Abstract
Initiation of reverse transcription in hepadnaviruses is accomplished by a unique protein-priming mechanism whereby a specific Y residue in the terminal protein (TP) domain of the viral reverse transcriptase (RT) acts as a primer to initiate DNA synthesis, which is carried out by the RT domain of the same protein. When separate TP and RT domains from the duck hepatitis B virus (DHBV) RT protein were tested in a trans-complementation assay in vitro, the RT domain could also serve, unexpectedly, as a protein primer for DNA synthesis, as could a TP mutant lacking the authentic primer Y (Y96) residue. Priming at these other, so-called cryptic, priming sites in both the RT and TP domains shared the same requirements as those at Y96. A mini RT protein with both the TP and RT domains linked in cis, as well as the full-length RT protein, could also initiate DNA synthesis using cryptic priming sites. The cryptic priming site(s) in TP was found to be S/T, while those in the RT domain were Y and S/T. As with the authentic TP Y96 priming site, the cryptic priming sites in the TP and RT domains could support DNA polymerization subsequent to the initial covalent linkage of the first nucleotide to the priming amino acid residue. These results provide new insights into the complex mechanisms of protein priming in hepadnaviruses, including the selection of the primer residue and the interactions between the TP and RT domains that is essential for protein priming.
INTRODUCTION
The Hepadnaviridae family includes the hepatitis B virus (HBV), a global human pathogen that chronically infects hundreds of millions and causes a million fatalities yearly, and related animal viruses such as the duck hepatitis B virus (DHBV) (40). Hepadnaviruses contain a small (∼3-kb), partially double-stranded DNA genome that is replicated through an RNA intermediate, called pregenomic RNA (pgRNA) by reverse transcription (39, 43). The virally encoded reverse transcriptase (RT) is unique among known RTs in its structure and function (14). RT is composed of four domains. The N-terminal TP (terminal protein) is conserved among all hepadnaviruses but absent from all other known RTs and is required for viral reverse transcription. The spacer domain, which does not have any known role in RT functions, connects TP to the central RT domain and the C-terminal RNase H domain. Both the RT and the RNase H domains share sequence homology with other RTs, including the RT active-site motif YMDD and the catalytic RNase H residues (4, 5, 35, 57).
A prerequisite for the initiation of reverse transcription in hepadnaviruses is the specific interaction between RT and a short RNA signal called ε located at the 5′ end of pgRNA (33, 51). RT-ε interaction is required to activate the polymerase activity of RT and ε also serves as the template for the initial stage of viral reverse transcription (13, 16, 44, 46, 47, 49). Instead of relying on an RNA primer to prime DNA synthesis, as is the case with most other RTs and DNA polymerases in general, the hepadnavirus RT uses a specific Y residue in the TP domain of the RT protein itself (Y96 in DHBV and Y63 in HBV) as a primer to initiate minus-strand DNA synthesis (24, 25, 50, 56, 59). During this so-called protein priming reaction, a short (3 to 4 nucleotides long) DNA oligomer is synthesized that is covalently attached to RT through the primer Y residue, using as a template an internal bugle of the ε RNA. Both the TP and the RT domains are essential for RT-ε interaction and protein priming but the RNase H domain is dispensable (10, 23, 33, 51). Also, isolated TP and RT domains, when combined together, are able to reconstitute a functional RT protein that is able to carry out protein priming in a trans-complementation reaction (1, 23, 25, 27). Protein priming can be further subdivided into two different stages, requiring distinct RT conformations, the initial covalent attachment of the first nucleotide of the minus-strand DNA (a dGMP in DHBV) to RT (to the primer Y) called initiation and the subsequent addition of the remaining 2 to 3 nucleotides to the initiating nucleotide called polymerization (27, 54).
DHBV RT-ε interaction and protein priming have been reconstituted in vitro using purified components. The full-length RT requires the assistance of the host cell chaperone proteins in order to establish and maintain a conformation that is competent to recognize the ε RNA and to initiate protein priming (9, 10, 13, 15, 17, 18, 41, 42). However, a truncated DHBV RT protein, MiniRT2, with deletion of the entire RNase H domain, the N-terminal third of the TP domain, and most of the spacer, retains ε RNA binding and protein priming activity but no longer requires the host chaperones (55). Recently, we reported that the divalent metal ions, Mg2+ versus Mn2+, have a dramatic effect on protein priming, including the overall priming efficiency, the template and nucleotide specificity, and the transition from initiation to polymerization, further attesting to the multiple RT conformational transitions that occur during protein priming (27).
In the present study, we found, surprisingly, that DHBV RT protein was able to initiate DNA synthesis using amino acid (S/T as well as Y) residues on both the TP and the RT domains in addition to the known Y96 site in the TP domain. We characterized the requirements for protein priming from these alternative or cryptic priming sites versus those from Y96 by using the trans-complementation priming reaction, as well as cis priming as carried out by MiniRT2 and the full-length DHBV RT. The discovery of cryptic priming sites has important implications on the selection of the primer residue, the RT conformational changes during protein priming, and potentially the role of protein priming in viral pathogenesis.
MATERIALS AND METHODS
Plasmids.
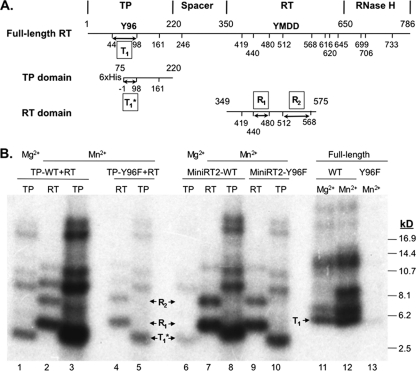
pGEX-MiniRT2 and pQE-MiniRT2 express a truncated DHBV RT protein, MiniRT2 (Fig. 1), that is fused to the glutathione S-transferase (GST) and an N-terminal hexahistidine (6×His) tag, respectively, as described previously (10). pGEX-TP and pGEX-RT were derived from pGEX-MiniRT2 and express the GST-tagged and truncated DHBV TP (residues 75 to 220) and DHBV RT (residues 349 to 575) domains, respectively (26). Similarly, pQE-TP and pQE-RT express the 6×His-tagged TP (residues 75 to 220) and RT (residues 349 to 575) domains, respectively, and were derived from pQE-MiniRT2 by removing the RT and TP domain coding sequences, respectively. pHis-MBP-TP expresses a maltose-binding protein (MBP)- and 6×His-tagged TP domain in a modified pMCGS7 vector (34). A Tev protease cleavage site is present between MBP and TP sequence in pHis-MBP-TP. pGEX-RT-YMHA was derived from pGEX-RT and contains two amino acid substitutions in the RT active site (changing the conserved YMDD motif into YMHA) (4, 54) that abolishes the polymerase activity of RT. The pQE-TP-Y96F and pQE-MiniRT2-Y96F were derived from pQE-TP and pQE-MiniRT2, respectively, by changing the Y at position 96 to phenylalanine (59). pHP and pHP-Y96F express the full-length DHBV RT (the wild type and the Y96F mutant, respectively) under the phage SP6 promoter in vitro (59). All mutations were verified by DNA sequencing.
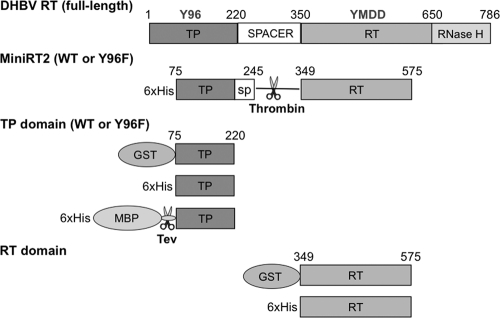
Fig. 1.
Schematic illustration of the DHBV RT protein and domain constructs. The primer Y residue (Y96) in the TP domain and the YMDD catalytic motif in the RT domain are denoted, as are the boundaries (in amino acid positions) of the truncated MiniRT2 protein and the TP and RT domain constructs, the thrombin cleavage site inserted into MiniRT2, the Tev protease cleavage site in His-MBP-TP, and the different affinity tags (GST, MBP, and 6×His).
Protein expression and purification.
DHBV full-length RT proteins (the wild type [WT] and Y96F mutant) were expressed in vitro using a coupled in vitro transcription and translation reaction kit, the TnT rabbit reticulocyte lysate (RRL) system (Promega), in the absence of any radiolabeling according to the manufacturer's instructions (9, 10, 50). GST-MiniRT2, -RT, and -TP were expressed in BL21DE3-CodonPlus-RIL cells and purified using the glutathione resin (10, 12). His-MiniRT2, -RT, -TP, and -MBP-TP were expressed in M15-(pREP4) cells and purified using Ni+ affinity resins under native conditions (10). For His-MBP-TP, the eluate from the Ni+ affinity resins was further purified using amylose affinity resins (New England Biolabs). To remove the His and MBP tags, purified His-MBP-TP was digested with the Tev protease. His-MiniRT2, -RT, and -TP proteins were also purified under denaturing conditions and refolded as described previously (55) with minor modifications. Briefly, the inclusion bodies were solubilized in 8 M urea, which was removed stepwise by washing the Ni+ resin in the refolding buffer (50 mM phosphate buffer [pH 8.0], 300 mM NaCl, 5 mM reduced glutathione [GSH], 0.5 mM oxidized GSH, 5 mM β-mercaptoethanol, and 0.1% NP-40) while the proteins were still bound.
In vitro protein priming.
Protein priming reactions were carried out as previously described (27, 50). Briefly, 1 pmol of purified RT or 5 μl of the in vitro translation reaction was mixed with 6 pmol DHBV ε RNA (10) or 1 μg of yeast tRNA, 1× EDTA-free protease inhibitor cocktail (Roche), 0.5 μl of [α-32P]dGTP (3,000 Ci/mmol and 10 mCi/ml) (or another labeled deoxynucleoside triphosphate [dNTP] or NTP, where indicated) per 10-μl reaction in TMnNK (10 mM Tris-HCl [pH 8.0], 1 mM MnCl2, 15 mM NaCl, 20 mM KCl) or, where indicated, TMgNK (same as TMnNK, except containing 2 mM MgCl2 instead of 1 mM MnCl2). NP-40 (0.2% [vol/vol], final concentration) was added to stimulate protein priming by the purified proteins and/or domains (55). For the trans-complementation assay, equimolar amounts (1 pmol each) of the TP and RT domains were used. For the initiation reaction (I) only a single labeled dNTP was used, whereas for the polymerization reaction (P) the other three unlabeled dNTPs (i.e., other than the labeled dNTP) or the indicated dNTP combinations or ddNTP, each at 10 μM, was also included. The protein priming reaction was carried out at 30°C for 2 h. The reaction products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantified by phosphorimaging.
Phosphoamino acid analysis.
Following SDS-PAGE, the gel was divided into two parts. Both parts were fixed with 10% acetic acid and 10% isopropanol mixture for 1 h with two changes. The gel pieces were rinsed in distilled H2O twice. One part was then treated with 3 M KOH at 55°C for 14 h (6, 7), and the other part was mock treated with water at the same temperature. After KOH or water treatment, the gel pieces were treated with 10% acetic acid and 10% isopropanol mixture for 2 h with three to four changes. Subsequently, both gel pieces were rinsed in water for 30 min and dried. The 32P-labeled bands were quantified by phosphorimaging.
Mapping of protein priming sites by thrombin and cyanogen bromide (CNBr) cleavage.
After protein priming, His-MiniRT2 was heat inactivated at 70°C for 10 min and then cooled to room temperature. To the heat-inactivated samples, 5 mM EDTA, 10 μg of E-64 protease inhibitor/ml, and 0.2 U of thrombin (GE Healthcare) per 50 ng of MiniRT2 were added, and the mixture was incubated overnight (ca. 15 h) at room temperature. For mock digestion, samples were incubated with phosphate-buffered saline (pH 7.4) instead of thrombin in parallel. The samples were then subjected to SDS-PAGE. The priming-labeled and thrombin-digested MiniRT2 protein, the TP and RT domains labeled as a result of trans-complementation priming, or the priming-labeled full-length DHBV (WT and Y96F mutant) RT proteins were transferred after SDS-PAGE to nitrocellulose membrane and exposed to X-ray film to localize the 32P-labeled bands. The labeled protein bands were excised from the membrane and incubated with 100 mg of CNBr (Sigma)/ml in 70% (vol/vol) formic acid (Sigma) (28) for 24 h at room temperature in the dark. At the end of digestion, the samples were centrifuged for 5 min, and the CNBr solution (containing 90 to 95% of the 32P signal) was transferred to a new tube and dried using a Speed-Vac to remove formic acid. The dried samples were dissolved in 40 μl of water and dried again. The dried samples were directly dissolved in SDS sample buffer, and the protein fragments were analyzed by 16% Tricine SDS-PAGE (48) and exposed to X-ray film.
RESULTS
Expression and purification of DHBV RT proteins and domains.
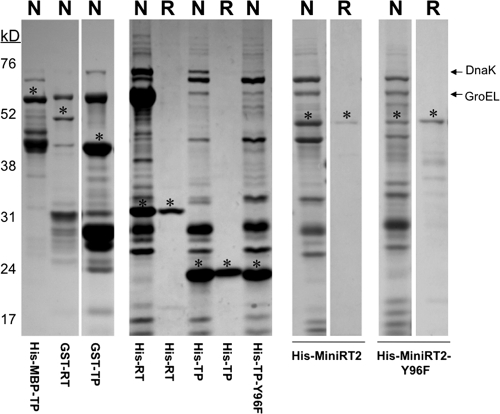
To facilitate protein expression and purification, we produced the DHBV MiniRT2 protein and the individual TP and RT domains as GST or 6×His fusion constructs and purified by glutathione or Ni+ affinity methods. In the case of 6×His-MBP-TP, the protein was purified with Ni+ affinity, followed by the amylose affinity method. Under native conditions MiniRT2 and the TP and RT domains copurified with GroEL and DnaK, two bacterial chaperone proteins known to bind the DHBV and HBV RT, as well as degradation products and other contaminants (Fig. 2, lanes N) (10, 12). In order to exclude the possibilities that the bacterial chaperone proteins and other contaminants might have influenced the results obtained, we also purified the 6×His-tagged constructs from inclusion bodies under denaturing conditions and refolded them. The protein samples that were purified under denaturing condition did not contain the chaperone proteins and were >95% pure (Fig. 2, lanes R). The low-molecular-mass species in the refolded MiniRT2 preparations were mostly degradation products.
Fig. 2.
MiniRT2 proteins and TP and RT domains purified from bacteria. The DHBV MiniRT2 proteins and the TP and RT domains were expressed in E. coli and purified by affinity methods. The His-tagged proteins/domains were purified under either native (N) or refolding (R) conditions. The GST-tagged domains were purified under native (N) conditions. His-MBP-TP was purified by a two-step (nickel followed by amylose affinity) method under native (N) conditions. The purified proteins and domains were analyzed by SDS-PAGE and Coomassie blue staining. The intact (undegraded) proteins or domains are denoted by the asterisks. The copurifying bacterial chaperone proteins, DnaK and GroEL are indicated. The protein molecular mass markers are indicated on the left in kilodaltons.
Both TP and RT domains served as a primer to initiate DNA synthesis.
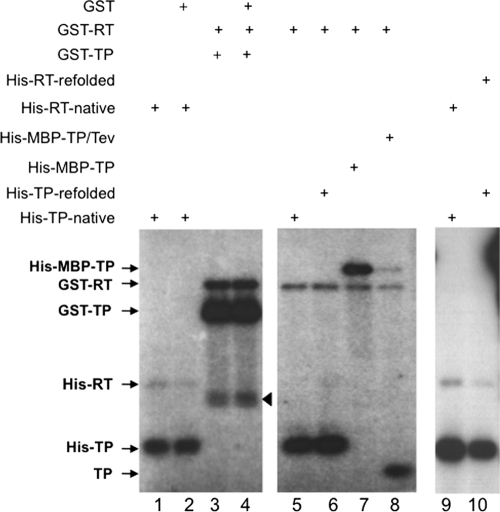
Independent TP and RT domains are known to trans-complement each other to carry out the protein priming reaction using the TP domain as a protein primer to initiate DNA synthesis (1, 25, 27). When the priming reactions were carried out in the presence of Mn2+, which is known to stimulate priming relative to Mg2+ (27), we found, surprisingly, that the RT domain, as well as TP, acted as a protein primer for initiating DNA synthesis (Fig. 3). The ability of the RT domain to serve as a protein primer for initiating DNA synthesis was independent of the nature of the tag on the RT or TP domain. Thus, 6×His-tagged (lanes 1, 2, 9, and 10) or GST-tagged (lanes 3 to 8) RT primed the initiation of DNA synthesis when trans-complemented with 6×His-tagged (lanes 1, 2, 5, 6, 9, and 10) or GST-tagged (lanes 3 and 4), 6×His and MBP double-tagged (lane 7), or untagged (lane 8) TP. Priming at both the TP and RT domains in the trans-complementation was nearly as efficient with refolded domains as with natively purified domains (lanes 9 and 10). On the other hand, unrelated proteins such as GST (lanes 2 and 4), bovine serum albumin, MBP, or rabbit serum proteins (data not shown) failed to serve as a protein primer for DNA synthesis when mixed with TP and RT domains.
Fig. 3.
In vitro protein priming by trans-complementation of purified RT and TP domains. Purified TP and RT domains were mixed together to reconstitute protein priming by trans-complementation, in the presence of [α-32P]dGTP. The domains used were natively purified (lanes 1, 2, 5, and 9) or refolded (lanes 6 and 10) His-tagged, GST-tagged (lanes 3 and 4), His- and MBP-double tagged (lane 7), or untagged (Tev cleaved, lane 8) TP domains and natively purified (lanes 1, 2, and 9) or refolded (lane 10) His-tagged or GST-tagged (lanes 3 to 8) RT domains. GST (1 μg) was also added to the reactions shown in lanes 2 and 4. The 32P-labeled TP and RT domains (arrows) as a result of protein priming were resolved by SDS-PAGE and detected by autoradiography. The arrowhead denotes degradation products from GST-TP and/or GST-RT.
Priming at either the TP or RT domain required both the TP and the RT domains and the ε RNA template.
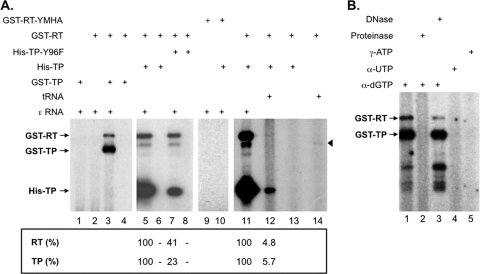
To verify that priming from both the RT and the TP domains was due to authentic priming activity, we assessed whether the trans-complementation reaction required the ε RNA template, the RT domain, and the TP domain, which are known to be required for priming at the authentic Y96 site. As shown in Fig. 4 A, initiation of DNA synthesis from either the TP or the RT domain indeed required all three components; in particular, the RT catalytic site was required since the reaction with the RT domain containing the YMHA mutation did not show any RT or TP priming (lanes 1 to 6, 9, 10, and 13). However, when the RT domain was trans-complemented with the TP-Y96F mutant, which lacked the authentic Y96 priming site, we could still detect the priming signal both on the RT and the mutant TP domain, albeit less efficiently (∼4-fold less priming from TP) than when the WT TP was used (lane 7 versus lane 5), indicating that another site(s) in TP, different from Y96, could also serve to initiate DNA synthesis. Interestingly, the Y96F mutation in TP also decreased the RT domain priming signal (by ∼2-fold) (see also Fig. 5A, Fig. 6C, and Fig. 7). Priming from the mutant Y96F TP still required the ε RNA (Fig. 4, lane 8). These results indicated that protein priming from the RT domain and from the site(s) in TP other than Y96 shared requirements similar to those from the known Y96 TP site. Also, priming from the authentic Y96 TP site seemed to stimulate priming from the RT domain. The putative sites for priming DNA synthesis in the RT domain and in TP (other than Y96) were termed cryptic priming sites since these sites were apparently not used effectively when Mg2+, instead of Mn2+, was present in the in vitro priming reaction (see below), nor are they utilized effectively in vivo (21, 25, 56, 59; see also the Discussion).
Fig. 4.
In vitro trans-complementation priming with WT or mutant TP and RT domains, different RNA templates, and nucleotide substrates. (A) Domains used in the priming reactions include the WT GST-TP (lanes 1, 3, and 4), His-TP (lanes 5, 6, and 10 to 13), His-TP-Y96F mutant (lanes 7 and 8), WT GST-RT (lanes 2 to 8, and 11 to 14), or GST-RT-YMHA mutant (lanes 9 and 10). The ε RNA (lanes 1 to 3, 5, 7, and 9 to 11) or tRNA (lanes 12 and 14) was added as the template for priming. The arrowhead denotes a degradation product from GST-RT. The quantifications of the TP and RT priming signals are indicated at the bottom, with the signals (TP and RT) shown in lane 5 or 11 set as 100% and the signals in lanes 6 to 8 normalized to lane 5 and those in lane 12 normalized to lane 11. (B) The trans-complementation reactions were performed with GST-TP, GST-RT, and the ε RNA. The 32P-labeled nucleotide substrate used was [α-32P]dGTP (lanes 1 to 3), [α-32P]UTP (lane 4), or [γ-32P]ATP (lane 5). The priming reaction products were further digested with proteinase K (lane 2) or DNase I (lane 3) before SDS-PAGE. Radiolabeled domains detected by autoradiography are indicated.
Fig. 5.
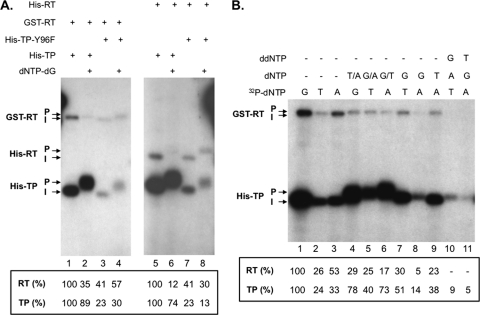
Priming initiation and DNA polymerization from the cryptic priming sites. (A) Trans-complementation priming reactions were performed using GST-RT (lanes 1 to 4) plus His-TP (lanes 1 and 2) or His-TP-Y96F (lanes 3 and 4) or using His-RT (lanes 5 to 8) plus His-TP (lanes 5 and 6) or His-TP-Y96F (lanes 7 and 8). For the initiation (I) reactions, [α-32P]dGTP alone was used (lanes 1, 3, 5, and 7); for the polymerization (P) reactions, the other three unlabeled dNTPs were also included. The quantifications of the TP and RT priming signals are indicated at the bottom, with the signals (TP and RT) shown in lane 1 or lane 5 set as 100% and the signals in lanes 2 to 4 normalized to lane 1 and those in lanes 6 to 8 normalized to lane 5. (B) Trans-complementation priming reactions were performed using GST-RT plus His-TP. For the initiation (I) reactions, the indicated [α-32P]dNTP alone was used (lanes 1 to 3); for the polymerization (P) reactions (lanes 4 to 11), other unlabeled dNTP or dNTPs, or ddNTP as indicated, were also included. The quantifications of the TP and RT priming signals are indicated at the bottom, with the signals (TP and RT) shown in lane 1 set as 100% and the signals in lanes 2 to 11 normalized to lane 1. Note the upward mobility shift of the labeled bands from the polymerization compared to the initiation reactions.
Fig. 6.
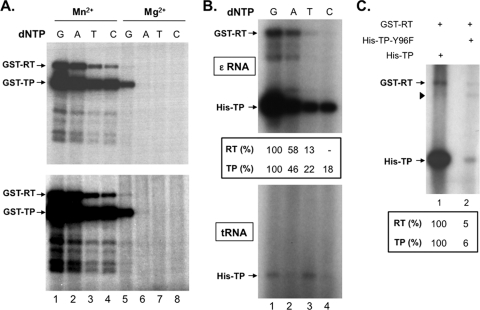
Nucleotide preferences for cryptic site priming initiation and effects of metal ions. (A) Trans-complementation priming reactions were performed using GST-TP, GST-RT, and the ε RNA in the presence of four different 32P-labeled dNTPs (G, lanes 1 and 5; A, lanes 2 and 6; T, lanes 3 and 7; and C, lanes 4 and 8) and either Mn2+ (lanes 1 to 4) or Mg2+ (lanes 5 to 8). The bottom panel is a longer exposure of the same reactions shown at the top. (B) Trans-complementation priming reactions were performed using GST-RT and His-TP in the presence of Mn2+ and four different 32P-labeled nucleotides (G, lane 1; A, lane 2; T, lane 3; and C, lane 4). Either ε RNA (top panel) or tRNA (bottom panel) was used as the template. The quantifications of the TP and RT priming signals are indicated at the bottom of the top image shown, with the signals (TP and RT) shown in lane 1 set as 100% and the signals in lanes 2 to 4 normalized to lane 1. The signals for the bottom image (with tRNA) were too weak to obtain reliable quantifications. (C) Trans-complementation priming reactions were performed using GST-RT, the ε RNA, and either His-TP (lane 1) and or His-TP-Y96F (lane 2) in the presence of Mg2+ and [α-32P]dGTP. The arrowhead denotes a degradation product from GST-RT. Note that 5-fold more proteins and ε RNA were used in the reactions shown in panel C to more clearly visualize cryptic site priming in the presence of Mg2+. The quantifications of the TP and RT priming signals are indicated at the bottom, with the signals (TP and RT) shown in lane 1 set as 100% and the signals in lane 2 normalized to lane 1.
Fig. 7.
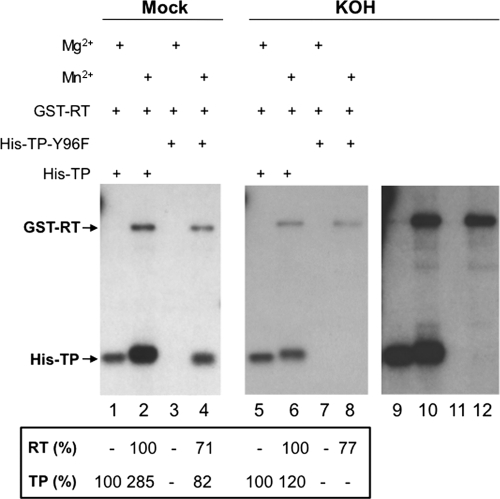
Differentiation of S and/or T versus Y priming sites via alkaline treatment. Trans-complementation priming reactions were carried out using GST-RT (lanes 1 to 8) plus His-TP (lanes 1, 2, 5, and 6) or His-TP-Y96F (lanes 3, 4, 7, and 8) in the presence of ε RNA and either Mg2+ (lanes 1, 3, 5, and 7) or Mn2+ (lanes 2, 4, 6, and 8). The reaction products were resolved by SDS-PAGE. The gel was then cut into two parts; one part was mock treated (lanes 1 to 4), and the other part was treated with 3 M KOH (lanes 5 to 8). Lanes 9 to 12 represent a longer exposure of lanes 5 to 8. The quantifications of the TP and RT priming signals are indicated at the bottom, with the TP signal shown in lane 1 or lane 5 set as 100% and the signals in lanes 2 to 4 normalized to lane 1 and those in lanes 6 to 8 normalized to lane 5. For the RT domain, the signal in lane 2 or lane 6 was set as 100% and the signals in lanes 3 and 4 normalized to lane 2 and those in lanes 7 and 8 normalized to lane 6.
Previously, we showed that MiniRT2, in the presence of Mn2+, was able to use tRNA as a template for protein priming, albeit at a lower efficiency than the authentic ε RNA (27). The full-length DHBV and HBV RT were also shown to synthesize DNA independent of the ε RNA, albeit inefficiently (25, 46, 51). We were thus interested in determining whether tRNA could serve as a template in trans-complementation to support protein priming from either the TP or the RT domain. As shown in Fig. 4A, tRNA indeed served as a template for initiating DNA synthesis on both the TP and RT domains though only at low efficiency (lane 12) (ca. 4% compared to the ε RNA), whereas complete omission of any RNA template eliminated priming at either the TP or RT domain (lanes 4, 6, 8, and 13). Interestingly, even when tRNA was used as the template, priming (from the RT domain) seemed to still require TP (lack of RT domain labeling in lane 14 versus the weak RT labeling in lane 12).
The nature of the TP and RT priming products were further verified by protease and DNase digestion of the reaction products and inclusion of different nucleotide substrate labels in the priming reactions (Fig. 4B). As expected from protein-primed initiation of DNA synthesis, the priming products from both the TP and the RT domains were degraded by protease (lane 2) but not by DNase (lane 3), and [α-32P]dGTP (lane 1; see also Fig. 6 below) but not [α-32P]UTP (lane 4) supported labeling of the TP or RT domain. Neither TP nor RT was labeled by using [γ-32P]ATP (lane 5), indicating that the domains were not labeled by a protein kinase reaction (Fig. 4B).
Cryptic priming sites supported DNA polymerization.
Since the cryptic sites in the TP and RT domains were able to initiate DNA synthesis, we were interested in determining whether these sites could also support subsequent DNA polymerization. We have recently shown that MiniRT2 was able to carry out DNA polymerization, i.e., the extension of the single dGMP attached to the protein to form a short (several nucleotides long) DNA oligomer in the presence of Mn2+ but not in the presence of Mg2+ (27, 54). Therefore, we performed trans-complementation in Fig. 5 A using His-TP and GST-RT (lanes 1 to 4) or His-RT (lanes 5 to 8) in the presence of Mn2+ and [α-32P]dGTP, as well as the other three unlabeled dNTPs. DNA polymerization indeed occurred on the RT domain, as well as the WT or mutant (Y96F) TP domain (lanes 2, 4, 6, and 8). As with the initiation reaction, DNA polymerization from the Y96F TP was also much less efficient compared to the WT TP (lanes 3, 4, 7, and 8), and the Y96F TP mutation again reduced (by ∼2-fold) priming initiation from the RT domain (lanes 3 and 7), as shown above in Fig. 4A. However, the Y96F mutation, compared to the WT TP, apparently increased (by ca. 2- to 3-fold) the polymerization signal from the RT domain (lane 4 versus lane 2; lane 8 versus lane 6) (see Discussion). With the WT TP, and particularly the RT domain, polymerization reactions, the unlabeled dNTPs apparently quenched the priming signals (lane 2 versus lane 1 and lane 6 versus lane 5; also see Fig. 5B below) by competing with the labeled dGTP for attachment to the RT priming site(s) due to the lowered dNTP selectivity with Mn2+ (see Fig. 6 below and Discussion).
Cryptic site priming shared the same nucleotide preference as the authentic site priming.
Previously, we showed that the nucleotide selectivity for initiating protein priming in the presence of Mn2+ was reduced compared to that with Mg2+ (when only dGTP was utilized) but dGTP was still preferred as the first nucleotide attached to the protein, followed by dATP, TTP, and dCTP (27). As shown in Fig. 6 A, the nucleotide selectivity for initiating protein priming from the RT domain was similar to that for priming from the TP domain or MiniRT2 (27). This result indicated that initiation of protein priming from the cryptic sites, as priming from the authentic Y96 site, likely utilized the authentic template, i.e., the internal bulge of the ε RNA, with the first base of the RNA template being a C residue.
To test whether the DNA oligomer synthesized during the polymerization reaction had the d(GTAA) sequence, as predicted from the ε RNA template sequence (UUAC), polymerization reactions were conducted by omitting selected dNTPs or adding the chain terminators ddNTPs (Fig. 5B). First, omitting dCTP did not affect DNA polymerization from either the TP or the RT domain (lanes 4 to 6), a finding consistent with the lack of dC in the DNA oligomer synthesized from either domain. The labeling of TP and RT with radioactive dCTP alone in Fig. 6 was apparently due to the misincorporation of dC opposite the template residue C as discussed above. Again, the unlabeled dNTPs quenched the RT domain polymerization signals (lane 4 versus lane 1 and lane 6 versus lane 3) by competing with the labeled nucleotides due to the lowered dNTP selectivity with Mn2+, as described in Fig. 5A above. Inclusion of dGTP, without dATP, allowed fairly efficient labeling of both TP and RT with radioactive TTP (lane 7) but not radioactive dATP (lane 8), which is consistent with the addition of T, but not dA, immediately following the initiation of dG. The labeled products under these conditions migrated between the initiation products (lanes 1 to 3) and full polymerization products (lanes 4 to 6), a finding consistent with truncated polymerization due to the omission of a dNTP substrate. Omitting dGTP, with inclusion of TTP, largely abolished the polymerization product labeled by radioactive dATP (lane 9), which is consistent with the incorporation of dA only after dG (and T, see above); the labeled TP and RT species under this condition was similar in signal intensity to those with radioactive dATP alone (lane 3) and migrated also like the initiation product, suggesting again misincorporation of dA at the first position rather than DNA polymerization. Furthermore, inclusion of ddGTP blocked labeling by TTP (lane 10) and of ddTTP blocked labeling by dATP (lane 11). Therefore, the results obtained with both omission of dNTPs and inclusion of ddNTPs were consistent with the notion that the DNA polymerization product synthesized, under these conditions, from priming sites in the TP (mostly from the cryptic site or sites, see Fig. 7 below) and RT (exclusively from the cryptic site or sites) domains had the sequence d(GTA(A)), the same as the polymerization product previously shown for the authentic Y96 site (50, 54), and thus was likely templated by the same ε internal bulge sequence (UUAC).
Since tRNA could serve as a (weak) template during trans-complementation (Fig. 4), we were interested in determining the nucleotide specificity of priming carried out in the presence of tRNA. As shown in Fig. 6B, the nucleotide specificity of protein priming in the presence of tRNA was clearly different from that using the ε RNA. When tRNA was used as the template, TTP and, to a slightly lesser extent, dGTP (lanes 1 and 3) were used to initiate priming from TP, whereas dATP or dCTP was used only barely or not at all (lanes 2 and 4). This result, together with the nucleotide selectivity observed above when ε was used as the template, further supported the notion that cryptic site priming, like the authentic Y96 site, was template directed.
Cryptic site priming could occur in the presence of Mg2+, albeit at reduced efficiency compared to that with Mn2+.
All the priming reactions described thus far used Mn2+ as the metal ion to support polymerase activity since Mn2+ was much more efficient in stimulating protein priming than Mg2+ (27). Since the metal ion that the RT protein uses in vivo is presumably Mg2+ rather than Mn2+ (27; see also the Discussion), we were interested in determining whether the cryptic priming sites in the TP and RT domains uncovered above using Mn2+ were able to initiate priming also in the presence of Mg2+. Indeed, the “cryptic” sites on both the TP and the RT domains were used to initiate protein priming in the presence of Mg2+ using the correct nucleotide (dGTP) substrate (Fig. 6A, lane 5, and Fig. 6C), albeit less efficiently compared to Mn2+. The Y96F mutation reduced the TP priming signal dramatically (to 6% of WT), and the RT priming signal was also reduced (by 5-fold) by the Y96F mutation (Fig. 6C), again suggesting that priming at the authentic Y96 site might stimulate priming from the RT domain, as shown in Fig. 4A and 5A.
The cryptic priming sites were S/T in the TP domain and Y, as well as S/T, in the RT domain.
Three different amino acid residues, S, T, and Y, each with a -OH group, are known to serve as primers to initiate DNA or RNA synthesis (38). Although the known priming residue to initiate viral minus-strand DNA synthesis in hepadnaviruses is a Y residue within the TP domain (25, 56, 59), the cryptic priming sites uncovered here could be one or more of the three known priming residues. It has been shown that the phosphate ester linkage in phosphoserine or phosphothreonine is susceptible to base hydrolysis, whereas that in phosphotyrosine is generally resistant to such treatment, although phosphothreonine ester linkages surrounded by proline on either side require more stringent and prolonged base treatment to disrupt (6, 7, 20, 29, 31). We therefore subjected the 32P-labeled RT and TP (WT or Y96F) domains, as a result of protein priming in the presence of either Mn2+ or Mg2+, to 3 M KOH treatment at 55°C for 14 h (which can hydrolyze even the phosphoserine or phosphothreonine linkages surrounded by proline) following resolution of the labeled proteins by SDS-PAGE as a way to determine the sites of linkage between the labeled nucleotide (dGMP) and the TP or RT domain (Fig. 7). Priming signals from S or T priming sites would be eliminated by this treatment, whereas those from Y would remain. Since the WT TP priming signal with Mg2+ was almost exclusively (ca. 94%) from Y96 (lane 3 versus lane 1; also see Fig. 6C above), we normalized the priming signals with or without the 3 M KOH treatment to that from the WT TP with Mg2+, which served as an internal control for the priming signal from Y with or without the KOH treatment. Any signal that was eliminated by the 3 M KOH, after the normalization, was thus attributed to priming at the S and/or T residue(s). The KOH treatment significantly reduced the priming signal from the WT TP with Mn2+, and the remaining signal was now equivalent to the TP priming signal with Mg2+ (lanes 5, 6, 9, and 10). This indicated that the stimulation of TP priming by Mn2+ relative to Mg2+ was mainly derived from priming at a cryptic S/T site(s) without affecting significantly priming at the authentic Y96 site. The priming signal from the Y96F TP mutant in the presence of Mn2+ was completely eliminated (lanes 8 and 12), indicating that the cryptic priming site(s) in the TP domain was a S and/or a T residue(s). However, the same KOH treatment only modestly reduced (∼2-fold) but did not eliminate the priming signal on the RT domain, indicating that Y, and possibly also S/T residues, served as cryptic priming sites in the RT domain (lanes 6, 8, 10, and 12).
The cryptic priming sites were used during cis priming by MiniRT2 and the full-length DHBV RT protein.
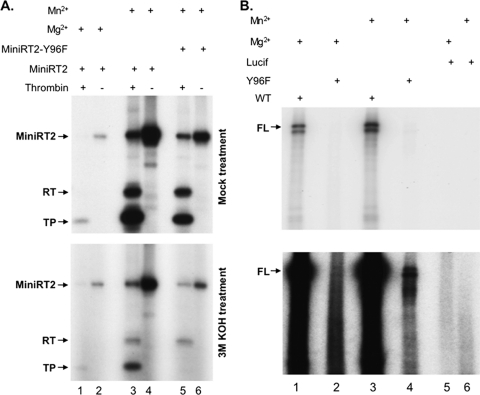
We were next interested in determining whether cryptic priming sites were also utilized during cis priming by MiniRT2 or the full-length RT. As shown in Fig. 8 A, the mutant MiniRT2 with the Y96F mutation was indeed able to carry out protein priming in the presence of Mn2+, although the priming signal was 2- to 3-fold lower than the WT MiniRT2 (top panel, lane 6 versus lane 4). This indicated that MiniRT2 was able to prime DNA synthesis from at least one cryptic site (on either the TP or RT domain or both). To determine whether the cryptic site(s) was located in the TP or RT domain, we took advantage of an engineered thrombin cleavage site (59) between the TP and RT domains in MiniRT2 and digested the primed MiniRT2 with thrombin. The released TP and RT domains, from either the WT or the Y96F MiniRT2, both showed priming signals (top panel, lanes 3 and 5), indicating that cryptic priming sites were located on both the TP and the RT domains, as observed above during trans-complementation. Again, the Y96F mutation decreased the priming signal not only from the TP domain (by 3- to 4-fold) but also from the RT domain (by ∼2-fold) as well (top panel, lane 5 versus lane 3).
Fig. 8.
Cryptic site priming by MiniRT2 and the full-length DHBV RT protein. Protein priming reactions were performed using purified and refolded His-MiniRT2 (A, lanes 1 to 4) or His-MiniRT2-Y96F (A, lanes 5 and 6) or in vitro-translated full-length WT DHBV RT (B, lanes 1 and 3) or the full-length Y96F mutant (B, lanes 2 and 4) or the negative control luciferase (B, Lucif, lanes 5 and 6), in the presence of Mg2+ (A, lanes 1 and 2; B, lanes 1, 2, and 5) or Mn2+ (A, lanes 3 to 6; B, lanes 3, 4, and 6). The MiniRT2 priming reaction products were then cleaved with thrombin to separate the TP and RT domains (A, lanes 1, 3, and 5) or mock digested (A, lanes 2, 4, and 6). Upon resolution via SDS-PAGE, the gel was either mock treated (A, top) or treated with 3 M KOH for 14 h at 55°C (A, bottom) to eliminate the priming signal at S and T, but not Y, sites. The priming reactions from the in vitro translated proteins were simply resolved by SDS-PAGE and detected by autoradiography (B, top, short exposure; bottom, long exposure). The labeled MiniRT2 protein and the TP and RT domains (A) or the full-length RT proteins (B, FL) are indicated.
KOH treatment of the WT MiniRT2 priming signal prior to thrombin cleavage showed that the Mn2+ priming signal was reduced (by ca. 2- to 3-fold) by the treatment, indicating that S and/or T, in addition to Y, were used to prime DNA synthesis by MiniRT2 (Fig. 8A, lane 4, bottom versus top panels). The MiniRT2 signal that remained after the treatment (i.e., priming from Y) was still significantly higher than the priming signal obtained with Mg2+ (Fig. 8A, bottom, lane 4 versus lane 2), suggesting that Mn2+ was able to stimulate priming at the authentic Y96 site in TP by MiniRT2 (i.e., cis priming), in contrast to the trans-complementation reaction where Mn2+ did not stimulate Y96 priming (Fig. 7) or that there was significant priming signal from Y residue(s) in the RT domain that contributed to the overall signal on MiniRT2. KOH treatment of thrombin cleaved MiniRT2 priming products showed that the priming signal remaining (i.e., priming from Y) from the thrombin-released TP was in fact stronger (by 6- to 7-fold) than the TP signal released from the Mg2+ reaction, indicating that Mn2+ was able to stimulate priming at Y residue (probably Y96, see below) in the TP domain in the cis priming reaction. The priming signal from the thrombin-released mutant (Y96F) TP domain from MiniRT2-Y96F was completely eliminated upon KOH treatment, supporting the notion that the only Y residue used for priming in the TP domain of MiniRT2 was the authentic Y96 site and the cryptic site(s) was either S and/or T, as was the case in the trans-complementation reaction shown above. Also, as with the trans-complementation reaction, the RT domain signal released from MiniRT2 or MiniRT2-Y96F was only partially removed after the KOH treatment, suggesting that both Y and S/T residues were used in the RT domain as cryptic priming sites.
Given that cryptic priming sites were used during cis priming by MiniRT2, we sought to determine whether cryptic priming sites might also be utilized during priming by the full-length RT protein. To express a priming active full-length DHBV RT, we used the well-established RRL in vitro translation system (9, 50) to produce both the WT full-length RT and the Y96F mutant version. As shown in Fig. 8B, in the presence of Mn2+, higher (∼2-fold) priming signal was obtained with the WT RT compared to that with Mg2+ (lane 3 versus lane 1) as we reported earlier (27). In addition, the Y96F mutant full-length RT was also able to carry out priming in the presence of Mn2+ (lane 4), albeit at a much reduced level (5% of the WT). A barely detectable priming signal by the Y96F full-length RT protein in the presence of Mg2+ was also present (lane 2). These results thus indicated that the full-length RT was able to use priming sites other than Y96 to initiate DNA synthesis.
Peptide mapping of cryptic priming sites in the TP and RT domains.
To help localize the cryptic priming sites, CNBr cleavage was carried out on primed TP and RT domains from the trans-complementation reaction, as well as the TP and RT domains released from the MiniRT2 cis priming reaction via thrombin digestion. As shown in Fig. 9 B, the WT TP domain primed (either in cis or in trans) in the presence of Mg2+ gave the expected cleavage product corresponding to a peptide containing the authentic Y96 priming site (T1*, 2.7 kDa, residues 75 to 98; lanes 1 and 6). The TP domain primed in the presence of Mn2+ also produced the same 2.7-kDa peptide (but stronger signals as expected) (lanes 3 and 8). With the mutant Y96F TP domain, CNBr cleavage of the Mn2+ priming reaction again produced the 2.7-kDa peptide (lane 5 and 10). These results together suggested that the cryptic priming site in the TP domain was most likely localized on the same CNBr peptide as Y96, i.e., S93, since it is the only S residue within the peptide and there is no T in this peptide. In contrast to this 2.7-kDa TP peptide for which the priming product using Mg2+ served as a precise control, the identification of the RT domain CNBr peptides was less definitive because peptides may not always migrate according to their size alone (see, for example, reference 36) and the covalent nucleotide attachment may also affect their mobility. Nevertheless, the two major and smallest peptides derived from the RT domain, designated R1 and R2 (lanes 2, 4, 7, and 9), were most consistent with the two predicted species containing residues 441 to 480 (predicted molecular mass of 4.5 kDa) and residues 513 to 568 (predicted molecular mass of 6.4 kDa), respectively. These CNBr peptides derived from the RT domain were detected regardless of the TP domain sequence (WT, lanes 2 and 7, or Y96F, lanes 4 and 9) provided in cis (lanes 7 and 9) or in trans (lanes 2 and 4). The identification of two major primed CNBr peptides in the RT domain and the reduction of the RT domain priming signal by KOH treatment (Fig. 7 and 8A) suggested that the RT domain contained multiple cryptic priming sites that included both Y and S/T residues.
Fig. 9.
Peptide mapping of cryptic priming sites. (A) Shown schematically on the top is the domain structure of the DHBV RT protein. The authentic priming site Y96 in the TP domain and the catalytic YMDD motif in the RT domain are highlighted, as are the boundaries of the domains (numerals in amino acid positions on top of the line). The truncated TP and RT domains used for the peptide mapping are shown below, with the truncated domain boundaries shown on top of the lines. The CNBr cleavage sites (in amino acid position of the full-length RT protein) are indicated at the bottom. The −1 number refers to the cleavage site on the vector sequence immediately N-terminal to the TP fusion site. (B) The RT and TP (either WT or Y96F) domains labeled from the trans-complementation reaction (lanes 1 to 5), the labeled domains released from MiniRT2 (WT or Y96F) after thrombin digestion (lanes 6 to 10), or the priming labeled full-length RT protein (WT or Y96F) (lanes 11 to 13) were subjected to CNBr digestion after transfer to nitrocellulose membrane. In each case, the priming reaction in the presence of Mg2+ was used to label the authentic Y96 priming site and help identify the Y96-containing peptide (lanes 1, 6, and 11). The other priming reactions all contained Mn2+ (lanes 2 to 5, 7 to 10, and 12 to 13). The peptide molecualr mass markers are indicated on the right in kilodaltons. T1, the TP peptide 45-98 containing the authentic Y96 site; T1*, the truncated TP peptide 75-98 containing the Y96 site; R1, the RT peptide from residues 441 to 480; R2, the RT peptide from residues 513 to 568.
CNBr cleavage of the full-length RT was also carried out to localize cryptic priming site(s) on the full-length protein. As shown in Fig. 9B, cleavage of the full-length WT RT primed in the presence of Mg2+ produced a 5.8-kDa peptide (T1, peptide 45-98) containing the authentic Y96 priming site as expected (lane 11). Cleavage of the full-length RT primed in the presence of Mn2+ produced the same 5.8-kDa band (lane 12) as obtained in the presence of Mg2+, with the expected stronger signal. Cleavage of the Y96F mutant full-length RT primed in the presence of Mn2+ also produced the 5.8-kDa peptide (with a much reduced signal, as expected [lane 13]), suggesting the existence of one or more cryptic priming sites within the TP peptide 45-98 and consistent with the above identification of S93 as a cryptic priming site using the truncated TP domain (T1*). The two RT domain peptides identified above using trans-complementation and MiniRT2 could not be identified using the full-length RT protein, suggesting that the putative cryptic priming sites in the RT domain might not be utilized in the context of the full-length protein. The other higher-molecular-mass species derived from the full-length RT protein, as well as from the individual TP and RT domains in Fig. 9B (mostly minor), did not appear to match predicted peptides based on complete CNBr digestion, and their identification was complicated by the possibility of incomplete CNBr cleavage and aberrant mobility of the peptides, as discussed above.
DISCUSSION
Hepadnaviruses are thought to utilize a specific Y residue in the TP domain of their RT protein to prime viral minus-strand DNA synthesis. Here, we have found that for the DHBV RT, additional priming sites, other than the authentic Y96 site, in both the TP and RT domains, could also be utilized to prime DNA synthesis in vitro. These so-called cryptic priming sites initiated DNA synthesis in the trans-complementation priming reaction, where separately purified TP and RT domains were combined together, and in the normal cis priming reaction whereby the truncated MiniRT2 and the full-length RT carried out priming when the TP and RT domains were covalently linked in cis. Priming from either the TP or the RT cryptic sites, as from Y96, required the RT domain (in particular, the RT active site), the TP domain, and the authentic viral RNA template ε. These cryptic priming sites not only allowed the covalent attachment of a nucleotide to the protein (nucleotidylation or the initiation step of protein priming) but could also support subsequent DNA polymerization to generate DNA oligomers as in authentic protein priming. One cryptic S priming site was localized to the TP domain, and both S (T) and Y residues appeared to be used as priming sites in the RT domain.
The identification of both Y and S/T residues as protein priming sites suggests that the RT active site is rather flexible in accommodating the amino acid residues used to prime DNA synthesis. Indeed, both Y and S are known to initiate DNA or RNA synthesis by other DNA or RNA polymerases, mostly from viruses (38), and even highly related viruses can use different amino acids (either Y or S) as substrates for the nucleotidylylation (i.e., the covalent attachment of a nucleotide to an amino acid residue in the protein) reaction (30). The fact that cryptic site priming was much more prominent in the presence of Mn2+ than in the presence of Mg2+ is consistent with our recent observation that Mn2+ may induce an RT conformation that is more catalytically active but less stringent in its selection of the template and nucleotide substrates (27). The results reported here further suggest that this conformation is also less stringent in its selection of primers. Further, we found here that Mn2+ did not significantly enhance priming at the authentic Y96 site in the trans-complementation reaction despite its dramatic stimulatory effect on cryptic site priming. However, Mn2+ did enhance Y96 priming, as well as cryptic site priming, by MiniRT2 (i.e., priming in cis), suggesting that priming site selection may be somewhat different between priming in trans and in cis. It would be difficult to precisely identify all cryptic priming sites, and the structural basis for selecting any particular priming site, whether authentic or cryptic, remains elusive. However, the fact that one likely TP priming site (S93) is close to the authentic Y96 site suggests that some local structural characteristics may be involved. That the full-length RT protein did not appear to use the same cryptic priming sites in the RT domain as those used by the truncated MiniRT2 (or the truncated TP and RT domains in trans) suggests that sequences distant from the priming sites and/or the overall RT protein structure can also influence cryptic priming site selection.
Although its relevance for viral replication in vivo remains uncertain (see below), priming from the cryptic sites may nevertheless provide opportunities to dissect aspects of protein priming that would otherwise be difficult to study. For example, it is well established that TP, in addition to the RT domain, is required for ε binding (11, 12, 33, 51) and harbors the primer Y residue (25, 56, 59). We have shown here that TP was still required for priming at the cryptic sites in the RT domain, when it did not serve as a primer and when tRNA, instead of ε RNA, served as the template. Assuming TP was not required for the RT domain to bind tRNA as a (presumably nonspecific or generic) template, these results suggest that TP may also serve as an allosteric activator of the RT catalytic activity or as a structural component of the catalytic activity itself.
Another intriguing finding was that TP priming was significantly reduced (by ∼4-fold) by the Y96F mutation in the trans-complementation reaction with Mn2+, which was much more than expected from just the loss of Y96 priming, since the latter contributed only ca. 30% of the total TP priming signal under the same conditions. Furthermore, the Y96F mutation also decreased priming from the RT domain. This result suggests that Y96 and, presumably, priming at this authentic site could activate the RT catalytically so that it could more efficiently catalyze priming at the cryptic sites. Also, the Y96F mutation apparently enhanced the DNA polymerization signal from the cryptic RT domain priming sites. As we reported earlier, the nucleotide selectivity during protein priming is reduced in the presence of Mn2+ compared to Mg2+ (27). In the case of the WT TP and RT domains, the unlabeled dNTPs added to the polymerization reaction could quench the labeling of the priming signal by competing, due to the low dNTP selectivity, with the labeled dGTP for attachment to the RT priming site(s). The Y96F mutation might have somehow increased the dNTP selectivity in the presence of Mn2+ and, hence, the apparently enhanced polymerization signal. Interestingly, the same mutation did not show such an enhancing effect on polymerization on the cryptic TP priming sites: it dramatically reduced the TP polymerization signal just as it did the TP initiation signal, suggesting that priming from the cryptic TP and RT sites may not be entirely the same, a notion also supported by the observation that the TP priming signal was consistently stronger than the RT priming signal, whether the authentic Y96 priming site was present or not.
The significance of protein priming at the cryptic sites identified here for either viral replication or pathogenesis is not yet clear. As alluded to earlier, it is currently uncertain whether cryptic site priming is relevant for viral DNA synthesis. Elimination of the authentic Y96 priming site (including the same Y96F mutation used here) in DHBV or the analogous Y63 site in HBV decreased viral DNA synthesis in cells to undetectable levels (21, 56), suggesting that cryptic site priming, if it occurs in vivo, is unable to sustain viral replication. This could be due to the low efficiency of cryptic site priming but could also be due to the lack of the stimulating effect in RT catalytic activity induced by priming at the authentic site, as discussed above. It is also possible that priming at the cryptic sites does not allow the subsequent template switch from ε to the 3′ end of pgRNA to continue minus-strand DNA synthesis, which normally occurs following protein priming (45, 49). The possibility that authentic site priming depends on priming first at the cryptic sites, which would be unprecedented for protein-primed DNA or RNA synthesis, has been excluded in the case of one cryptic site in the RT domain (see the accompanying report by Beck and Nassal [1a]). This possibility formally remains in the case of the cryptic site(s) in the TP domain and possibly other site(s) in the RT domain. However, the very low priming efficiency of the cryptic sites in the presence Mg2+, the presumptive metal ion used in cells by RT, makes this unlikely. We also note that TP S93 is highly conserved among hepadnaviruses, but it can also be a residue other than S, T, or Y. On the other hand, low-level DNA synthesis independent of the authentic priming site, by the full-length DHBV RT expressed in vitro (51) and HBV RT packaged within nucleocapsids (21), has been reported before and interpreted as primer independent, similar to RNA polymerases. The discovery of cryptic priming sites suggests that the DNA synthesis activity detected in such cases might actually have been due to protein-primed DNA synthesis using the cryptic sites. Interestingly, elimination of the normal priming site (Y3) in the poliovirus protein primer, VPg, which is normally used to prime viral RNA-dependent RNA synthesis, did not completely prevent viral replication; a very low level of RNA replication persisted, primed by a nearby cryptic priming site (T4) on VPg, allowing for the selection of revertant viruses (3). By analogy, the DHBV or HBV mutant with the authentic primer Y eliminated may sustain very low levels of replication. In a spreading infection system such as the liver in vivo, such low levels of replication could lead to the selection of revertants as well.
The identification of cryptic priming sites on RT also suggests that RT may be able to use other proteins, viral or host, as primers to initiate DNA synthesis. As mentioned above, the RNA-dependent RNA polymerase of poliovirus also uses the protein primer VPg, which is not covalently linked to the polymerase, to initiate viral RNA synthesis (32). Similar to what we found for the DHBV RT protein, the poliovirus RNA polymerase could also use priming site(s) on the polymerase protein itself to initiate RNA synthesis, i.e., self uridylylation or the covalent attachment of UMP to the RNA polymerase (37). Although uridylylation of the poliovirus RNA polymerase was found to be dispensable for viral replication, the viral RNA polymerase was found to uridylylate also cellular proteins. Furthermore, certain cellular proteins are modified by covalent attachment of nucleotides (nucleotidylation) during normal cellular metabolism and signaling (19). It is thus conceivable that the hepadnavirus RT may also modify cellular proteins with covalent nucleotide or DNA oligomer attachment. We have tested a number of different proteins in our in vitro priming assay and found that none tested thus far, including GST, bovine serum albumin, MBP, and rabbit serum proteins, could serve as a protein primer for the DHBV RT in vitro. One prerequisite for a cellular protein to be subjected to such a modification would likely be a close interaction with RT so that it can gain access to the RT active site. The RT protein is known to interact with a number of cellular proteins, including molecular chaperone proteins (13, 15), the helicase DDX3 (52), and the translation factor eIF4E (22). The RT protein is also thought to be cytotoxic when overexpressed and has been reported to affect cellular functions such as interferon signaling and gene expression (2, 8, 14, 53). Perhaps, the formation of nucleotide abducts of cellular proteins, i.e., RT-mediated protein priming from these cellular proteins, can contribute to the cellular effects of the RT protein and thus could play a role in cytotoxicity and viral pathogenesis, especially if substantial amounts of the RT protein are unpackaged (58).
ACKNOWLEDGMENTS
We thank Christina Adams and Laurie Mentzer for excellent technical assistance.
This study was supported by a Public Health Service grant from the National Institutes of Health.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Beck J., Nassal M. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a. Beck J., Nassal M. 2011. A Tyr residue in the reverse transcriptase domain can mimic the protein-priming Tyr residue in the terminal protein domain of a hepadnavirus P protein. J. Virol. 85:7742–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cao F., Tavis J. E. 2006. Suppression of mRNA accumulation by the duck hepatitis B virus reverse transcriptase. Virology 350:475–483 [DOI] [PubMed] [Google Scholar]
- 3. Cao X., Wimmer E. 1995. Intragenomic complementation of a 3AB mutant in dicistronic polioviruses. Virology 209:315–326 [DOI] [PubMed] [Google Scholar]
- 4. Chang L. J., Hirsch R. C., Ganem D., Varmus H. E. 1990. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J. Virol. 64:5553–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y., Robinson W. S., Marion P. L. 1994. Selected mutations of the duck hepatitis B virus P gene RNase H domain affect both RNA packaging and priming of minus-strand DNA synthesis. J. Virol. 68:5232–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper J. A., Sefton B. M., Hunter T. 1983. Detection and quantification of phosphotyrosine in proteins. Methods Enzymol. 99:387–402 [DOI] [PubMed] [Google Scholar]
- 7. Duclos B., Marcandier S., Cozzone A. J. 1991. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 201:10–21 [DOI] [PubMed] [Google Scholar]
- 8. Foster G. R., et al. 1991. Expression of the terminal protein region of hepatitis B virus inhibits cellular responses to interferons alpha and gamma and double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 88:2888–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu J. 2004. Studying DHBV polymerase by in vitro transcription and translation. Methods Mol. Med. 95:259–269 [DOI] [PubMed] [Google Scholar]
- 10. Hu J., Anselmo D. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74:11447–11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu J., Boyer M. 2006. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80:2141–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J., Flores D., Toft D., Wang X., Nguyen D. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 78:13122–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu J., Lin L. 2009. RNA-protein interactions in hepadnavirus reverse transcription. Front. Biosci. 14:1606–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu J., Seeger C. 1996. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 275:195–208 [DOI] [PubMed] [Google Scholar]
- 15. Hu J., Seeger C. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 93:1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J., Seeger C. 1997. RNA signals that control DNA replication in hepadnaviruses. Semin. Virol. 8:205–211 [Google Scholar]
- 17. Hu J., Toft D., Anselmo D., Wang X. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu J., Toft D. O., Seeger C. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Itzen A., Blankenfeldt W., Goody R. S. 2011. Adenylylation: renaissance of a forgotten posttranslational modification. Trends Biochem. Sci. 36:221–228 [DOI] [PubMed] [Google Scholar]
- 20. Kemp B. E. 1980. Relative alkali stability of some peptide o-phosphoserine and o-phosphothreonine esters. FEBS Lett. 110:308–312 [DOI] [PubMed] [Google Scholar]
- 21. Kim H. Y., et al. 2004. Oligomer synthesis by priming deficient polymerase in hepatitis B virus core particle. Virology 322:22–30 [DOI] [PubMed] [Google Scholar]
- 22. Kim S., Wang H., Ryu W. S. 2010. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J. Virol. 84:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanford R. E., Kim Y. H., Lee H., Notvall L., Beames B. 1999. Mapping of the hepatitis B virus reverse transcriptase TP and RT domains by transcomplementation for nucleotide priming and by protein-protein interaction. J. Virol. 73:1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lanford R. E., Notvall L., Beames B. 1995. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J. Virol. 69:4431–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lanford R. E., Notvall L., Lee H., Beames B. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin L., Hu J. 2008. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J. Virol. 82:2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin L., Wan F., Hu J. 2008. Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions. J. Virol. 82:5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo K. X., Hurley T. R., Sefton B. M. 1991. Cyanogen bromide cleavage and proteolytic peptide mapping of proteins immobilized to membranes. Methods Enzymol. 201:149–152 [DOI] [PubMed] [Google Scholar]
- 29. Martensen T. M. 1982. Phosphotyrosine in proteins. Stability and quantification. J. Biol. Chem. 257:9648–9652 [PubMed] [Google Scholar]
- 30. Olspert A., Peil L., Hebrard E., Fargette D., Truve E. 2011. Protein-RNA linkage and posttranslational modifications of two sobemovirus VPgs. J. Gen. Virol. 92:445–452 [DOI] [PubMed] [Google Scholar]
- 31. Parker P. J., Embi N., Caudwell F. B., Cohen P. 1982. Glycogen synthase from rabbit skeletal muscle. State of phosphorylation of the seven phosphoserine residues in vivo in the presence and absence of adrenaline. Eur. J. Biochem. 124:47–55 [DOI] [PubMed] [Google Scholar]
- 32. Paul A. V., van Boom J. H., Filippov D., Wimmer E. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280–284 [DOI] [PubMed] [Google Scholar]
- 33. Pollack J. R., Ganem D. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin H., et al. 2008. Construction of a series of vectors for high throughput cloning and expression screening of membrane proteins from Mycobacterium tuberculosis. BMC Biotechnol. 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Radziwill G., Tucker W., Schaller H. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reiser K. M., Last J. A. 1983. Anomalous electrophoretic behavior of a cyanogen bromide peptide from type III collagen. Connect. Tissue Res. 12:1–16 [DOI] [PubMed] [Google Scholar]
- 37. Richards O. C., et al. 2006. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J. Virol. 80:7405–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salas M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39–71 [DOI] [PubMed] [Google Scholar]
- 39. Seeger C., Mason W. S. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seeger C., Zoulim F., Mason W. S. 2007. Hepadnaviruses, p. 2977–3030In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott/The Williams & Wilkins Co., Philadelphia, PA [Google Scholar]
- 41. Stahl M., Beck J., Nassal M. 2007. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J. Virol. 81:13354–13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stahl M., Retzlaff M., Nassal M., Beck J. 2007. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. 35:6124–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Summers J., Mason W. S. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415 [DOI] [PubMed] [Google Scholar]
- 44. Tavis J. E., Ganem D. 1996. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tavis J. E., Ganem D. 1995. RNA sequences controlling the initiation and transfer of duck hepatitis B virus minus-strand DNA. J. Virol. 69:4283–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tavis J. E., Massey B., Gong Y. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 72:5789–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tavis J. E., Perri S., Ganem D. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walker J. M., Judd R. 2002. SDS-polyacrylamide gel electrophoresis of peptides, p. 73–79 In The protein protocols handbook. Humana Press, Inc., Totowa, NJ [Google Scholar]
- 49. Wang G. H., Seeger C. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang G. H., Seeger C. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663–670 [DOI] [PubMed] [Google Scholar]
- 51. Wang G. H., Zoulim F., Leber E. H., Kitson J., Seeger C. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68:8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang H., Kim S., Ryu W. S. 2009. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 83:5815–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H., Ryu W. S. 2010. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PLoS Pathog. 6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X., Hu J. 2002. Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses. J. Virol. 76:5857–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang X., Qian X., Guo H. C., Hu J. 2003. Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase. J. Virol. 77:4471–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weber M., et al. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiong Y., Eickbush T. H. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yao E., Gong Y., Chen N., Tavis J. E. 2000. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J. Virol. 74:8648–8657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zoulim F., Seeger C. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]