Abstract
From infection studies with cultured chicken cells and experimental mammalian hosts, it is well known that influenza viruses use the nonstructural protein 1 (NS1) to suppress the synthesis of interferon (IFN). However, our current knowledge regarding the in vivo role of virus-encoded NS1 in chickens is much more limited. Here, we report that highly pathogenic avian influenza viruses of subtypes H5N1 and H7N7 lacking fully functional NS1 genes were attenuated in 5-week-old chickens. Surprisingly, in diseased birds infected with NS1 mutants, the IFN levels were not higher than in diseased birds infected with wild-type virus, suggesting that NS1 cannot suppress IFN gene expression in at least one cell population of infected chickens that produces large amounts of the cytokine in vivo. To address the question of why influenza viruses are highly pathogenic in chickens although they strongly activate the innate immune system, we determined whether recombinant chicken alpha interferon (IFN-α) can inhibit the growth of highly pathogenic avian influenza viruses in cultured chicken cells and whether it can ameliorate virus-induced disease in 5-week-old birds. We found that IFN treatment failed to confer substantial protection against challenge with highly pathogenic viruses, although it was effective against viruses with low pathogenic potential. Taken together, our data demonstrate that preventing the synthesis of IFN is not the primary role of the viral NS1 protein during infection of chickens. Our results further suggest that virus-induced IFN does not contribute substantially to resistance of chickens against highly pathogenic influenza viruses.
INTRODUCTION
The nonstructural protein 1 (NS1) of influenza A virus serves multiple functions in the viral life cycle. It regulates viral polymerase activity, and it modulates cellular signaling pathways (for a review, see reference 14). It is well established that NS1 can suppress innate immune responses in infected cells by blocking intracellular signaling pathways that lead to the synthesis of type I and type III interferon (IFN). Specifically, NS1 inhibits ubiquitinylation of RIG-I by TRIM25, thereby preventing efficient induction of IFN in infected cells (9, 12, 28, 35). The NS1 proteins of most influenza viruses block global posttranscriptional processing of cellular mRNAs, including mRNAs encoding IFN and IFN-induced proteins (20, 27, 33, 34). NS1 further inhibits specific antiviral effector proteins, such as 2′–5′ oligoadenylate synthetase (OAS) (29) and double-stranded RNA (dsRNA)-activated protein kinase (PKR) (23, 30). In addition, NS1 was shown to modulate host cell apoptosis (39, 55), stimulate phosphoinositol 3-kinase signaling (13), and regulate viral polymerase activity (2, 30).
Current knowledge regarding the in vivo functions of NS1 is mainly based on studies in mice with targeted deletions of several key components of the IFN system. Viruses expressing defective NS1 proteins are generally strongly attenuated in hosts with an intact IFN system but retain a high degree of virulence in hosts with defective IFN systems (11, 31, 37), suggesting that the enhanced IFN-inducing potential is the major cause of attenuation of NS1-deficient viruses. Others have shown that a mutant influenza A virus with C-terminally truncated NS1 is highly attenuated in chickens and that this mutant virus induces far more IFN in cultured cells than wild-type virus (3). Similarly, a virus expressing an NS1 variant with reduced metabolic stability that induced enhanced levels of IFN in cultured cells was found to be attenuated in chickens (23). These observations led to the conclusion that NS1 might exhibit similar profound effects on cellular functions in mammals and birds. However, confirmatory in vivo data that would support this view are not available.
The IFN system of birds is functionally conserved, although most key factors of the avian IFN system usually exhibit only low sequence homology to the corresponding factors in mammals (49). The chicken genome contains multiple intronless IFN genes that are strongly induced in response to virus infection and which seem to represent the avian homologues of mammalian alpha interferon (IFN-α) (43, 45). A single intronless IFN gene might represent the homologue of mammalian IFN-β (17, 45). Further, chickens carry genes encoding IFN-γ (6, 53) and IFN-λ (19).
In the current work, we performed in vivo studies to assess the biological properties of highly pathogenic avian influenza A viruses (HPAIV) with NS1 defects. We found that attenuation of the mutant viruses in chickens was not associated with enhanced activation of type I IFN in infected organs. Furthermore, we observed that highly pathogenic avian influenza A viruses exhibit a relatively high degree of IFN resistance in the chicken, a natural host, suggesting that in this species, the main role of NS1 may not be suppression of IFN synthesis but may rather involve other functions.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed in compliance with the German animal protection law (TierSchG). The animals were housed and handled in accordance with good animal practice as defined by FELASA (http://www.felasa.eu/guidelines.php) and the national animal welfare body, GV- SOLAS (http://www.gv-solas.de/index.html). All animal experiments were approved by the local animal welfare committees.
Cells.
Chicken embryo fibroblasts (CEF) were prepared from 11-day-old embryos by trypsin digestion. CEF and CEC-32 reporter cells were maintained in Dulbecco's modified Eagle high-glucose medium (DMEM) containing 8% fetal bovine serum and 2% chicken serum. CEF cultures were used between passages 3 and 10. Madin-Darby canine kidney (MDCK), MDCK-NS1-GFP (green fluorescent protein) (22), and 293T cells were maintained in DMEM containing 10% fetal bovine serum.
Tracheal organ cultures.
Tracheal organ cultures were made as described previously (4). Briefly, embryos from 19-day-old fertilized chicken eggs were removed aseptically from their shells, and the tracheas were dissected. Excess tissue was removed, and 0.5-mm transverse ring sections were cut using a microtome blade. The rings were placed in capped tissue culture tubes filled with 600 μl of prewarmed 199 Hanks' medium (Biochrom AG, Germany). The tubes were placed in a roller drum and incubated at 37°C. Survival of the cultures was determined by observing cilia activity. Only rings with full cilia activity were used for further experiments.
Mice.
B6.A2G-Mx1 mice were used for the IFN induction studies, and BALB.A2G-Mx1 mice were used for all other experiments. Both mouse strains (48) were bred at the Institute for Virology, Freiburg, Germany. Six- to 8-week-old animals were used for all infection experiments. Briefly, groups of 5 mice were anesthetized by intraperitoneal injection of ketamine (100 μg per gram body weight) and xylazine (5 μg per gram body weight), and the animals were infected intranasally with the indicated doses of the various influenza A viruses in 50 μl of phosphate-buffered saline (PBS) containing 0.3% bovine serum albumin (BSA). Animals were euthanized if severe symptoms developed or body weight loss reached 30%.
Chickens.
Four- to 5-week-old commercial specific-pathogen-free (SPF) white leghorn chickens (Valo) were obtained from Lohmann Tierzucht GmbH, Cuxhaven, Germany. Groups of birds were housed in enriched cages. The chickens were infected by the indicated routes. Intratracheal inoculation was performed with virus suspended in 300 μl of cell culture medium using a blunt-end canula. At the indicated time points or when severe symptoms occurred, the birds were euthanized following ketamin/xylazin anesthesia. For virus titration and IFN bioassay, heparin blood was taken from the jugular vein.
Clinical scores.
During infection experiments, the chickens were monitored twice daily for the presence of clinical signs. Symptoms were recorded as clinical scores from 0 to 3. A score of 0 represented the absence of any clinical disease. Scores of 0.5, 1, and 2 represented mild to severe disease, as judged by the presence of diarrhea, apathy, or hemorrhages of the feet. The highest score of 3 was assigned to dead animals.
Viruses.
HPAIV strain A/Cygnus cygnus/Germany/R65/2006 (designated R65) and its recombinant equivalent were used as prototype H5N1 viruses (52). A variant of A/seal/Mass/1/80 adapted to grow in CEF (designated SC35) was used as the prototype H7N7 strain (38). Specific deletions were introduced into segment 8 using a PCR-based cloning strategy (36). Mutants R65-delNS1 and SC35-delNS1 contain a deletion in segment 8 that removes the NS1 open reading frame almost completely. SC35-trunc NS1 contains a deletion in segment 8 which terminates the NS1 open reading frame at amino acid position 126. R65-truncNS1 is a reassortant virus that contains segments 1 to 7 of R65 and a truncated segment 8 of SC35, which codes for amino acids 1 to 126 of NS1. The NS2 open reading frame was not affected by the deletions. The presence of the introduced changes and the absence of unwanted mutations were confirmed by sequencing. R65-PR8(123) is a reassortant virus that contains segments 4 to 8 from R65 and segments 1 to 3 from standard laboratory H1N1 strain A/PR/8/34. For virus rescue, plasmids were transfected into a mixture of 293T and MDCK-NS1-GFP cells (22). Stocks of recombinant viruses were prepared in either MDCK-NS1-GFP or standard MDCK cells. In addition, the HPAIV isolate A/FPV/Rostock/1934 H7N1 (FPV Rostock); the low-pathogenic AIV (LPAIV) isolates A/chicken/United Arabian Emirates/R66/2002 H9N2 (R66), A/tern/Turkmenistan/18/1972 H3N3 (Turk-18), and A/turkey/Ontario/6118/1968 H8N4 (Ont6118); and the laboratory H1N1 strains WSN and A/PR/8/34 (PR8) were used for growth kinetics in CEF and tracheal organ cultures. Stocks of these viruses were prepared in embryonated SPF chicken eggs. All work with HPAIV was performed in biological safety level 3 (BSL3) facilities.
Virus titrations and growth curves.
Titers were determined in 96-well plates using either MDCK-NS1-GFP or standard MDCK cells as described previously (26). Briefly, cells were infected with 50 μl per well of serial 10-fold virus dilutions in PBS containing 0.3% BSA. After 1 h of incubation at room temperature, the virus was removed, and 100 μl per well of Avicel overlay medium was added. After 24 h of incubation at 37°C in 5% CO2 to allow plaque formation, the cells were fixed, permeabilized, and stained with a monoclonal antibody specific for influenza A virus nucleoprotein (AbD Serotec, Germany) for 1 h, followed by 1 h of incubation of a peroxidase-labeled anti-mouse antibody (Dako, Denmark). Afterward, the cells were incubated with the peroxidase substrate True-Blue (KPL) for a maximum of 30 min, and stained virus plaques (foci) were counted. Virus titers are expressed as focus-forming units (FFU) per ml.
For virus growth curves in CEF, cells were treated for 24 h with the indicated concentrations of chicken IFN-α before infection at a multiplicity of infection (MOI) of 0.001 in PBS containing 0.3% BSA for 1 h. Afterward, the cells were washed at least two times with PBS, and cell culture medium containing the indicated concentrations of chicken IFN-α was added. For growth of strains R66, Turk-18, and PR8, the medium was supplemented with 0.5 μg/ml acetylated trypsin (Sigma-Aldrich). Every 12 h, aliquots of the supernatant were removed and stored at −80°C until titration on either MDCK-NS1-GFP or standard MDCK cells. For virus growth curves in tracheal organ cultures, tissue samples were treated for 8 h with the indicated concentrations of chicken IFN-α before infection with the indicated viral doses in 199 Hanks' medium (Biochrom AG, Germany) containing 0.2% BSA for 1 h. After removal of the inoculum, the rings were cultured in 750 μl medium supplemented with the indicated concentrations of chicken IFN-α. Aliquots of supernatant were removed every 12 h postinfection and stored at −80°C until titration on MDCK cells.
Preparation of tissue extracts.
Tissue samples from mice and chickens were removed and immediately frozen in liquid nitrogen. Lung, spleen, and brain homogenates were prepared by suspending the tissue in 1 ml of PBS using FastPrep-24 equipment (MP Biomedicals, France) or a TissueLyser II (Qiagen, Germany). Tissue debris was removed by low-speed centrifugation, and samples were frozen at −80°C until further processing.
Production of recombinant chIFN-α.
Chicken IFN-α (chIFN-α) was purified from Escherichia coli as described previously (40). Biological activity was determined by comparing the antiviral activity against vesicular stomatitis virus (VSV)-GFP with a known IFN standard.
Bioassay for chicken type I IFN.
IFN activity in supernatants from infected CEF, chicken plasma, and organ homogenates from infected birds was analyzed using a bioassay as described previously (41). Briefly, CEC-32 reporter cells containing a luciferase gene under the chicken Mx promoter were incubated with sample dilutions or an IFN-α standard. After 6 h of incubation at 37°C, the cells were lysed with 1× cell culture lysis reagent (Promega), and luciferase activity was measured using luciferase substrate (Promega) and a Berthold Sirius Single Tube Luminometer or in 96-well plates using a Tecan infinite M200.
Measuring mouse IFN-β.
Mouse lung homogenates were analyzed for IFN-β levels using a commercial enzyme-linked immunosorbent assay (ELISA) (PBL; InterferonSource, Piscataway, NJ) according to the manufacturer's protocol.
Northern blot analysis.
To detect chicken IFN-α transcripts, RNA samples were analyzed as previously described (43, 45).
Real-time reverse transcription (RT)-PCR to quantify influenza virus.
Tissue samples and swabs from infected birds were harvested, immediately snap-frozen in liquid nitrogen, and stored at −80°C until viral RNA isolation was performed using the viral RNA isolation NucleoSpin Multi-96 Virus Kit (Macherey-Nagel AG, Oensingen, Switzerland) according to the manufacturer's protocol.
Influenza A virus-specific RNA was quantified with a one-step real-time RT-PCR detecting the viral M gene, essentially as described previously (47). Briefly, a conserved fragment of the M gene was amplified with primers FLU _panAmod_F (5′-AGATGAGYCTTCTAACCGA-3′) and FLU_panAmod_R (5′-GCAAAGACATCTTCAAGTYTC-3′) in combination with the 5′ 6-carboxyfluorescein (FAM)/3′ 6-carboxytetramethylrhodamine (TAMRA)-labeled probe FLU_panA-P (identical to probe M+64 lsqb]47]). Quantitative RT-PCR (50 cycles) was carried out with a QuantiTectProbe RT-PCR kit (Qiagen) on a 7500 Fast Real-Time PCR system (Applied Biosystems). Mean threshold cycle (CT) values were determined from triplicate amplifications of each sample. For simple presentation in figures, CT values were subtracted from the number 50, and the 50 − CT values were plotted.
Immunohistochemistry.
To detect the nucleoprotein of R65, tissue samples were fixed in 4% phosphate-buffered neutral formaldehyde and embedded in paraffin. Endogenous peroxidase in 3- to 4-μm sections was blocked with hydrogen peroxide (3%) in methanol before antigen retrieval by microwave irradiation in citrate buffer (10 mM, pH 6.0). Nonspecific background was blocked with normal goat serum. Sections were incubated with rabbit-anti-nucleoprotein (NP) serum (1:750 in Tris-buffered saline [TBS], pH 7.6), followed by biotinylated goat anti-rabbit IgG1 (Vector, Burlingame, CA; 1:200 in TBS) and the Vectastain Elite ABC Kit. Peroxidase activity was developed using the Dako AEC substrate-chromogen system (Dako, Carpinteria, CA). The sections were counterstained with Mayer's hematoxylin and sealed with aqueous medium (Aquatex; Merck, Darmstadt, Germany).
RESULTS
NS1 mutants of an H5N1 HPAIV induce more type I IFN than the parent virus in cultured chicken embryo cells and lungs of infected mice.
Wild-type R65 (R65-wt) and mutant R65-delNS1 lacking the complete coding region of the NS1 gene were generated by reverse genetics. For unknown reasons we failed to generate R65 mutants expressing C-terminally truncated NS1 protein. However, we managed to rescue a reassortant virus (designated R65-truncNS1) that carries seven segments of R65 and a modified segment 8 from the H7N7 virus strain SC35. This segment codes for a C-terminally truncated NS1 protein comprising amino acids 1 through 126 (21). Mutant R65-delNS1 replicated in chicken embryo cells (CEF), but it grew substantially less well than wild-type R65 and reached titers that were about 1,000-fold reduced (Fig. 1 A). R65-delNS1 induced about 150 units per ml of biologically active type I IFN in these cells, whereas wild-type R65 induced hardly any IFN (Fig. 1B). Mutant virus R65-truncNS1 exhibited an intermediate phenotype with regard to growth and IFN-inducing capacity in CEF (Fig. 1A and B).
Fig. 1.
Growth and IFN induction of NS1 mutants of R65 in chicken embryo fibroblasts and mouse lungs. (A) CEF were infected with the indicated viruses at an MOI of 0.001. Virus titers in the cell culture supernatants were determined at the indicated times postinfection. Mean values and standard deviations (SD) of three independent experiments are shown. The origin of the y axis was set to the detection limit of the titration (101.3 FFU/ml). (B) CEF were infected with the various viruses at an MOI of 0.5 and analyzed for IFN activity in the supernatants at 24 h postinfection using a bioassay. The mean values and SD of three independent experiments are shown. (C and D) Mx1+/+ mice were infected intranasally with 5 × 104 FFU of the indicated viruses per animal. At 24 h postinfection, the animals (n = 5 to 7) were killed, and virus titers (C) and IFN-β levels (D) in the lungs were determined by titration on MDCK cells and ELISA, respectively. The asterisk indicates that the viral titer in the sample was below the detection limit (101.3 FFU/ml).
In lungs of infected mice, R65-delNS1 failed to grow productively (Fig. 1C). Nevertheless, at 24 h postinfection, IFN-β levels in the lungs of mice infected with R65-delNS1 were much higher than in the lungs of mice infected with the same dose of wild-type R65 (Fig. 1D). Mutant virus R65-truncNS1 again showed an intermediate phenotype. It replicated substantially less well in mouse lungs than wild-type R65 (Fig. 1C), but it induced more IFN-β than wild-type R65. Thus, in both CEF and mouse lungs, the R65 NS1 mutants behaved as expected from previous studies with other influenza A viruses carrying partial or complete NS1 deletions (21, 36, 46).
NS1 mutants are attenuated in 5-week-old chickens.
To investigate the in vivo consequences of the NS1 gene defects in a natural host for HPAIV, groups of 5-week-old chickens were infected intratracheally with 6 × 105 FFU of either wild-type R65 or the two NS1 mutants of R65. The parent virus was highly pathogenic, and all infected chickens died or had to be euthanized due to severe disease within 48 h postinfection (Fig. 2 A). In contrast, R65-delNS1 was strongly attenuated, and all chickens survived the challenge without clinical symptoms. R65-truncNS1 was highly pathogenic, like wild-type R65, but disease progression was slower, with four of five chickens developing severe symptoms between 60 and 120 h postinfection (Fig. 2A). R65 carrying unmodified segment 8 of SC35, which served as an additional control in this experiment, killed all chickens within 60 h (data not shown).
Fig. 2.
Growth behavior of wild-type R65 and NS1 mutants in 5-week-old chickens. Chickens were infected by the intratracheal route with 6 × 105 FFU per animal of the indicated viruses. The physical status of the animals was determined daily, and moribund chickens were sacrificed. (A) Survival curves (5 animals per group). (B) Virus titers in lung and spleen were determined at the indicated times. The origin of the y axis was set to the detection limit of the titrations (103.2 and 103.4 FFU/g organ, respectively). The asterisks indicate that the viral titers of samples were below the detection limit. na, not applicable. (C) Immunohistochemical staining of viral nucleoprotein in lungs of chickens infected with R65-wt (left) and R65-delNS1 (right) at 12 and 30 h postinfection. Paraffin-embedded tissue samples were used for analysis. Brown staining reveals the presence of viral antigen.
Organs of infected chickens were collected at 12, 24, and 72 h postinfection and analyzed for the presence of infectious virus. The wild-type virus replicated to high titers in the lung (Fig. 2B) and was detected in the spleen as early as 12 h postinfection, indicating quick systemic spread. Wild-type R65 reached the brains of infected chickens within 24 h (data not shown), which might explain the quick fatal course of the infection. Mutant R65-truncNS1 also replicated efficiently in chicken lungs but disseminated to the spleen with substantial delay (Fig. 2B), mirroring its attenuated phenotype. In chickens infected with R65-delNS1, in which NS1 is deleted entirely, no infectious virus was detected in lung or spleen at any time point analyzed (Fig. 2B). However, immunohistochemical analysis of lung tissue clearly revealed scattered patches of virus antigen-positive cells at 12 h postinfection (Fig. 2C, top right). Interestingly, at a later time point, only very few infected individual cells were found in the lung (Fig. 2C, bottom right), suggesting efficient initial infection but restricted viral spread. In contrast, with wild-type R65, the numbers of viral-antigen-positive cells increased in all lung areas between 12 and 30 h postinfection (Fig. 2C, left). These results collectively showed that NS1 is essential for efficient virus replication and dissemination in chickens.
NS1 mutants do not induce enhanced synthesis of type I IFN in chickens.
To monitor innate immune responses of infected chickens, virus-induced IFN was quantified using the previously described bioassay for chicken type I IFN (41). Substantial levels of IFN were measured as early as 12 h postinfection in the lungs, spleens, and blood plasma of R65-infected chickens (Fig. 3 A). Mutant R65-truncNS1 induced similar or slightly reduced levels of IFN compared to wild-type virus in all organs tested (Fig. 3A). Surprisingly, even at 72 h postinfection, when R65-truncNS1 had replicated to high titers in the lungs of infected birds (Fig. 2B) and the animals were severely ill (Fig. 2A), IFN titers in lung, spleen, and plasma were not higher than in diseased birds infected with R65-wt (Fig. 3A). Only low levels of IFN were detected in the lungs and other organs of chickens infected with R65-delNS1 (Fig. 3A), although the virus induced very high levels of IFN in cultured chicken cells and lungs of mice (Fig. 1B and D). Northern blot analysis of RNA samples from infected chicken lungs (Fig. 3B) supported the finding that readily detectable levels of IFN-α are present in the lungs of chickens infected with the wild-type virus but not in the lungs of chickens infected with mutant R65-delNS1.
Fig. 3.
IFN-inducing capacities of wild-type R65 and NS1 mutants in 5-week-old chickens. (A) Chicken IFN activities in the lungs, spleens, and plasma of animals from the experiment shown in Fig. 2 were determined at the indicated times using a bioassay (n = 4 to 7). (B) Northern blot analysis of RNA samples (10 μg per lane) from the lungs of chickens infected with either R65-wt or R65-delNS1. RNA samples from the lungs of uninfected birds served as controls. The blot was hybridized to radiolabeled DNA derived from the chicken IFN-α1 gene. Ethidium bromide staining of the 18S rRNA was used as a loading control.
To evaluate the possibility that the NS1-deficient mutant virus induced only transient synthesis of IFN at early times, we determined the IFN levels in chickens intratracheally infected with 106 PFU of R65-wt or R65-delNS1 for only 3 and 6 h. Under such conditions, our bioassay measured no elevated levels of IFN in organs or plasma of animals infected with either of the two viruses (data not shown). These results clearly indicated that, contrary to what was predicted from experiments with chicken cell cultures and mice, R65 mutants with defective NS1 are not powerful inducers of IFN in chickens. Rather, the IFN levels observed in infected chickens correlated directly with virus titers, irrespective of whether wild-type or mutant viruses were present in the tissues. Thus, the NS1 protein of R65 did not appear to efficiently suppress IFN synthesis in virus-infected chickens, although it strongly suppressed IFN synthesis in mouse lungs and cultured chicken embryo fibroblasts.
NS1 mutants of an H7N7 virus also do not induce more IFN than the wild-type virus in chicken lungs.
In light of the unexpected observations described above that wild-type R65 induced similar or even higher levels of IFN in chickens than mutants with partial or complete NS1 deficiency, it was of great interest to determine whether in vivo synthesis of IFN might also not be suppressed by NS1 of other HPAIV. To address this question, we studied the H7N7 virus strain SC35 (8, 38). If applied to 5-week-old chickens by the intratracheal route at a dose of 106 FFU per animal, wild-type SC35 induced severe disease within 4 to 6 days, whereas the NS1-deficient mutant SC35-delNS1 was strongly attenuated (Fig. 4 A). Wild-type SC35 disseminated efficiently and was detected in both pharyngeal and cloacal swabs from all examined animals on day 2 postinfection (Fig. 4B). In contrast, SC35-delNS1 was found only in pharyngeal swabs collected at day 1 postinfection (Fig. 4B). Quantitative RT-PCR revealed that wild-type SC35 and SC35-delNS1 were both abundantly present in the lungs of infected birds at 20 h postinfection, but at 66 h postinfection, wild-type SC35 had grown to much higher levels than the NS1-deficient mutant (Fig. 4C). At 66 h and to a lesser extent also at 20 h postinfection, type I IFN levels were substantially higher in the lungs of wild-type SC35-infected than SC35-delNS1-infected chickens (Fig. 4D). These data demonstrate that the NS1-deficient SC35 mutant has no higher IFN-inducing capacity in chickens than wild-type virus, confirming our main conclusion from similar studies with R65 discussed above.
Fig. 4.
Growth behavior and IFN-inducing capacities of wild-type SC35 and NS1 mutants in 5-week-old chickens. The animals were infected by the intratracheal route with 106 FFU per animal of either wild-type SC35 (SC35-wt) or NS1-deficient SC35 (SC35-delNS1). The physical status of the animals was determined daily. Chickens were killed in cases of severe clinical disease. (A) Survival curves (5 animals per group). (B) Viral RNA in pharyngeal and cloacal swabs at the indicated times postinfection. Titers are expressed as 50 − CT. The asterisks indicate samples with viral RNA levels below the detection limit. (C an D) Kinetics of viral RNA accumulation (C) and IFN titers (D) were assessed in the lungs of birds infected with the indicated viruses.
R65 exhibits a relatively high degree of IFN resistance in chicken embryo cells and tracheal organ cultures.
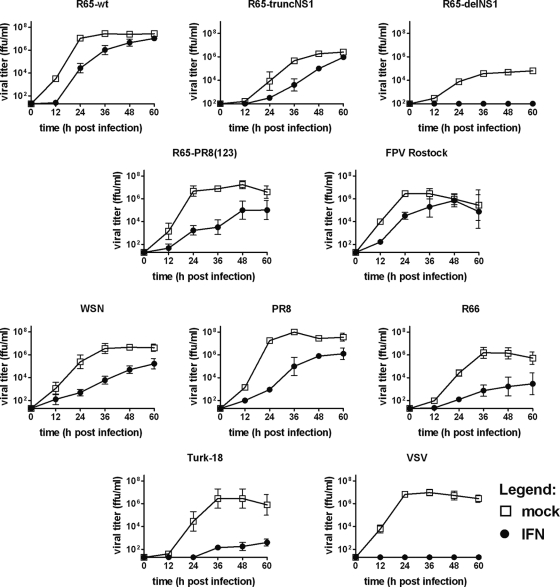
The above-described experiments showed that influenza viruses can be highly pathogenic in chickens in spite of the fact that they induce high levels of type I IFN. This observation suggested that such viruses might exhibit a high degree of resistance to the antiviral activity of IFN. We tested this possibility by first determining the IFN sensitivities of various influenza viruses in CEF. Cultures pretreated with medium containing or lacking 1,000 units per ml of chicken IFN-α were infected at an MOI of 0.001, and culture supernatants harvested at the indicated times were assayed for infectivity. We found that growth of R65-wt was delayed in the presence of IFN, but peak titers of both treated and untreated cultures were indistinguishable (Fig. 5). Similarly, R65-truncNS1 and the HPAIV strain FPV-Rostock (H7N1) exhibited relatively high degrees of resistance to IFN treatment. These viruses reached comparably high peak titers in IFN-treated and untreated cultures. In contrast, the LPAIV strain R66 (H9N2) and Turk-18 (H3N3) displayed a substantially higher degree of IFN sensitivity that was comparable to that of WSN and PR8, two H1N1 laboratory strains originating from human isolates (Fig. 5). R65-delNS1, which grows only poorly in untreated CEF, failed to grow productively if IFN was present in the culture medium. VSV, which served as a positive control, was also strongly affected by IFN, demonstrating that the preparation of recombinant chicken IFN-α used for these assays was highly active (Fig. 5).
Fig. 5.
IFN sensitivities of various influenza A virus strains in CEF cultures. CEF cultured in plain medium or in the presence of 1,000 U/ml of chicken IFN-α were infected with the indicated viruses at an MOI of 0.001. Viral growth was monitored by titrating samples of culture supernatants harvested at the indicated times. For each virus, mean titers (±standard error of the mean) of two to nine independent experiments are presented. The origin of the y axis was set to the detection limit of the titrations.
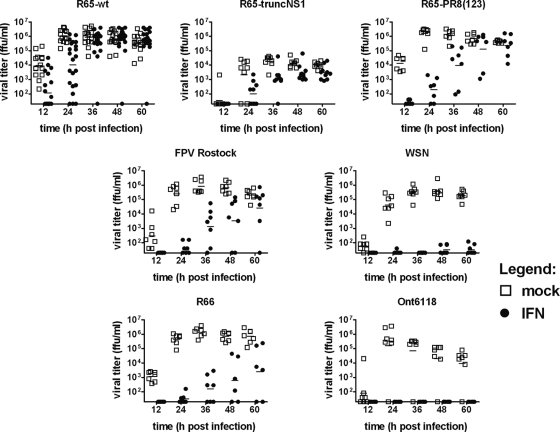
To further assess the apparent IFN resistance of R65-wt, we next infected tracheal organ cultures from chicken embryos. R65-wt replicates vigorously in these tissue explants, as was evident from the fact that infectious virus accumulated quickly in the culture supernatant (Fig. 6). If infected at a dose of 100 FFU per tracheal ring, substantial growth of R65-wt was already observed at 12 h postinfection, and maximal viral titers in the culture supernatant (∼106 FFU per ml) were reached at approximately 24 h postinfection (Fig. 6). In the presence of 1,000 units per ml of chicken IFN-α, the growth of R65-wt was delayed by some 12 h, but the viral titers of treated and untreated cultures were virtually identical at late time points (Fig. 6). Under such conditions, WSN was highly susceptible to chicken IFN-α (Fig. 6). The LPAIV strain Ont6118 (H8N4) exhibited a similar high degree of IFN sensitivity in tracheal organ cultures, whereas strains R66 and FPV Rostock showed low and moderate degrees of IFN resistance, respectively (Fig. 6). Interestingly, R65-truncNS1 exhibited a high degree of resistance to the action of IFN in this system (Fig. 6). Overall, the virulence phenotypes of the various viruses correlated well with IFN resistance.
Fig. 6.
IFN sensitivities of various influenza A virus strains in chicken tracheal organ cultures. Tracheal organ cultures (6 or 7 individual cultures for each experimental condition) kept in medium containing or lacking 1,000 U/ml of chicken IFN-α were infected with 100 PFU of R65-wt, R65-truncNS1, R65-PR8(123), and FPV Rostock or 1,000 PFU of WSN, R66, and Ont6118. Virus titers in individual culture supernatants were determined at the indicated times postinfection. Each dot represents the result of one individual culture. The origin of the y axis was set to the detection limit of the titrations.
To exclude the remote possibility that R65 and other HPAIV strains might quickly acquire mutations conferring IFN resistance, we performed parallel growth kinetics in CEF with pairs of viruses originating from 48-hour supernatants of IFN-treated or untreated CEF and tracheal organ cultures, respectively. The IFN sensitivities of “IFN-experienced” and “IFN-nonexperienced” R65 stocks were indistinguishable, rendering this possibility rather unlikely (see Fig. S1 in the supplemental material).
R65 is resistant to IFN in chickens, but not mice.
Recent studies with mice showed that a single dose of IFN-α applied by the intranasal route can protect from lethal infection with highly virulent H5N1 virus (50). To verify that R65 is not exceptional with regard to IFN sensitivity, we performed similar protection experiments using IFN-competent (Mx1+/+) mice. Groups of mice were treated intranasally with a moderate dose (2 × 104 units per mouse) of IFN-α or with PBS as a control, and at 8 h posttreatment, the mice were challenged with a lethal dose (3 × 105 FFU per mouse) of R65-wt. As shown in Fig. 7 A, all control animals developed severe respiratory disease and had to be killed between 4 and 5 days postinfection. In contrast, all five IFN-treated mice remained healthy during the complete 8-day observation period. At 48 h postinfection, the virus could readily be reisolated from lung tissue of untreated, but not IFN-treated, mice (Fig. 7B), demonstrating that R65 exhibits a high degree of IFN sensitivity in mice, similar to previously tested H5N1 strains (50). Since an earlier study had suggested that some H5N1 virus strains are resistant to IFN in cultured mammalian cells (44), we performed inhibition studies in human A549 cells. Titers of R65 in supernatants of IFN-treated A549 cells at 48 h postinfection at an MOI of 0.001 were approximately 100-fold lower than in untreated control cells (data not shown), indicating that R65 is well controlled by the mammalian IFN system.
Fig. 7.
Inhibition of R65 by IFN-α in mice. Groups of Mx1+/+ mice (n = 5) were treated intranasally with a buffer solution (mock) or with 2 × 104 units of human hybrid IFN-αB/D, which is highly active in mice (16). At 8 h after IFN treatment, the animals were challenged with 3 × 105 FFU per mouse (∼3 times the lethal dose) of wild-type R65. (A) The physical status of the animals was determined daily, and animals were killed if severely ill or if weight loss approached the critical value of 25 to 30%. (B) Other groups of animals were killed at 48 h postinfection, and viral titers in the lung were determined. The origin of the y axis was set to the detection limit of the titrations. The asterisk indicates that the viral titers of samples were below the detection limit.
To test the IFN sensitivity of R65 in a natural host, in a first experiment, groups of 5-week-old chickens were simultaneously treated oculonasally with 5 × 106 units and intratracheally with 5 × 106 units of chicken IFN-α or with corresponding amounts of a control protein. Six hours later, the animals were challenged with 106 PFU of R65-wt by the same routes. IFN-treated and control animals all became severely ill and died or had to be killed by day 3 postinfection (data not shown). We considered several technical reasons for the lack of IFN-mediated protection, including the possibilities that local delivery of IFN to the lung by intranasal and intratracheal application might not be optimal, that multiple treatments with IFN might be necessary to maintain an antiviral state in chickens for several days, and that the virus challenge dose might have been too high.
To account for these possibilities, we designed a second experiment with altered application routes, repeated IFN administrations, and challenge by transmission from virus-infected animals. Chickens were treated with 107 units of chicken IFN-α or a control preparation at 8 and 2 h before and at 12 h after virus exposure, as depicted in Fig. 8 A. The animals were infected by exposure to infected chickens that excreted R65-wt virus (designated “seeder birds” in Fig. 8A), thus mimicking a more natural course of infection. To verify that the IFN was applied correctly and reached the circulation of the treated birds, we measured IFN levels in plasma samples taken 15 min after the second IFN injection. Plasma IFN levels were above 1,000 units per ml in most treated birds (Fig. 8B). Nevertheless, we observed no significant difference in survival between IFN-treated and mock-treated birds (Fig. 8C), and all animals died within 9 days after being cohoused with seeder birds.
Fig. 8.
IFN treatment failed to block transmission of R65 during cohousing with infected birds. (A) Experimental setup. Uninfected birds were treated by the intravenous route with a total of 107 units of chicken IFN-α or with a control protein (MxA [10]) at 8 and 2 h prior to exposure to seeder birds and once more at 12 h after first exposure to seeder birds. Twelve hours before cohousing, the seeders were infected with a lethal dose of R65-wt (106 PFU per bird) so that they shed infectious virus when grouped together with the uninfected birds. (B) Plasma IFN levels in treated birds. Fifteen minutes after the second treatment of the birds with either IFN or control protein, blood was drawn from each animal and plasma IFN levels were measured using a bioassay. Each dot represents the value from one individual bird. (C) Survival curves of IFN-treated (n = 8) and control-treated (n = 8) birds.
The remote possibility remained that exogenously applied chicken IFN-α might not reach the relevant cell types of infected chickens and that the lack of IFN-mediated protection against R65-wt could be explained on this basis. To evaluate this possibility we searched for a suitable control virus that shares biological properties with R65 and can be administered by the same route but differs in IFN sensitivity. LPAIV strains might have been an option, but these viruses do not grow well in experimentally infected birds, and it is difficult to monitor their growth and the course of disease. Therefore, we created virus R65-PR8(123), which contains genomic RNA segments 1 to 3 (encoding the viral polymerase subunits PB2, PB1, and PA) from laboratory strain PR8 and segments 4 to 8 from R65. R65-PR8(123) is substantially less pathogenic in chickens than R65-wt: it induces severe disease in 4-week-old chickens, but it does not readily kill like R65-wt (unpublished data).
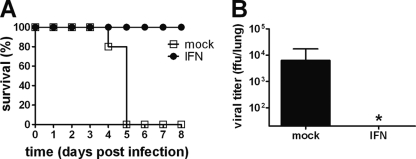
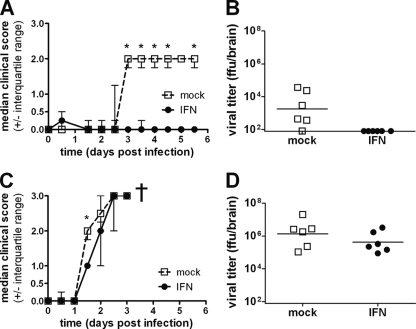
R65-PR8(123) was clearly more sensitive to IFN in CEF than R65-wt (Fig. 5), although this difference was rather moderate in tracheal organ cultures (Fig. 6). To analyze the in vivo IFN sensitivity of R65-PR8(123), 4-week-old birds were treated with 107 units of chicken IFN-α by the intravenous route at 8 and 2 h prior to infection and by the intramuscular route at 12, 36, 60, 84, and 108 h postinfection. A control group of birds was treated at the same times with the same dose of an irrelevant control protein. R65-PR8(123) was applied by the oculonasal route at a dose of 2 × 106 PFU per bird. The control birds started to show signs of severe disease on day 3 postinfection. Symptoms included apathy, untended plumage, and hemorrhagic lesions on the feet. No such symptoms were observed in the group of birds that was treated with IFN, with the exception of one bird transiently showing ruffled feathers and mild apathy (Fig. 9 A). No virus could be reisolated from the brains of IFN-treated birds at day 5.5 postinfection, when the experiment was terminated, whereas virus isolation was successful in 5 of 6 animals from the control group (Fig. 9B), indicating that the IFN treatment had been highly effective. As expected from the results presented above, under identical experimental conditions, the IFN treatment was again not effective against R65-wt. Control-treated and IFN-treated birds both became severely ill within 3 days postinfection with R65-wt (Fig. 9C), and the virus grew to high titers in the brains of both groups of birds (Fig. 9D).
Fig. 9.
Repeated IFN treatment strongly inhibits an attenuated variant of R65 in chickens but is ineffective against wild-type R65. (A) Kinetics of disease development in IFN-treated (n = 6) and mock-treated (n = 6) birds following infection with 2 × 106 PFU of mutant virus R65-PR8(123). Animals were examined at 12-h intervals, and clinical scores were assigned as described in Materials and Methods. The graph depicts the median clinical scores of the groups. The asterisks indicate significant differences between groups (comparison of medians by Mann-Whitney U test or Wilcoxon signed-rank test; P < 0.05). (B) Viral titers in brain homogenates of birds infected with R65-PR8(123) at day 5.5 postinfection. (C) Kinetics of disease development in IFN-treated (n = 6) and mock-treated (n = 6) birds following infection with 2 × 106 PFU of R65-wt. The animals were examined at 12-h intervals, and clinical scores were assigned as described in Materials and Methods. The graph depicts the median clinical scores of the groups. The asterisk indicates a significant difference between groups. (D) Viral titers in brain homogenates of birds infected with R65-wt. The animals were killed when severely ill between days 2.5 and 3 postinfection. Each dot represents the result for one animal. The origin of the y axis was set to the detection limit of the titrations (101.9 FFU/brain).
DISCUSSION
Only a few previous reports addressed the roles of NS1 proteins of avian influenza A viruses in their natural hosts (3, 7, 21, 24, 51, 56), and it has remained unclear whether NS1 modulates different cellular signaling pathways in mammals and birds. A previous study suggested that highly pathogenic avian H5H1 influenza viruses might lack the ability to activate IFN genes in chickens (5). Our experiments with the HPAIV strain R65 do not support this view, and the reason for this discrepancy remains unclear. By Northern blot analysis and by testing for biological activity, we have demonstrated that wild-type R65 is a very potent inducer of IFN in infected chickens. Further, our experiments with viruses carrying partial or complete deletions of the NS1 gene led to the conclusion that, unlike in mice, NS1 does not suppress the synthesis of type I IFN in chickens.
We believe we have excluded all trivial explanations of why the wild-type virus was a strong IFN inducer and why NS1 may not suppress IFN synthesis in infected chickens. One could argue that the inability of R65-delNS1 to induce high levels of IFN in chickens may be due to its failure to replicate efficiently in birds. However, this virus is a very strong inducer of IFN in mouse lungs, in which it also cannot replicate well. Further, our analysis demonstrated that the IFN genes were also not expressed strongly in chickens at early times postinfection with R65-delNS1, when a substantial number of cells in the lung were positive for viral antigen. Above all, R65-truncNS1, which has a less severe NS1 defect than R65-delNS1, is also not a better inducer of IFN-α in chickens than wild-type R65. It should be noted that at 72 h postinfection R65-truncNS1 had grown to almost as high titers in chicken lungs as wild-type R65 at 24 h postinfection (Fig. 2B). Thus, if viruses with partial or complete NS1 defects were able to stimulate IFN synthesis more efficiently than R65-wt, the IFN levels in organs of chickens infected with R65-truncNS1 at 72 h postinfection should have been higher than the IFN levels in organs of chickens infected with R65-wt at 24 h postinfection. However, this was clearly not the case (Fig. 3A). Rather, the IFN titers correlated fairly well with viral titers irrespective of the virus used for infection, in agreement with another recent report (32).
Another argument could be that R65 is not a representative influenza A virus and that NS1 failed to suppress IFN synthesis in chickens due to our challenge virus choice. However, we excluded this possibility by showing that NS1-deficient mutants of an unrelated virus, the H7N7 strain SC35, were also no better inducers of IFN than wild-type SC35. Thus, taken together, our data demonstrate that virus-induced IFN synthesis in chickens is not strongly influenced by the NS1 protein of HPAIV, although such restriction can be observed in cultured chicken cells.
Recent work demonstrated that chickens lack RIG-I (1), which is an important virus sensor in mammals that is targeted by NS1 proteins of influenza A viruses. Our experiments showed that the NS1 proteins of R65 and SC35 were able to suppress IFN synthesis rather efficiently in cultured chicken cells (Fig. 1), excluding the possibility that the lack of RIG-I in chickens can explain our results. Obviously, RIG-I is functionally compensated for by another intracellular sensor in chicken cells that recognizes invading viruses.
To account for our seemingly contradictory finding that NS1 of HPAIV repressed IFN synthesis in cultured chicken cells but not in infected birds, we hypothesize that several cell types are able to synthesize IFN in response to influenza virus challenge but the cell type responsible for the vast majority of IFN synthesis in virus-infected chickens does not respond to NS1. This insensitivity could result from a very early blockage of the viral replication cycle in the unidentified cell type, so that NS1 cannot be synthesized in sufficient quantities. Alternatively, specialized cells, such as plasmacytoid dendritic cells, might sense influenza viruses via Toll-like receptors, as suggested recently (32). This view is supported by the observation that wild-type H5N1 viruses can induce high levels of type I IFN in splenocyte but not fibroblast cultures (32). Finally, it remains possible that the in vivo situation is far more complex and that IFN synthesis by infected cells is mainly determined by the interplay between IFN-inducing and IFN-suppressing viral particles, as suggested previously (25, 42), and that our wild-type and mutant viruses differ in this respect.
The second unexpected result from our study is that certain avian influenza viruses exhibited a relatively high degree of resistance to the antiviral action of type I IFN. Given the fact that HPAIV strains, such as R65, are good inducers of IFN, it is probably not surprising that they exhibit a high degree of IFN resistance. If they did not, they would probably be less virulent than they actually are. The HPAIV strains R65 and, to a lesser extent, FPV Rostock exhibited a high degree of IFN resistance in our experiments with CEF and tracheal organ cultures, whereas the LPAIV strain Ont6118 and the mammalian virus strain WSN were strongly inhibited by IFN in tracheal organ cultures. Interestingly, the LPAIV strain R66, which induced substantial morbidity in vivo, was clearly less susceptible to IFN than Ont6118 (Fig. 6). Thus, the IFN resistances of the various virus strains seemed to correlate with in vivo virulence. It should be noted that our results are consistent with historical experiments that led to the discovery of IFN (18), in which it was found to inhibit the replication of a human influenza A virus strain related to WSN and PR8 in chicken cells.
We believe we have excluded all trivial explanations for the lack of efficient inhibition of R65 by IFN-α in vivo. The possibility that the administered IFN did not reach the organs in which the virus replicates is highly unlikely because we found high serum IFN levels in treated birds (data not shown). Further, the reassortant virus R65-PR8(123), which is less pathogenic than wild-type R65, showed a high degree of IFN sensitivity in an in vivo setting that revealed the pronounced IFN resistance of R65 (Fig. 9). At present, we cannot fully explain the in vivo IFN resistance of wild-type R65. The growth kinetics of R65-PR8(123) and wild-type R65 did not differ in CEF cultures. Possibly, the various virus-permissive cell types in the chicken do not respond to IFN uniformly. Our in vivo results could be ascribed to differences in cell tropism of the two viruses. R65-PR8(123) might prefer to replicate in particular cells that are highly sensitive to IFN, whereas R65-wt might have an extended tropism and might also efficiently replicate in at least one cell population that is fairly resistant to the antiviral action of IFN in chickens. It remains unknown how the polymerases of the two viruses might determine the cell tropism.
Taken together, our results are in accordance with earlier findings with a low-pathogenic avian influenza A virus strain that the NS1 protein is an important viral virulence factor in chickens (3). We show here that NS1 is also essential for virulence of highly pathogenic influenza A viruses. Most importantly, our study indicates that, unlike what has been assumed from studies in mice and other mammals, the main function of the NS1 protein in the chicken is not suppression of IFN synthesis. Others have recently shown that NS1 can efficiently suppress Fas-mediated apoptosis of infected chicken macrophages (54). Further, NS1 proteins of HPAIV can hyperactivate phosphoinositol 3-kinase signaling (15), which inhibits host apoptotic responses. It remains to be investigated whether these or other activities of NS1 are of critical importance for the virulence of avian influenza viruses in chickens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Silke Rautenschlein and Henning Petersen for help with the tracheal organ cultures, Markus Mordstein for help with the mouse experiments, and Annette Ohnemus for excellent technical assistance.
This work was supported by grants from the European Commission (INN-FLU) and the German Ministry for Education and Research (FluResearchNet).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Barber M. R., Aldridge J. R., Jr., Webster R. G., Magor K. E. 2010. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. U. S. A. 107:5913–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burgui I., Aragon T., Ortin J., Nieto A. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263–3274 [DOI] [PubMed] [Google Scholar]
- 3. Cauthen A. N., Swayne D. E., Sekellick M. J., Marcus P. I., Suarez D. L. 2007. Amelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 81:1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherry J. D., Taylor-Robinson D. 1970. Large-quantity production of chicken embryo tracheal organ cultures and use in virus and mycoplasma studies. Appl. Microbiol. 19:658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daviet S., et al. 2009. Induction of Mx and PKR failed to protect chickens from H5N1 infection. Viral Immunol. 22:467–472 [DOI] [PubMed] [Google Scholar]
- 6. Digby M. R., Lowenthal J. W. 1995. Cloning and expression of the chicken interferon-gamma gene. J. Interferon Cytokine Res. 15:939–945 [DOI] [PubMed] [Google Scholar]
- 7. Dundon W. G., Milani A., Cattoli G., Capua I. 2006. Progressive truncation of the Non-Structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res. 119:171–176 [DOI] [PubMed] [Google Scholar]
- 8. Gabriel G., et al. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gack M. U., et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao S., et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506 [DOI] [PubMed] [Google Scholar]
- 11. Garcia-Sastre A., et al. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 12. Guo Z., et al. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36:263–269 [DOI] [PubMed] [Google Scholar]
- 13. Hale B. G., Jackson D., Chen Y. H., Lamb R. A., Randall R. E. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hale B. G., Randall R. E., Ortin J., Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 15. Heikkinen L. S., et al. 2008. Avian and 1918 Spanish influenza A virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling. J. Biol. Chem. 283:5719–5727 [DOI] [PubMed] [Google Scholar]
- 16. Horisberger M. A., de Staritzky K. 1987. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J. Gen. Virol. 68:945–948 [DOI] [PubMed] [Google Scholar]
- 17. Hughes A. L., Roberts R. M. 2000. Independent origin of IFN-alpha and IFN-beta in birds and mammals. J. Interferon Cytokine Res. 20:737–739 [DOI] [PubMed] [Google Scholar]
- 18. Isaacs A., Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267 [PubMed] [Google Scholar]
- 19. Karpala A. J., et al. 2008. Molecular cloning, expression, and characterization of chicken IFN-lambda. J. Interferon Cytokine Res. 28:341–350 [DOI] [PubMed] [Google Scholar]
- 20. Kochs G., Garcia-Sastre A., Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kochs G., et al. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403–1409 [DOI] [PubMed] [Google Scholar]
- 22. Kochs G., et al. 2009. Strong interferon-inducing capacity of a highly virulent variant of influenza A virus strain PR8 with deletions in the NS1 gene. J. Gen. Virol. 90:2990–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z., et al. 2006. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 80:11115–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Long J. X., Peng D. X., Liu Y. L., Wu Y. T., Liu X. F. 2008. Virulence of H5N1 avian influenza virus enhanced by a 15-nucleotide deletion in the viral nonstructural gene. Virus Genes 36:471–478 [DOI] [PubMed] [Google Scholar]
- 25. Marcus P. I., Rojek J. M., Sekellick M. J. 2005. Interferon induction and/or production and its suppression by influenza A viruses. J. Virol. 79:2880–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matrosovich M., Matrosovich T., Garten W., Klenk H. D. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melen K., et al. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mibayashi M., et al. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Min J. Y., Krug R. M. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Min J. Y., Li S., Sen G. C., Krug R. M. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236–243 [DOI] [PubMed] [Google Scholar]
- 31. Mordstein M., et al. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moulin H. R. High interferon type I responses in the lung, plasma and spleen during highly pathogenic H5N1 infection of chicken. Vet. Res. 42:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nemeroff M. E., Barabino S. M., Li Y., Keller W., Krug R. M. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 34. Noah D. L., Twu K. Y., Krug R. M. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386–395 [DOI] [PubMed] [Google Scholar]
- 35. Opitz B., et al. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9:930–938 [DOI] [PubMed] [Google Scholar]
- 36. Quinlivan M., et al. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richt J. A., Garcia-Sastre A. 2009. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 333:177–195 [DOI] [PubMed] [Google Scholar]
- 38. Scheiblauer H., Kendal A. P., Rott R. 1995. Pathogenicity of influenza A/Seal/Mass/1/80 virus mutants for mammalian species. Arch. Virol. 140:341–348 [DOI] [PubMed] [Google Scholar]
- 39. Schultz-Cherry S., Dybdahl-Sissoko N., Neumann G., Kawaoka Y., Hinshaw V. S. 2001. Influenza virus ns1 protein induces apoptosis in cultured cells. J. Virol. 75:7875–7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schultz U., Rinderle C., Sekellick M. J., Marcus P. I., Staeheli P. 1995. Recombinant chicken interferon from Escherichia coli and transfected COS cells is biologically active. Eur. J. Biochem. 229:73–76 [DOI] [PubMed] [Google Scholar]
- 41. Schwarz H., Harlin O., Ohnemus A., Kaspers B., Staeheli P. 2004. Synthesis of IFN-beta by virus-infected chicken embryo cells demonstrated with specific antisera and a new bioassay. J. Interferon Cytokine Res. 24:179–184 [DOI] [PubMed] [Google Scholar]
- 42. Sekellick M. J., Carra S. A., Bowman A., Hopkins D. A., Marcus P. I. 2000. Transient resistance of influenza virus to interferon action attributed to random multiple packaging and activity of NS genes. J. Interferon Cytokine Res. 20:963–970 [DOI] [PubMed] [Google Scholar]
- 43. Sekellick M. J., Ferrandino A. F., Hopkins D. A., Marcus P. I. 1994. Chicken interferon gene: cloning, expression, and analysis. J. Interferon Res. 14:71–79 [DOI] [PubMed] [Google Scholar]
- 44. Seo S. H., Hoffmann E., Webster R. G. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950–954 [DOI] [PubMed] [Google Scholar]
- 45. Sick C., Schultz U., Staeheli P. 1996. A family of genes coding for two serologically distinct chicken interferons. J. Biol. Chem. 271:7635–7639 [DOI] [PubMed] [Google Scholar]
- 46. Solorzano A., et al. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535–7543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spackman E., et al. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Staeheli P., Dreiding P., Haller O., Lindenmann J. 1985. Polyclonal and monoclonal antibodies to the interferon-inducible protein Mx of influenza virus-resistant mice. J. Biol. Chem. 260:1821–1825 [PubMed] [Google Scholar]
- 49. Staeheli P., Puehler F., Schneider K., Gobel T. W., Kaspers B. 2001. Cytokines of birds: conserved functions—a largely different look. J. Interferon Cytokine Res. 21:993–1010 [DOI] [PubMed] [Google Scholar]
- 50. Tumpey T. M., et al. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81:10818–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang L., et al. 2008. Characterization of influenza virus variants with different sizes of the non-structural (NS) genes and their potential as a live influenza vaccine in poultry. Vaccine 26:3580–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weber S., et al. 2007. Molecular analysis of highly pathogenic avian influenza virus of subtype H5N1 isolated from wild birds and mammals in northern Germany. J. Gen. Virol. 88:554–558 [DOI] [PubMed] [Google Scholar]
- 53. Weining K. C., Schultz U., Munster U., Kaspers B., Staeheli P. 1996. Biological properties of recombinant chicken interferon-gamma. Eur. J. Immunol. 26:2440–2447 [DOI] [PubMed] [Google Scholar]
- 54. Xing Z., et al. 2009. Differential regulation of antiviral and proinflammatory cytokines and suppression of Fas-mediated apoptosis by NS1 of H9N2 avian influenza virus in chicken macrophages. J. Gen. Virol. 90:1109–1118 [DOI] [PubMed] [Google Scholar]
- 55. Zhirnov O. P., Konakova T. E., Wolff T., Klenk H. D. 2002. NS1 protein of influenza A virus down-regulates apoptosis. J. Virol. 76:1617–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu Q., et al. 2008. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J. Virol. 82:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.