, 21, 23) and rapidly spread worldwide. This virus, which was named CPV-2 to distinguish it from the unrelated parvovirus minute virus of canines (MVC), is thought to have emerged from a related virus, feline panleukopenia (FPV) (23), possibly via some wildlife intermediate, such as a fox (44). This initial species jump was mapped to amino acid mutations in VP2 compared to FPV, which resulted in a gain of receptor affinity for the canine host. Subsequently, antigenic variants (CPV types 2a, 2b, and 2c) have been described based largely on mouse monoclonal antibody reactivity, and the mutations responsible have been mapped to specific residues in VP2 (CPV-2a, Met-87-Leu, Ile-101-Thr, Ala-300-Gly, Asp-305-Tyr, and Val-555-Ile; CPV-2b, Asp-426-Asn and Ile-555-Val reversion; and CPV-2c, Asp-426-Glu) (6). This has led to several experimental studies showing that CPV vaccines based on CPV-2 or CPV-2b are able to cross-protect against all four antigenic types, including the newly emerged CPV-2c (38, 40). Mutations in VP2 have also been shown to influence the host range (27), hemagglutination spectrum (30), and affinity of receptor binding (26).
Currently, original CPV type 2 is thought not to circulate in the general dog population, although it is present within certain live vaccines. The distributions of CPV types 2a, 2b, and 2c seem to differ across different regions of the world (11, 14, 16, 19, 31, 35, 45). It has recently been suggested that the initial rapid global spread of CPV-2 was a feature of the naïve dog population that it gained access to and is in contrast to the current, more endemic phase of disease, where the international range of new strains is more restricted (20).
While these studies are collectively improving our knowledge of the spatial and temporal dynamics of CPV transmission, they are based generally on relatively unstructured sampling strategies of national collections and/or are limited to using only partial VP2 gene sequence analysis or typing by key amino acid mutations (7, 10, 12, 16, 22, 46). In this paper, we have used a cross-sectional study of clinically ill dogs and full VP2 sequence analysis to investigate in depth the evolution and spread of virus at national and local levels, and in relation to the global situation, to specifically test the hypothesis that currently there is limited international spread of CPV.
MATERIALS AND METHODS
Samples.
Fecal samples were obtained from 25 People's Dispensary for Sick Animals (PDSA) Petaid hospitals across the mainland United Kingdom (Fig. 1) from 373 clinically ill dogs presenting with diarrhea of unknown etiology that required more than conservative treatment. Clinical information and signalment were supplied with the samples and presented elsewhere (18). In addition, 16 CPV-positive samples from 11 different locations within the United Kingdom (1 postmortem sample, 12 from a commercial diagnostic laboratory, and 3 potential vaccine breakdowns) and viruses from the six commercial vaccines currently used within the United Kingdom (coded A to F) were also obtained. All samples and vaccines were collected over a 2-year period (2006-2008). Samples were stored at −80°C until used.
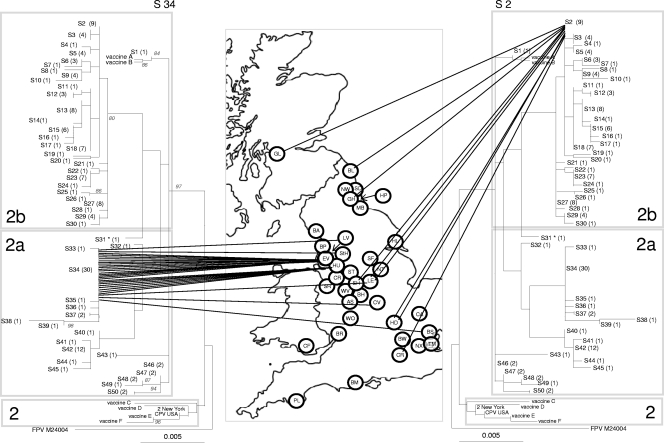
Fig. 1.
Phylogenetic analysis of the CPV VP2 DNA sequences in the United Kingdom. Highlighted are the geographical distribution of two different sequence types, S34 (left) and S2 (right). The phylogenetic tree is based on maximum likelihood with bootstrap support for individual nodes indicated at appropriate nodes (for clarity they are only included on the left-hand tree). In all trees, each sequence type is labeled with its number followed by the number of identical sequences within that group (e.g., S3 (4), indicating that sequence type 3 contains 4 identical sequences). On the map, a circle represents the approximate location in the United Kingdom of each individual hospital where a CPV-positive sample was obtained (for key to hospital origin codes see Table 1). Lines connecting sequences to their geographical origin link the map to the phylogeny. The classification of viruses as CPV-2, -2a, and -2b, as indicated by the shading on each tree, are a classification based on key amino acid mutations (see text). To save space, the same tree is shown in mirror image on either side of the map.
Amplification and sequence of VP2.
DNA was extracted as described previously (13) with a slight modification. Briefly, samples were homogenized (10%, wt/vol) in phosphate-buffered saline (PBS) and centrifuged for 15 min at 9,300 rpm. The supernatant was boiled for 15 min, chilled on ice, and then centrifuged again as previously for 5 min. The supernatant was stored at 4°C. Primers were chosen to amplify the full VP2 region, using both full genomes of CPV and FPV, and VP2 sequences available on GenBank, aligned using CLUSTAL as implemented in MEGA4 (42). Two primers, EF (2748 to 2765) (GCCGGTGCAGGACAAGTA) and JS2R (24) (4818 to 4799) (CAACCCACACCATAACAACA) (all primer sequences numbered based on reference 33), were used to obtain the full VP2 sequence. In order to minimize PCR inhibition, DNA extraction supernatant was diluted 1 in 10 before being amplified by PCR (36). Amplification was carried out in 25-μl reactions, consisting of 12.5-μl extensor PCR master mix (Abgene), 8.5 μl of molecular water (Sigma), (12.5 pmol of primer EF, 12.5 pmol primer JS2R) (24) and 2 μl DNA. Negative controls were processed alongside fecal samples throughout all stages (1 negative control [water or PBS] per 2 fecal samples). All samples were processed in the order of arrival at the laboratory and not batched according to location. The PCR cycling conditions were 1 min at 94°C, followed by 40 cycles of denaturation at 94°C, annealing at 52°C, and extension at 72°C, followed by a final extension phase of 68°C for 10 min.
Amplicons were purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. Sequencing of full VP2 was generated in three overlapping fragments using the two PCR primers (EF and JS2R) and four internal sequencing primers MF (TACCATCTCATACTGGAACTAGTGG [3441 to 3466]), ER (TGTTCCTGTAGCAAATTCATCACC [3581 to 3558]), 555F ([6] CAGGAAGATATCCAGAAGGA [4003 to 4022]), and MR (GTATAGTTAATTCCTGTTTTACCTCC [4140 to 4118]). All sequences were aligned into a double-stranded consensus sequence using Chromas Pro 1.41 (Technelysium Pty Ltd.). All external primer sites were removed, giving a final consensus sequence of 1,755 bp. All sequences were homozygous, with no evidence to suggest dual or mixed infections.
Sequence analysis.
Consensus sequences were aligned by ClustalW. These alignments included sequences available for feline panleukopenia virus (GenBank accession number M24004) and also included published sequences for two CPV-2 strains (M23255 and U22186). The most appropriate evolution model was predicted using MODELTEST as implemented in Topali (25). The final model for nucleotide substitutions chosen was the TrN model (43), which was used to infer bootstrapped maximum likelihood trees using PHYML implemented on the ATGC bioinformatics platform (20). Amino acid trees were drawn using MEGA4, rooted using FPV, and drawn using the Dayhoff PAM matrix. The final alignment was screened for evidence of recombination and selection using GARD and SLAC available through the Datamonkey web server (32).
To seek correlations between the geographical origin of a particular sequence and its position within the phylogeny, a posterior set of trees was obtained through Bayesian Markov chain Monte Carlo (MCMC) analysis using BEAST, v1.4 (17). This analysis implemented the most-favored model identified in the earlier step (TrN + γ + I; where γ is the gamma-distributed rate heterogeneity and I is the proportion of invariable sites); a comparison of alternative MCMC models (HKY + γ + I; GTR + γ + I) by Bayes' factor (41) confirmed that the TrN model was also the most appropriate in a Bayesian MCMC context. We also compared the fit of the strict and “relaxed” (uncorrelated exponential distribution [UCED]) molecular clock models; the UCED model provided a better fit. Similarly, the constant population-size model was preferred over an exponential-growth model (available on request). The MCMC trace was inspected in TRACER, v1.5 (A. Rambaut and A. J. Drummond [http://beast.bio.ed.ac.uk/Tracer]), for convergence. This posterior set of trees was subjected to Bayesian tip-associated significance testing implemented by BaTS (28). In addition, Simpson's equitability index (E) was also used as a measure of population diversity (4, 39): , where Pi is the proportion of identical sequence types (S) within the population and S is the total number of distinct sequence types within the population. An E value of 1 equates to maximum diversity (all sequences different), whereas E tends toward zero as the diversity decreases and the number of sequence types increases. In order to test for geographical clustering, we examined the proportion of each sequence type in each sampling location. Since many sites were negative for a given sequence we used a nonparametric analysis and performed a separate Fisher exact test for each sequence type, using Bonferroni's method to correct for multiple tests. Analyses used Stata 11 (StataCorp, College Station, TX), and significance was set at a P value of <0.05.
United Kingdom sequences were compared to worldwide sequences using the BLAST software (2).
Analysis of clinical signs.
As signalment and clinical details were available with all the samples, we were able to evaluate possible associations between CPV type (CPV-2a, -2b, or -2c) and clinical outcome (death or survival), severity of clinical signs (as reported by the clinician), breed, age, sex, color, presence of vomiting, and presence of hemorrhagic diarrhea.
RESULTS
Of the 373 samples obtained from the PDSA, 255 (68%) were PCR positive (Table 1). Of these, a consensus VP2 sequence was obtained for 134 samples, selected to include one or more sequences from each of the 25 hospitals that submitted a positive sample. A sequence was also obtained from the 16 samples obtained from other sources (postmortem, commercial lab, and potential vaccine breakdowns) and from the six vaccines (A to F) currently used in the United Kingdom.
Table 1.
Origin of samples and the samples contained within each DNA sequence typea
| PDSA PetAid hospital |
Single 2b sequence type no. | No. of samples |
Single 2a sequence type no. | No. of samples | Ratio of 2a seqences | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2b sequence type no.: |
2a sequence type no.: |
||||||||||||||||||||||||
| Code | Hospital | Prevalence | 2 | 3 | 5 | 6 | 9 | 12 | 13 | 15 | 18 | 23 | 27 | 29 | 34 | 37 | 42 | 46 | 47 | 48 | 50 | ||||
| ST | Stoke | 79% (23/29) | 20, 30, 1 | 1 | 2 | 3* | 1 | 1 | 11 | 0.09 | |||||||||||||||
| BW | Bow | 51% (22/43) | 11, 24 | 1 | 3* | 3* | 39 | 10 | 0.1 | ||||||||||||||||
| WV | Wolverhampton | 52% (9/17) | 16 | 4* | 31 | 6 | 0.17 | ||||||||||||||||||
| BH | Birmingham | 71% (23/32) | 14, 17, 26 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 12 | 0.25 | |||||||||||||
| LE | Leicester | 73% (11/15) | 2 | 2 | 1 | 1 | 43 | 7 | 0.29 | ||||||||||||||||
| NX | New Cross | 39% (11/28) | 4, 28 | 2 | 1 | 2 | 7 | 0.29 | |||||||||||||||||
| EV | Everton | 85% (49/57) | 10, 7 | 3 | 3 | 2 | 15* | 33 | 26 | 0.62 | |||||||||||||||
| CV | Coventry | 82% (14/17) | 3 | 1 | 4* | 45 | 9 | 0.67 | |||||||||||||||||
| CF | Cardiff | 76% (10/13) | 21 | 1 | 4* | 6 | 0.67 | ||||||||||||||||||
| HU | Huyton | 82% (24/29) | 1 | 10* | 36 | 12 | 0.92 | ||||||||||||||||||
| BM | Bournemouth | 33% (3/9) | 1 | 40 | 2 | 1 | |||||||||||||||||||
| GL | Glasgow | 45% (5/11) | 1 | 44 | 2 | 0.5 | |||||||||||||||||||
| NW | Newcastle | 50% (1/2) | 49 | 1 | 1 | ||||||||||||||||||||
| BR | Bristol | 61% (8/13) | 1 | 1 | 0 | ||||||||||||||||||||
| BS | Basildon | 63% (7/11) | 4* | 1 | 5 | 0.2 | |||||||||||||||||||
| GH | Gateshead | 66% (4/6) | 1 | 1 | 0 | ||||||||||||||||||||
| MB | Middlesboro | 66% (4/6) | 1 | 2 | 3 | 0.67 | |||||||||||||||||||
| PL | Plymouth | 66% (2/3) | 1 | 1 | 1 | ||||||||||||||||||||
| NT | Nottingham | 71% (5/7) | 25 | 1 | 0 | ||||||||||||||||||||
| HL | Hull | 75% (6/8) | 4* | 4 | 0 | ||||||||||||||||||||
| TM | Thamesmead | 75% (3/4) | 1 | 1 | 1 | ||||||||||||||||||||
| CR | Croydon | 80% (4/5) | 1 | 1 | 0 | ||||||||||||||||||||
| SF | Sheffield | 80% (4/5) | 2 | 2 | 0 | ||||||||||||||||||||
| HD | Hendon | 100% (2/2) | 2 | 2 | 0 | ||||||||||||||||||||
| SD | Sunderland | 100% (1/1) | 1 | 1 | 1 | ||||||||||||||||||||
| Subtotal for PDSA samples | 68% (255/373) | 16 | 7 | 4 | 4 | 3 | 4 | 3 | 7 | 6 | 7 | 7 | 6 | 3 | 28 | 1 | 11 | 2 | 2 | 2 | 2 | 9 | 134 | 0.43 | |
| Subtotal for others | 100% (16/16) | 19 HP | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 32 BA | 16 | 0.5 | |||||||||||||
| (non-PDSA | 8 CW | SH | LV | WO | LV | StH | LV | CA | 38 CA | ||||||||||||||||
| samples) | 22 AS | BL | StH | LV | 41 BP | ||||||||||||||||||||
| 35 StH | |||||||||||||||||||||||||
| Total no. of each sequence type | 18 | 9 | 4 | 4 | 3 | 4 | 8 | 3 | 6 | 7 | 7 | 8 | 4 | 30 | 2 | 12 | 2 | 2 | 2 | 2 | 13 | 150 | |||
| No. of hospitals per sequence type | 9 | 1 | 2 | 1 | 2 | 5 | 2 | 3 | 4 | 2 | 6 | 3 | 7 | 2 | 5 | 2 | 1 | 2 | 2 | ||||||
Origin of samples are shown, with the code on the left (for positions of these codes relating to United Kingdom geography, see Fig. 1 and 2) and percent prevalence (no. sampled/no. testing positive). Sequence types found more than once are indicated across the top (and correspond to those on the tree in Fig. 1 and 2). Sequence types found only once are also shown, as are the total number of sequences from an area and the proportion of these which are 2a. The numbers and origins of samples of non-PDSA origin are indicated, as are the total numbers of sequences per sequence type. The number of hospitals where a sequence type was isolated is also shown. Sequence types which showed significant evidence of clustering in certain areas are indicated with an *, though numbers were small (see text). Origin of non-PDSA samples: HP, Hartlepool; CW, Crewe; LV, Liverpool; StH, St. Helens; BL, Blyth; WO, Worcester; CA, Cambridge; BA, Barrow; BP, Blackpool; SH, Shrewsbury.
Prevalence of CPV types in the United Kingdom.
Based on key amino acid mutations, the 150 viruses were typed as 2a (43%; n = 65) or 2b (57%; n = 85) (Table 1). No type 2 or 2c was found. For those hospitals for which more than five sequences were available, the proportion of 2a/2b sequences ranged from 92% type 2a in Huyton (11 of 12) to 91% type 2b in Stoke (10 of 11) (Table 1), suggesting that types 2a and 2b may have different distributions in different areas of the United Kingdom. The vaccines consisted of two CPV-2b vaccines (A and B) and four CPV-2 vaccines (C to F) (Fig. 1 and 2).
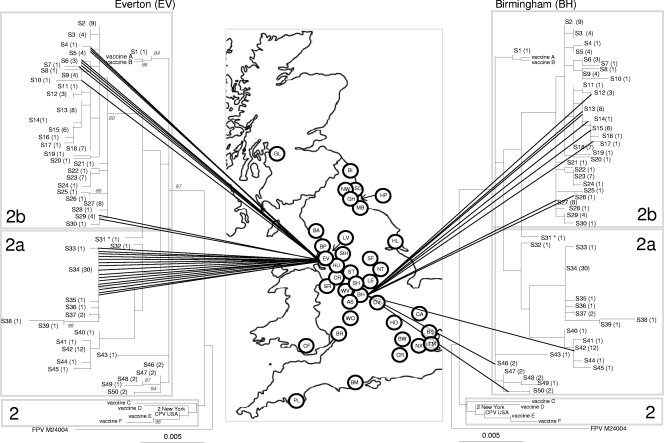
Fig. 2.
Phylogenetic analysis of the CPV VP2 DNA sequences in the United Kingdom. The genetic diversity of CPV in Everton, on the left (lowest diversity as measured by Simpson's index and BaTS), and Birmingham, on the right (highest diversity as measured by Simpson's index and BaTS), are shown.The phylogenetic tree is based on maximum likelihood with bootstrap support for individual nodes indicated at appropriate nodes (for clarity they are only included on the left-hand tree). In all trees, each sequence type is labeled with its number followed by the number of identical sequences within that group (e.g., S3 (4), indicating that sequence type 3 contains 4 identical sequences). On the map, a circle represents the approximate location in the United Kingdom of each individual hospital where a CPV-positive sample was obtained (for key to hospital origin codes see Table 1). Lines connecting sequences to their geographical origin link the map to the phylogeny. The classification of viruses as CPV-2, -2a, and -2b, as indicated by the shading on each tree, are a classification based on key amino acid mutations (see text). To save space, the same tree is shown in mirror image on either side of the map.
DNA sequence analysis.
Considerable diversity was found within the VP2 DNA sequences that we obtained, with 50 genetically distinct sequences, or sequence types (S), identified among the 150 clinical samples sequenced (Fig. 1 and 2 and Table 1). Thirty-one of the 50 different sequence types were found only once, whereas some sequence types were quite common; for example, 8 sequence types contained more than 5 sequences, and the most common (S34) comprised 30 sequences (Table 1 and Fig. 1). Of the 50 different sequence types, 20 comprised type 2a viruses and the remaining 30 were type 2b viruses (Table 1; Fig. 1), suggesting that type 2b might be more variable in the United Kingdom.
The majority of sequences clustered separately depending on their type (2a or 2b). However, neither CPV-2a or -2b was monophyletic: S31 which typed as a 2a virus based on key amino acid substitutions, grouped phylogenetically within the 2b virus sequences (Fig. 1). In addition, S1 (type 2b) clustered close (2- to 8-nucleotide [nt] substitutions) to S46 to 50 (type 2a). This may suggest possible 2a/2b intermediates or parallel evolution. No evidence of recombination or positive selection was found within this data set (data not presented).
Spatial range of sequence types in the United Kingdom.
In order to explore the spatial range of individual sequence types, we calculated the number of hospitals in which each of these sequences was found (Table 1). Some sequence types were geographically restricted, as exemplified by S34, which was found at seven hospitals clustered mostly in the Northwest of England, including Liverpool, Everton, Huyton, and St. Helens (left side of Fig. 1). Other sequence types were geographically dispersed as exemplified by S2 which was again found in seven different hospitals ranging, from the North (Glasgow) to the South (Croydon and Hendon) (right side of Fig. 1). There was statistical support (P < 0.05) for geographical clustering for S34 (Everton and Huyton), S3 (Hull), S23 (Basildon and Bow), and S42 (Coventry and Cardiff). However, for many areas, sample numbers were small.
The finding of evidence of clustering for some sequence types in particular geographic locations was confirmed using Simpson's index of diversity (Table 2). For example, Everton had the highest number of viral sequences (n = 26) but showed the lowest level of DNA (E = 0.1) and second-lowest amino acid diversity (0.08). In other areas, for example, in Birmingham, sequence diversity was much higher (E = 0.86 and 0.3 for DNA and amino acid sequences, respectively) (Table 2). Results of BaTS analysis also suggested in some cases that there was an association between the hospital of isolation and phylogenetic clustering (Everton, Stoke, Bow, and Coventry), whereas in other hospitals there was no evidence for this (Birmingham, Leicester, New Cross, and Cardiff) (Table 2). These extremes of diversity for Everton and Birmingham can also be represented phylogenetically (Fig. 2) and are shown in Fig. S1 to S4 in the supplemental material for the other hospitals in Table 2.
Table 2.
Support for geographical clusteringa
| Type and/or city | No. of sequences (N) | DNA |
Protein |
P (BaTS) | ||
|---|---|---|---|---|---|---|
| S | E | S | E | |||
| 2a | 65 | 20 | 0.2 | 10 | 0.04 | |
| 2b | 85 | 30 | 0.54 | 14 | 0.02 | |
| Birmingham | 12 | 11 | 0.86 | 5 | 0.3 | 1 |
| Stoke | 12 | 8 | 0.8 | 5 | 0.04 | 0.04 |
| Leicester | 7 | 5 | 0.64 | 4 | 0.37 | 1 |
| New Cross | 7 | 5 | 0.64 | 3 | 0.33 | 1 |
| Bow | 10 | 6 | 0.45 | 4 | 0.19 | 0.03 |
| Coventry | 9 | 4 | 0.33 | 4 | 0.33 | 0.05 |
| Cardiff | 6 | 3 | 0.33 | 3 | 0.33 | 1 |
| Huyton | 12 | 3 | 0.12 | 3 | 0.12 | 0.06 |
| Everton | 26 | 7 | 0.1 | 3 | 0.08 | 0.01 |
Simpson's index of diversity, where S is the number of different sequence types and E is the equitability index (where 1 is maximum diversity). This is shown for CPV types 2a and 2b and for hospitals with more than five sequences. P values calculated by BaTS (significant at <0.05), suggesting correlation between phylogeny and PDSA origin, are also indicated.
Simpson's index also showed that at the DNA level, the 2b viruses had a higher level of diversity than the 2a viruses (E = 0.54 and 0.20, respectively) (Table 2). However, these relative diversities were reversed at the amino acid level, due to the merging of the majority of the 2b viruses into a single amino acid sequence (amino acid group A) (Fig. 3).
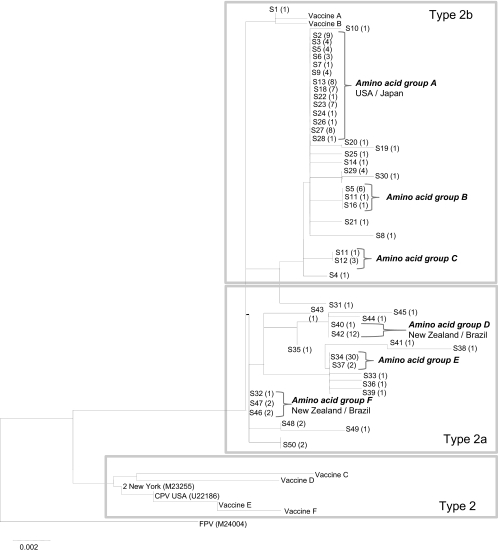
Fig. 3.
A neighbor-joining amino acid tree of the same samples used for Fig. 1. Samples are labeled with the same sequence type number as that used in the DNA tree. Brackets are used to indicate large amino acid groups which are referred to in the text (marked A to F). For A, D, and F, where these sequences have been identified both in the United Kingdom (this study) and in other countries (based on GenBank data), the other countries are listed next to the taxon name.
Comparison of United Kingdom and worldwide sequences.
Almost all the sequence types that we identified were unique to the United Kingdom at the DNA level, with only S40 being found outside the United Kingdom, in China (GenBank accession number EF666069). The DNA distances between the United Kingdom 2b viruses and those from the rest of the world (as represented by GenBank) were relatively high (average, 7.23; standard deviation [SD], 1.4), compared to those for the United Kingdom 2a sequences (average, 3.75; SD, 2.8) (data not presented). Formal evidence for geographical clustering was sought using an expanded data set of 617 sequences (accession numbers available on request) from 21 countries using an alignment of 1,386 bases of VP2. A maximum likelihood phylogeny for these sequences suggests that some of the United Kingdom sequences are concentrated in certain areas of the tree (see Fig. S5 in the supplemental material). Results of BaTS analysis on this data set confirmed that there was strong evidence for geographical clustering within this expanded world phylogeny (association index and parsimony score < 0.001), with significant (P < 0.01) clustering observed for 14 of the 21 countries represented, including the United Kingdom (data not presented).
Amino acid variability.
When nucleotide sequences were translated, the 50 United Kingdom DNA sequence types converted to 30 different predicted amino acid sequences (14 type 2b, 16 type 2a), six of which contained more than one DNA sequence type (A to F) (Fig. 3). The most prevalent was amino acid sequence A, which contained 59 DNA sequences from 14 different DNA sequence types, with an average of 3.8 mutations between them (data not presented). In total, 57% of the viruses had predicted amino acid sequences identical to ones also found elsewhere in the world (A, D, F, and another comprising only one DNA sequence type) (Fig. 3), and 32% were different only by one amino acid.
When full VP2 consensus sequences were analyzed, two specific mutations, Val-139-Iso (n = 35) and Arg-274-Lys (n = 8), were identified only within the United Kingdom samples compared to those from the rest of the world via GenBank. The mutation at position 139 is responsible for the formation of nucleotide sequence type 34.
Clinical outcome in relation to sequence type.
There appeared to be no association between clinical signs and sequence either at the level of virus type (CPV-2a or -2b), DNA sequence type, or at the level of individual specific amino acid mutations (data not shown). Neither type 2 nor type 2b vaccine virus was detected in fecal samples, although one type 2b field virus (S1) was only two bases different from vaccines A and B (Fig. 1 and 2).
DISCUSSION
Spatial and temporal dynamics of CPV-2 spread, based largely on unstructured sampling techniques, may lead to a potentially biased view of the diversity and types of virus circulating both within and between countries. Here, we have used a cross-sectional sampling strategy over 2 years, together with sequence analysis of the full CPV capsid gene (VP2, 1,755 bp) to investigate the spread and evolution of CPV-2 at both local and national levels and to relate this to the global transmission of the virus.
We have found considerable diversity of CPV strains obtained from across the United Kingdom, with 50 different DNA sequence types identified within the 150 viruses analyzed. Interestingly, apart from one sequence previously identified in China, all VP2 sequence types appeared to be unique to the United Kingdom. Phylogenetic analysis and Bayesian tip-associated significance testing of an expanded set of published global sequences showed some evidence for geographical clustering at an international level, suggesting that currently there are limited opportunities for global transmission, as has previously been suggested by others (21). Despite this observation, sequences from individual countries, as exemplified by the United Kingdom, were generally not monophyletic, implying that national diversity is produced by a combination of local evolution occasionally supplemented by importation of new sequence types. The relative significance of these two processes remains to be determined.
There was less diversity at the amino acid level than at the DNA level, with only 32 unique amino acid sequences identified. In contrast to the DNA sequences, the amino acid sequences were similar in the majority of instances (57% identical) (Fig. 3) to those found elsewhere in the world. This apparent relative stability of the virus at the amino acid level is likely due to high structural and functional requirements of the capsid gene in this small virus and, in contrast to previous studies (21), was associated with a lack of evidence for positive selection in VP2. Two unique amino acid mutations were, however, identified within the United Kingdom samples. Residue 139 was mutated from valine to isoleucine in 35 samples and forms part of the beta barrel inside the virus (34). Residue 274 was mutated from arginine to lysine in eight samples and is found on the 3-fold spike, a region of high antigenicity (30). At this point, we have no evidence to indicate whether these mutations have any biological significance.
The variability in the full VP2 gene sequence enabled us to investigate in depth the molecular epidemiology of these viruses at the national level. We showed that within the background of countrywide diversity, there was also evidence of significant geographic clustering in some areas, as exemplified by S34 (Fig. 1), which not only was the most predominant virus in the Liverpool region (i.e., Everton, Huyton, St. Helens, and Liverpool) but was also rarely found elsewhere. As this sequence type was present over a period of 2 years, it suggests that we were not observing a short-term epidemic. Such spatial clustering suggests that some CPV sequence types may have restricted opportunities for spread, even within countries. This geographical restriction of certain virus types highlights the importance of rigorous, epidemiologically representative sampling strategies for the study of viral molecular epidemiology.
Of the potential vaccine breakdowns which were included in our study, none were identical to the sequences obtained from vaccine used in the United Kingdom, suggesting that live vaccines are not causing disease in this population. However, one sequence (S1) was only two nucleotides different in the VP2 region from two type 2b vaccines (vaccines A and B in Fig. 1 and 2), and several viruses classified as antigenic type 2a (S46 to S50) were also phylogenetically close to these vaccine sequences (equivalent to 2- to 8-nt substitutions). Whether these field viruses represent the ancestor strains for these vaccines or whether S1 may have evolved from the vaccine either in that individual dog or in the wider dog population is unknown. In addition, all of the potential vaccine breakdowns were distinct and showed no consistent mutations, suggesting that there is no group of viruses circulating within the United Kingdom which is specifically capable of evading vaccine-induced immunity. This is in agreement with a range of studies that have shown that currently available vaccines protect against the full range of antigenic types identified to date (38, 40).
The traditional typing of the viruses based on key amino acid positions of CPV types 2a, 2b, and 2c showed that only CPV types 2a and 2b were circulating within the United Kingdom and is in agreement with a previous study (10). Decaro et al. (14) found only one 2c virus circulating within the United Kingdom, and this remains the only 2c virus identified in the United Kingdom to date. Since many countries have high levels of CPV-2c circulating (15), it might be expected that CPV-2c would have reached the United Kingdom by now, unless the virus has some restrictions on its global spread (21). No samples of ancestral CPV type 2 were found in our study, confirming that it no longer appears to be circulating as an important cause of disease.
Although the majority of viruses clustered on the basis of these key amino acid mutations, neither 2a or 2b viruses appeared to be monophyletic. Indeed there was some evidence to suggest that in the United Kingdom at least, the 2a/2b phenotype is not that stable and may have evolved on several occasions, as indicated by the S46-to-S50 group, S31, and S32 to S45. Although 2a and 2b (and indeed 2c) clearly do have some antigenic differences based largely on monoclonal antibody reactivity (29), such lack of stability, together with no clear evidence for clinical differences, may suggest that this classification system needs revising. In this regard, it will be important to further explore the historical diversification of this virus by obtaining sequences for older viruses. In addition, as neither the 2a or 2b type is monophyletic, results by real-time PCR typing generally targeting a small part of the VP2 gene may give a false impression of the epidemiology of this virus (12).
Supplementary Material
ACKNOWLEDGMENTS
S.R.C. is in receipt of a BBSRC case award, partly funded by Intervet Schering Plough Animal Health.
We are very grateful to P. J. Noble and A. Jones for informatics support with the BaTS analysis.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Agbandje M., Parrish C. R., Rossmann M. G. 1995. The structure of parvoviruses. Semin. Virol. 6:299–309 [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Appel M. J., Scott F. W., Carmichael L. E. 1979. Isolation and immunisation studies of a canine parvo-like virus from dogs with haemorrhagic enteritis. Vet. Rec. 105:156–159 [DOI] [PubMed] [Google Scholar]
- 4. Begon M., Harper J. L., Townsend C. R. 1990. Ecology: individuals, populations and communities, 2nd ed. Blackwell Scientific Publications, Cambridge, MA [Google Scholar]
- 5. Berns K. I. 1990. Parvovirus replication. Microbiol. Rev. 54:316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buonavoglia C., et al. 2001. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 82:3021–3025 [DOI] [PubMed] [Google Scholar]
- 7. Costa A. P., Leite J. P., Labarthe N. V., Cubel Garcia R. C. N. 2005. Genomic typing of canine parvovirus circulating in the State of Rio de Janeiro, Brazil from 1995 to 2001 using polymerase chain reaction assay. Vet. Res. Comm. 29(8):735–743 [DOI] [PubMed] [Google Scholar]
- 8. Cottam E. M., et al. 2008. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathogens 4:e1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coyne K. P., Gaskell R. M., Dawson S., Porter C. J., Radford A. D. 2007. Evolutionary mechanisms of persistence and diversification of a calicivirus in an endemically infected natural host population. J. Virol. 81:1961–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies M. 2008. Canine parvovirus strains identified from clinically ill dogs in the United Kingdom. Vet. Rec. 163:543–544 [DOI] [PubMed] [Google Scholar]
- 11. Decaro 2005. Clinical and virological findings in pups naturally infected by canine parvovirus type 2 Glu-426 mutant. J. Vet. Diagn. Invest. 17:133–138 [DOI] [PubMed] [Google Scholar]
- 12. Decaro N., et al. 2005. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the faeces of dogs. Vet. Microbiol. 105:19–28 [DOI] [PubMed] [Google Scholar]
- 13. Decaro N., et al. 2006. First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. J. Vet. Med. B Infect. Dis. Vet. Public Health 53:468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decaro N., et al. 2007. The molecular epidemiology of canine parvovirus, Europe. Emerg. Infect. Dis. 13:1222–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Decaro N., et al. 2009. Genetic analysis of canine parvovirus type 2c. Virology 385:5–10 [DOI] [PubMed] [Google Scholar]
- 16. de Ybañez R. R., Vela C., Cortes E., Simarro I., Casal J. I. 1995. Identification of types of canine parvovirus circulating in Spain. Vet. Rec. 136:174–175 [DOI] [PubMed] [Google Scholar]
- 17. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Godsall S. A., Clegg S. R., Stavisky J., Radford A. D., Pinchbeck G. 2010. Epidemiology of canine parvovirus and coronavirus in dogs presented with severe diarrhoea to PDSA PetAid hospitals. Vet. Rec. 167:196–201 [DOI] [PubMed] [Google Scholar]
- 19. Greenwood N. M., Chalmers W. S., Baxendale W., Thompson H. 1996. Comparison of isolates of canine parvovirus by monoclonal antibody and restriction enzyme analysis. Vet. Rec. 138:495–496 [DOI] [PubMed] [Google Scholar]
- 20. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 21. Hoelzer K., Shackelton L. A., Parrish C. R., Holmes E. C. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J. Gen. Virol. 89:2280–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapil S., et al. 2007. Canine parvovirus types 2c and 2b circulating in North American dogs in 2006 and 2007. J. Clin. Microbiol. 45:4044–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelly W. R. 1978. An enteric disease of dogs resembling feline panleucopaenia. Aust. Vet. J. 54:593. [DOI] [PubMed] [Google Scholar]
- 24. Meers, Kyaw-Tanner J. M., Bensink Z., Zwijnenberg R. 2007. Genetic analysis of canine parvovirus from dogs in Australia. Aust. Vet. J. 85:392–396 [DOI] [PubMed] [Google Scholar]
- 25. Milne I., et al. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palermo L. M., Hafenstein S. L., Parrish C. R. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80:8482–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parker J. S., Parrish C. R. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parker J., Rambaut A. R., Pybus O. G. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. MEEGID 8:239–246 [DOI] [PubMed] [Google Scholar]
- 29. Parrish C. R., Carmicael L. E. 1986. Characterisation and recombination mapping of an antigenic and host range mutation of canine parvovirus. Virology 148:121–132 [DOI] [PubMed] [Google Scholar]
- 30. Parrish C. R., Burtonboy G., Carmichael L. E. 1988. Characterization of a non-hemagglutinating mutant of canine parvovirus. Virology 163:230–232 [DOI] [PubMed] [Google Scholar]
- 31. Pereira C. A., Monezi T. A., Mehnert D. U., D'Angelo M., Durigon E. L. 2000. Molecular characterization of canine parvovirus in Brazil by polymerase chain reaction assay. Vet. Microbiol. 75:127–133 [DOI] [PubMed] [Google Scholar]
- 32. Pond S. L., Frost S. D. W. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 33. Rambaut A., et al. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reed A. P., Jones E. V., Miller T. J. 1988. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 62:266–7635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sagazio P., Tempesta M., Buonavoglia D., Cirone F., Buonavoglia C. 1998. Antigenic characterization of canine parvovirus strains isolated in Italy. J. Virol. Methods 73:197–200 [DOI] [PubMed] [Google Scholar]
- 36. Schunck B., Kraft W., Truyen U. 1995. A simple touch-down polymerase chain reaction for the detection of canine parvovirus and feline panleukopenia virus in feces. J. Virol. Methods 55:427–433 [DOI] [PubMed] [Google Scholar]
- 37. Shackelton L. A., Parrish C. R., Truyen U., Holmes E. C. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 102:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siedek E. M., Schmidt H., Munyira P., Raue R. 2007. Vanguard 7 protects against challenge with virulent canine parvovirus antigenic type 2c (CPV-2c). Proceedings of the International Parvovirus Meeting 2007. Monopoli, Italy [Google Scholar]
- 39. Simpson E. H. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 40. Spibey N., Greenwood N. M., Sutton D., Chalmers W. S., Tarpey I. 2008. Canine parvovirus type 2 vaccine protects against virulent challenge with type 2c virus. Vet. Microbiol. 128:48–55 [DOI] [PubMed] [Google Scholar]
- 41. Suchard M. A., Weiss R. E., Sinsheimer J. S. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 42. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 43. Tamura K., Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 3:512–526 [DOI] [PubMed] [Google Scholar]
- 44. Truyen U. 1999. Emergence and recent evolution of canine parvovirus. Vet. Microbiol. 69:47–50 [DOI] [PubMed] [Google Scholar]
- 45. Wang H. C., Chen W. D., Lin S. L., Chan J. P., Wong M. L. 2005. Phylogenetic analysis of canine parvovirus VP2 gene in Taiwan. Virus Genes 31:171–174 [DOI] [PubMed] [Google Scholar]
- 46. Yilmaz Z., Pratelli A., Torun S. 2005. Distribution of antigen types of canine parvovirus type 2 in dogs with hemorrhagic enteritis in Turkey. Turk. J. Vet. Anim. Sci. 29:1073–1076 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.