Abstract
Cholesterol plays an essential role in the life cycle of several enveloped viruses. Many of these viruses manipulate host cholesterol metabolism to facilitate their replication. HIV-1 infection of CD4+ T cells activates the sterol regulatory element-binding protein 2 (SREBP2) transcriptional program, which includes genes involved in cholesterol homeostasis. However, the role of SREBP2-dependent transcription in HIV-1 biology has not been fully examined. Here, we identify TFII-I, a gene critical for HIV-1 transcription in activated T cells, as a novel SREBP2 target gene. We found TFII-I expression increased after HIV-1 infection or activation of human primary CD4+ T cells. We show that inhibition of SREBP2 activity reduced TFII-I induction in response to these stimuli. More importantly, small interfering RNA (siRNA)-mediated gene silencing of either SREBP2 or TFII-I significantly reduced HIV-1 production in CD4+ T cells. We also found that TFII-I potentiates Tat-dependent viral gene expression, consistent with a role at the level of HIV-1 transcription. Collectively, our results demonstrate for the first time that HIV-1 transcription in T cells is linked to cholesterol homeostasis through control of TFII-I expression by SREBP2.
INTRODUCTION
A number of studies indicate that cholesterol plays an important role in HIV-1 replication (31). The virus appears to bud from cholesterol-rich membrane domains in the plasma membrane, and cholesterol in the membrane of both virus and target cells is required for fusion and entry (13, 22). The dependence of HIV-1 on cholesterol is further substantiated by the observation that HIV-1 Nef profoundly impacts the cholesterol content of virus particles through effects on cellular cholesterol homeostasis (30, 34). Among other actions mediated by Nef, it inhibits cellular cholesterol efflux by down-modulating ABCA1 (20).
Optimal replication of HIV-1 in primary T cells requires cell activation (7). Both T cell activation and HIV-1 infection are known to stimulate transcription of the full spectrum of genes required for cholesterol biosynthesis (2, 30). Interestingly, an oxysterol (25-hydroxycholesterol) known to suppress the induction of the cholesterol biosynthetic pathway by blocking the activation of sterol response element binding protein 2 (SREBP2), the master controller of cholesterol biosynthesis, also inhibits HIV-1 replication (17, 23). Several studies have shown that modulating the cholesterol content of cells through inhibitors of synthesis or enhancement of efflux can profoundly affect HIV-1 infection and replication (13, 18). Additionally, del Real et al. found that lovastatin, a potent inhibitor of the rate-limiting enzyme for cholesterol biosynthesis, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoAR), inhibited HIV-1 infection at the level of virus entry (5). This effect appeared to be related to inhibition of geranylgeranylation and effects on Rho activation. Based on other studies, the effects observed could also be attributed in part to the effect of lovastatin on cellular cholesterol content and lipid rafts. In the del Real study an unexpected observation was the finding that while lovastatin reduced HIV-1 entry, HIV-1 LTR transcription was increased. The latter observation suggested that HIV-1 LTR transcription was potentially linked to cholesterol homeostasis.
TFII-I is a multifunctional transcription factor that plays an important role in transcription of HIV-1 genes in activated T cells (14). Here we demonstrate for the first time that the TFII-I gene contains functional sterol response elements and that expression of the TFII-I protein is controlled by SREBP2. Inhibition of SREBP2 activity by small interfering RNA (siRNA) reduced expression of TFII-I and restricted HIV-1 replication. Furthermore, activation of T cells results in increased expression of TFII-I that could be suppressed by inhibiting activation of the SREBP2 pathway. Our results demonstrate that HIV-1 transcription in T cells is linked to cholesterol homeostasis through control of TFII-I expression by SREBP2.
MATERIALS AND METHODS
Cell culture.
The 293T cell line was maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone), 100 μg/ml streptomycin, and 100 U/ml penicillin. Jurkat cells, U1 cells, and peripheral blood mononuclear cells (PBMC) were propagated in RPMI supplemented with 10% FBS (HyClone), 100 μg/ml streptomycin, and 100 U/ml penicillin. PBMC were obtained by Ficoll-Paque (Amersham) density centrifugation from several healthy blood donors (New York Blood Center). Total CD4+ T and naïve CD4+ T cells were isolated from PBMC by negative selection using magnetic bead sorting (Miltenyi Biotec) and were cultured in RPMI supplemented with 10% FCS (HyClone), 100 μg/ml streptomycin, and 100 U/ml penicillin. CD4+ T cells treated with phytohemagglutinin (PHA) were stimulated in the presence of interleukin-2 (IL-2) (20 U/ml; Roche Applied Science).
Flow cytometry.
Flow cytometry was performed by first fixing cells with phosphate-buffered saline (PBS) containing 2% paraformaldehyde and then permeabilizing cells with PBS buffer containing 1% BSA, 5% normal goat serum (NGS), and 0.2% saponin, followed by staining with appropriate antibodies (Abs). Cell viability was determined by using either aqua or far red Live/Dead fixable dead cell stains (Invitrogen). Infection of CD4+ T cells, activation status, and TFII-I expression levels were determined by flow cytometry using fluorescein isothiocyanate (FITC)-conjugated anti-p24 (Coulter), FITC-conjugated anti-CD25 (BD Biosciences), and anti-TFII-I monoclonal antibodies (MAbs) (Santa Cruz), respectively. The primary MAbs were detected with Alexa Fluor 647-conjugated goat anti-mouse polyclonal antibody. Virus production was measured by anti-p24gag enzyme-linked immunosorbent assay (ELISA). Cells were stained with polyclonal SREBP1 (Santa Cruz) and SREBP2 (Cayman Chemical) Abs and detected with R-phycoerythrin-conjugated goat F(ab′)2 anti-rabbit IgG from Invitrogen to determine SREBP expression levels. Compensation was performed with single-stained cells. Data were collected on FACSCalibur or FACSAria (BD Biosciences) instruments and analyzed with FACSDiva (BD Biosciences) or Flowjo (Tree Star) software. For immunofluorescence staining, cells were fixed with 2% paraformaldehyde in PBS and permeabilized with PBS containing 5% NGS, 1% BSA, and 0.25% Triton X-100. Cells were than stained with indicated primary and secondary Abs.
Transfections.
Transient transfections were performed using Lipofectamine 2000 (Invitrogen). For overexpression of HA-SREBP2, 293T cells (5 × 106) were seeded in 10-cm dishes, transfected at 50 to 80% confluence, and harvested 48 h after transfection. The siRNAs were nucleofected into primary CD4+ T cells using an AMAXA Nucleofector apparatus (program U-14 or V-024; 3 μg of siRNA per 1 × 107 cells).
Luciferase assays.
293T cells (2 × 104 cells/well) were seeded into 96-well plates 16 to 18 h before transfection. Cells were transfected with 100 ng of luciferase reporter plasmid, 10 ng of the control Renilla luciferase expression plasmid pRL-CMV (Promega), and various amounts of SREBP1 or SREBP2 expression plasmids. Cells were then disrupted with passive lysis buffer (Promega), and luciferase activity was measured using a reporter assay kit (Promega) 36 h after transfection. Transfections were performed in triplicate, and results were normalized to the Renilla luciferase signal. Each experiment was performed at least three times.
Immunoblotting.
Cells were washed with ice-cold PBS and then lysed on ice for 30 min with buffer containing 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 7.5), and protease inhibitors (Roche). Protein lysates were clarified by centrifugation at 15,200 × g at 4°C for 20 min. Protein lysates were fractionated by SDS-PAGE, transferred to nitrocellulose, and detected by immunoblotting with an anti-TFII-I MAb at a 1:1,000 dilution, anti-SREBP2 MAb at a 2-μg/ml dilution, or anti-NF-κB MAb at 1:1,000. Immunoreactive proteins were detected using horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG and an ECL assay (Amersham).
Nuclear extracts.
Preparation of nuclear extract from 293T cells overexpressing HA-SREBP2 (5a) was done as described by Dignam et al. Briefly, cells were washed with ice-cold PBS, pelleted by centrifugation at 1,000 × g at 4°C for 10 min, resuspended in hypotonic buffer A (10 mM Tris [pH 8.0], 1.5 mM MgCl2, 1 mM KCl, and 0.5 mM dithiothreitol [DTT]), and incubated on ice for 10 min. Cells were homogenized with 15 strokes of a tight-fitting Dounce homogenizer, and nuclei were pelleted. Nuclear protein was extracted by adding NaCl to a final concentration of 300 mM, and the cells were incubated on ice for an additional 30 min. Nuclear lysates were clarified by centrifugation at 25,000 × g for 30 min at 4°C. Aliquots of nuclear lysates were stored at −80°C.
EMSAs.
Assays were performed using biotin-labeled DNA obtained from Invitrogen Inc. Oligonucleotides were annealed by heating to 95°C for 5 min and slowly cooling to room temperature prior to use in electrophoretic mobility shift assays (EMSAs). The LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL) was used for EMSAs. Briefly, 2 μg of nuclear extracts was incubated in binding buffer containing 50 mM Tris (pH 8.0), 1 mM DTT, and 5 mg of poly(dI-dC) along with 10 fmol of duplexed biotin-labeled probe in a total volume of 20 μl. Competition assays were carried out by adding the indicated molar excess of unlabeled duplexed probes immediately prior to the addition of labeled probe. Supershift assays were performed by preincubating anti-HA polyclonal Ab (Abcam) with nuclear extracts in binding buffer on ice for 3 h prior to the addition of duplexed oligonucleotides. The reaction mixtures were incubated for 30 min at room temperature. DNA-protein complexes were resolved on 4% polyacrylamide gel (37.5:1, acrylamide:bis) in 0.5× Tris-borate-EDTA buffer for 1 h at 100 V and 4°C. The complexes were transferred to nylon membranes (Zetaprobe) at 380 mA for 30 min and detected according to the manufacturer's protocol.
Real-time PCR.
RNA was extracted from CD4+ T cells by using the total RNA purification kit (NORGEN Biotek Corporation, Thorold, ON, Canada). Residual contaminating DNA was removed from RNA by using the Turbo DNA-free kit (Ambion). In brief, 0.1 vol of 10× Turbo DNase buffer and 1 μl (2 U) of Turbo DNase were added to 50 μl of RNA and incubated for 30 min at 37°C. Then, 5 μl of DNase inactivation reagent was added to the reaction mixture described above, mixed well, and incubated at room temperature for 2 min with occasional mixing. After centrifugation at 10,000 × g for 1.5 min, the supernatant was transferred to a fresh tube.
Real-time reverse transcription (RT)-PCR analysis.
The RNA samples (500 ng each) were subjected to real-time PCR by using the qScript cDNA synthesis kit (Quanta Biosciences) for a reverse transcription reaction and PCR using iQ SYBR green Supermix (Bio-Rad). Reaction mixtures contained 200 nM primer pairs and 2 to 4 μl of 6-fold-diluted cDNA in a final volume of 25 μl. After initial incubation of 95°C for 10 min, 40-cycle amplifications were performed at 95°C for 15 s and 60°C for 45 s. The primer pairs used were as follows: HIV-RNA, 5′-TGTGTGCCCGTCTGTTGTGT-3′ and 5′-GAGTCCTGCGTCGAGAGAGC-3′; GAPDH, 5′-GAAGGTGAAGGTCGGAGT-3′ and 5′-GAAGATGGTGATGGGATTTC-3′.
DNA constructs and siRNAs.
The luciferase reporter plasmid for the low-density lipoprotein receptor (LDLR) promoter was described previously (29). Expression constructs pCMV5-HA-SREBP1 and pCMV5-HA-SREBP2 were generously provided by J. Shyy (University of California, Riverside, CA). To generate the luciferase reporter plasmid containing the TFII-I promoter upstream regulatory element sequence (pTFII-I-USE), the distal region (−1921 to −1593) of the gene was amplified by PCR using human genomic DNA as a template and cloned into the pGL3 promoter vector (Promega). The sequence of reporter constructs was verified by DNA sequence analysis.
The pZsGreen1-DR-LTR reporter construct was generated by first subcloning a 719-bp XhoI-HindIII LAI 3′ long terminal repeat (LTR) fragment derived from pBlue3′LTR-luc (AIDS Research and Reference Reagent Program) into XhoI/HindIII-digested pBluescript KS(+). The LTR fragment was liberated from this construct by digesting with XhoI and BamHI and subcloned into XhoI/BamHI-digested pZsGreen1-DR (Clontech). The pCIS2-TFII-I expression construct was previously described and kindly provided by Sonye Danoff and Stephen Desiderio, Johns Hopkins School of Medicine. The HIV-1 X4 envelope expression construct was generously provided by Robert Siliciano, Johns Hopkins School of Medicine. The reagents pYF-JRCSF and pNL4-3ΔE-EGFP were obtained through the AIDS Research and Reference Reagent Program.
The following siRNAs were obtained (Ambion identification number): SREBP2 (s29), TFII-I (289315), and negative-control siRNA (nontargeting siRNA).
Virus preparation and viral infection.
Virus stocks were prepared by transfecting 293T cells with HIV-1 plasmid DNA using Lipofectamine 2000 reagent (Invitrogen). X4 HIV-1 envelope-pseudotyped virus was produced as previously reported (35). Culture supernatants were collected 48 h after transfection, centrifuged at 1,000 × g to remove cell debris, filtered through a 0.45-μm-pore-size filter, and concentrated by ultracentrifugation at 100,000 × g through a cushion of 20% sucrose in PBS. The pelleted virus was resuspended in RPMI with 10% FBS, aliquoted, and stored at −80°C. The viral titer was measured by anti-p24gag ELISA. Forty-eight hours after nucleofection with siRNA, CD4+ T cells were infected with virus (200 ng of p24 per 5 × 105 cells) by spinoculation (1,200 × g, 2 h), followed by a 2-h incubation at 37°C, and were washed three times to remove input virus. Twenty-four hours after infection, cells were stimulated with the indicated amount of plate-bound anti-CD3 and anti-CD28 (1 μg/ml) MAbs (BD Biosciences) for 72 h to induce virus expression. Cells were treated with 25-hydroxycholesterol (Sigma, 0.5 μg/ml) to determine its effect on infected cells as indicated.
RESULTS
TFII-I promoter contains a functional sterol response element.
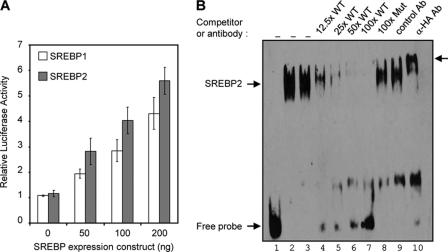
To explore the possibility that TFII-I expression is linked to cellular cholesterol homeostasis, we first examined potential regulatory sequences in the upstream promoter region of the TFII-I gene by utilizing the Database of Transcriptional Start Sites (DBTSS) (http://dbtss.hgc.jp/). Sequence analysis of the TFII-I promoter region revealed the presence of four putative sterol regulatory elements (SRE) upstream from the transcription start site (Table 1; see Fig. S1 in the supplemental material). Three putative SREs were located within a stretch of 330 bp upstream from the transcription start site (see Fig. S1). We hypothesized that the presence of multiple SREs in the TFII-I promoter would result in SREBP-dependent transcription of TFII-I. To test this hypothesis, we first cloned the distal region of the human TFII-I promoter, containing three putative SREs upstream of a minimal simian virus 40 (SV40) promoter in the PGL3-promoter vector. Next, we cotransfected 293T cells with the reporter constructs, along with plasmids encoding constitutively active forms of both SREBP1a and SREBP2. We found that cotransfection of either SREBP1a or SREBP2 with the reporter constructs containing the TFII-I promoter-derived sequence induced transcription in a DNA concentration-dependent manner (Fig. 1A). In comparison, the negative-control PGL3 vector demonstrated minimal responsiveness to SREBP overexpression (data not shown). This result indicated that active SREBPs induced transcription through TFII-I-derived SREs and indicated that at least one of the SRE motifs was functional and capable of mediating SREBP-dependent transcription.
Table 1.
Alignment of TFII-I promoter SRE sequences with the prototypical SRE sequence derived from the LDLR promoter
| SRE | Sequencea |
|---|---|
| TFII-I SRE1 | CTGACCCCTC |
| TFII-I SRE2 | ACGACTCCAC |
| TFII-I SRE3 | ATCGCGCCAC |
| LDLR | ATCACCCCAC |
Conserved nucleotide residues are indicated in bold.
Fig. 1.
TFII-I expression is regulated by SREBPs. (A) TFII-I-USE-Luc reporter was cotransfected into 293T cells with the indicated amount of constitutively active SREBP-1 (pCMV5-HA-SREBP1) or SREBP2 (pCMV5-HA-SREBP2). Data represented are the mean results from triplicate samples, and the error bars are standard deviations of the means. (B) Annealed biotinylated LDLR SRE probe (lane 2) or probes containing TFII-I SRE3 were incubated without (lane 1) or with (lanes 3 to 10) nuclear extract isolated from 293T cells overexpressing an active HA-tagged-SREBP2. Specificity of binding was confirmed by performing competition experiments. The indicated molar excess of an unlabeled LDLR SRE probe was added immediately before adding biotinylated TFII-I SRE3 probe. The identity of the shifted species was determined by preincubating nuclear extracts with control IgG (lane 9) or anti-HA polyclonal Ab (lane 10). Supershifted species are indicated by the arrow on the right. The mobilities of the SREBP2 shifted species and free probe are also indicated.
SREBP2 binds to a TFII-I promoter SRE.
Results of the reporter assays suggest that SREBPs bind to SRE motifs within the TFII-I promoter. To confirm this, we performed EMSAs using biotinylated double-stranded oligonucleotide probes containing SRE sequences from the TFII-I promoter or the prototype SRE from the human LDLR promoter as a positive control for SREBP binding (Fig. 1B and Table 1). For DNA binding studies, we used nuclear extract purified from 293T cells overexpressing HA-tagged SREBP2. SREBP2 in the nuclear extracts bound specifically to the labeled LDLR-SRE probe as expected but also bound equally well to labeled TFII-I SRE3 probe (Fig. 1B, lanes 2 and 3). Binding to the SRE3 probe was specifically inhibited in a dose-dependent manner by competition with unlabeled wild-type (WT) LDLR-SRE probe (Fig. 1B, lanes 4 to 7) but not with a 100-fold molar excess of unlabeled LDLR-SREm probe containing a mutation that blocks SREBP binding (Fig. 1B, lane 8). The binding specificity of HA-tagged SREBP2 and the identity of the shifted species were confirmed by the supershift induced by preincubation of nuclear extract with anti-HA polyclonal Ab but not a control polyclonal Ab (Fig. 1B, lane 9 and 10). We observed minimal or no SREBP2 binding to probes containing only SRE1 or SRE2 (data not shown). These results strongly support the notion that TFII-I is an SREBP2 target gene.
TFII-I expression is enhanced by CD4+ T cell activation.
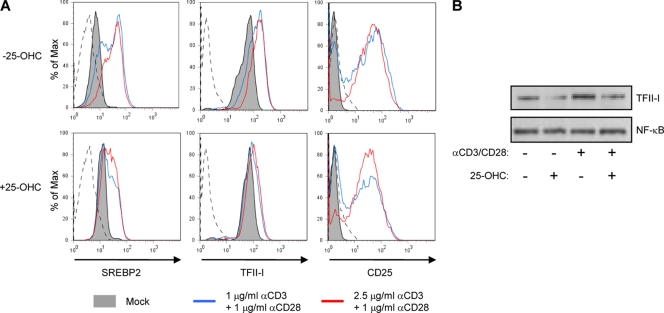
T cell activation increases de novo cholesterol biosynthesis and cholesterol-dependent target gene transcription via SREBP2. Our identification of TFII-I as an SREBP-dependent gene allows us to predict that TFII-I expression should also be upregulated by T cell activation. To address this prediction, we stimulated purified CD4+ T cells with plate-immobilized anti-CD3 and soluble anti-CD28 Abs for 72 h and assessed the level of TFII-I, SREBP2 and CD25 by 3-color flow cytometry (Fig. 2A). As with other SREBP2-dependent genes, TFII-I protein expression was enhanced in activated T cells. T cell activation resulted in nearly a 3-fold increase in the TFII-I mean fluorescence intensity (MFI) when treated cells were compared to controls (Fig. 2, top row). In titration experiments, the level of enhancement of TFII-I expression was found to be dependent on the concentration of anti-CD3/CD28 MAbs. SREBP2, itself the product of a sterol-responsive gene, was also induced significantly in anti-CD3/CD28-stimulated T cells. This is consistent with an earlier report showing that cholesterol synthesis and SREBP2-dependent transcription increase in activated T cells (2). The activation status of the stimulated T cells was confirmed by increased expression of CD25 after anti-CD3/CD28 treatment.
Fig. 2.
T cell activation induces TFII-I expression. (A) Primary CD4+ T cells were left untreated (gray filled) or were stimulated with plate-bound anti-CD3 (1 mg/ml [blue line] or 2.5 mg/ml [red line]) and soluble anti-CD28 in the presence (bottom) or absence (top) of 0.5 mg/ml of 25-OHC for 72 h. Representative histograms of flow cytometry analysis indicate levels of endogenous SREBP2 and TFII-I. The activation status of cells was confirmed by staining with anti-CD25. (B) Cells treated as described in the legend to panel A were lysed and analyzed for TFII-I and SREBP2 expression by Western blot analysis. Normalized protein loading was confirmed by immunoblotting with an anti-NF-κB MAb.
Oxysterols can suppress SREBP2 target gene transcription in response to T cell activation (2). Therefore, we assessed the impact of treatment with 25-hydroxycholesterol (25-OHC) on TFII-I induction by endogenous SREBP2 in anti-CD3/CD28-activated CD4+ T cells (Fig. 2, bottom row). We observed that 25-OHC abrogated induction of TFII-I in CD4+ T cells but did not inhibit activation as assessed by the induction of CD25+ expression (Fig. 2A). We performed Western blot analysis to confirm that changes in TFII-I MFI in flow cytometry experiments correlated with changes in levels of protein expression and were not due to nonspecific effects of treatments on cells that influence Ab binding. Consistent with flow cytometry, Western blots showed that anti-CD3/CD28 stimulation enhances TFII-I expression (Fig. 2B, lane 3). As observed in earlier experiments, inhibition of SREBP2 with 25-OHC suppresses induction of TFII-I and SREBP2 by T cell stimulation (Fig. 2B, lane 4). There was no effect on NF-κB p65 subunit protein expression in cells treated with 25-OHC (Fig. 2B). These results indicate that TFII-I upregulation in activated T cells is SREBP2 dependent.
Endogenous SREBPs regulate TFII-I expression.
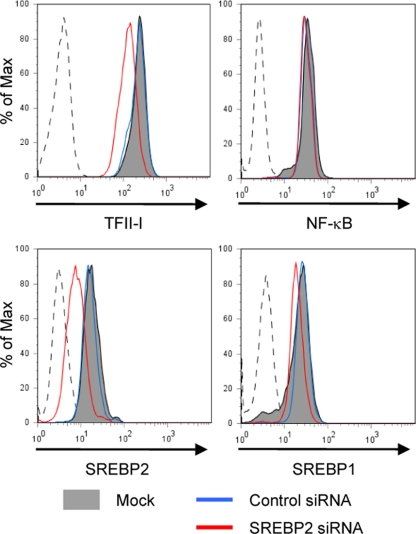
SREBP2 is the master regulator of cholesterol biosynthetic genes and inhibition of SREBP2 activity in cells suppresses cholesterol synthesis. Thus, a reduction in cellular cholesterol levels may mediate transcriptional changes observed upon SREBP2 depletion. In order to analyze SREBP2 function independent of its effect on cholesterol biosynthesis, we performed siRNA-mediated silencing of SREBP2 in U1 cells. Because they are derived from the U937 cell line, devoid of 3-ketosteroid reductase activity, these cells are cholesterol auxotrophs and depend on an exogenous cholesterol source for growth (3). Further, U1 cells are easily transfected and so are also amenable to genetic studies by siRNA-mediated silencing. To determine whether endogenous SREBP2 controls TFII-I expression, U1 cells were transfected with siRNAs directed against SREBP2. We assessed relative levels of expression of TFII-I in siRNA-transfected cells by performing flow cytometry after intracellular staining for TFII-I. As predicted, U1 cells transfected with SREBP2-specific siRNA expressed lower levels of SREBP2 protein than mock-transfected or nonsilencing control siRNA-transfected cells (Fig. 3). The specificity of siRNA-mediated silencing of SREBP2 is confirmed by minimal changes in TFII-I protein expression in mock-treated and control siRNA-transfected cells (Fig. 3). Further, expression of NF-κB p65 subunit, another factor that can modulate HIV-1 transcription, was not affected by SREBP2 siRNA. The viability of U1 cells transfected with SREBP2 siRNA was indistinguishable from that of mock- or control siRNA-transfected cells (data not shown). These data showed that down-modulating SREBP2 expression resulted in decreased expression of TFII-I independent of SREBP2's role in cholesterol regulation.
Fig. 3.
SREBPs modulate levels of TFII-I in vivo. Flow cytometric analysis of intracellular TFII-I levels in U1 cells 48 h after transfection with indicated siRNAs. Staining with TFII-I MAb demonstrated that cells transfected with SREBP2 siRNA (red line) had lower levels of TFII-I than those that were transfected with nonsilencing siRNA (blue line) or mock transfected (gray filled).
HIV-1 infection induces TFII-I expression in CD4+ T cells.
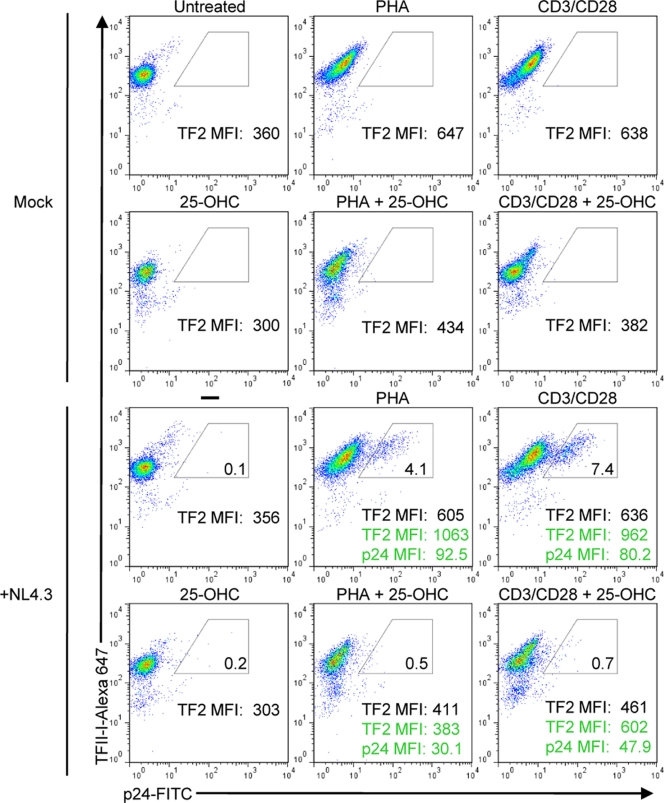
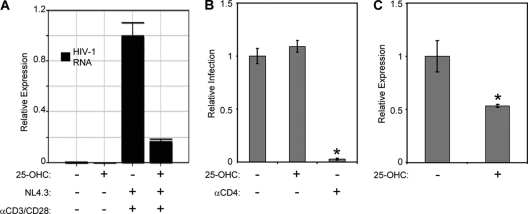
Cholesterol synthesis and SREBP2-dependent genes are induced upon HIV-1 infection of CD4+ T cells (30). Thus, we predicted that HIV-1 infection would induce TFII-I expression. To test this hypothesis, we first infected naïve primary CD4+ T cells with HIV-1 NL4-3 and then stimulated cells with either PHA and IL-2 or anti-CD3 and anti-CD28 antibodies for 3 days to induce HIV-1 expression before performing two-color flow cytometry to assess intracellular Gag and TFII-I protein levels. A recent study showed that these mitogens activate signaling pathways that are responsible for inducing HIV-1 transcription (4). We observed a nearly 2-fold increase in TFII-I MFI in stimulated cells (TFII-I + PHA MFI, 647; TFII-I + CD3/CD28 MFI, 638) compared to that in unstimulated cells (TFII-I MFI = 360). A recent study showed that these mitogens activate signaling pathways that are responsible for inducing HIV-1 transcription (4). As predicted, TFII-I expression was further enhanced in gated HIV-infected cells (TFII-I + PHA MFI, 1,063; TFII-I + CD3/CD28 MFI, 962) compared to uninfected cells (Fig. 4). We examined the role of SREBP2 in mediating the induction of TFII-I by using 25-OHC to inhibit endogenous SREBP2 activity in HIV-1-infected T cells. T cells were first pretreated with 25-OHC and then infected with HIV-1 NL4-3 and cultured in medium containing 25-OHC. As observed with T cell activation-dependent increase in TFII-I expression, 25-OHC treatment of CD4+ T cells abrogated induction of TFII-I in HIV-1-infected cells stimulated with PHA (TFII-I MFI, 1,063 versus 383) or anti-CD3/CD28 (TFII-I MFI, 962 versus 602) (Fig. 4). Further, 25-OHC treatment inhibited HIV-1 infection in PHA-stimulated cells by more than 87% and anti-CD3/CD28-stimulated CD4+ T cells by 91%, compared to results for untreated cells. Interestingly, 25-OHC treatment also reduced HIV-1 gag levels in infected PHA-stimulated (p24 MFI, 92.5 versus 30.1) and CD3/CD28-stimulated (p24 MFI, 80.2 versus 47.9) cells. This observation supports the notion that SREBP2-dependent transcription plays a role in HIV-1 replication. These levels of inhibition of HIV infection correlate well with the degree of suppression of TFII-I expression. To confirm that 25-OHC treatment of infected cells resulted in reduced HIV-1 replication, we measured HIV-1 RNA by performing real-time PCR on total RNA isolated from HIV-1 NL4-3-infected CD4+ T cells that were first infected and then stimulated with anti-CD3/CD28 Ab in the presence or absence of 25-OHC. As shown in Fig. 5A, 25-OHC treatment reduced HIV-1 RNA in the infected cultures by more than 80%. Since some oxysterols have been demonstrated to disrupt lipid raft structures, which are critical for viral fusion to target cells, 25-OHC could also potentially inhibit this early step of HIV-1 replication (16). To investigate this possibility, we pretreated anti-CD3/CD28-activated primary CD4+ T cells with 25-OHC, prior to infecting with HIV-1 X4 envelope-pseudotyped GFP reporter virus, which is capable of only a single round of infection. As a positive control for inhibition of infection, we used an anti-CD4 antibody that blocks gp120 binding (11). After 2 days of culture in the presence of inhibitors, we assessed relative levels of infection by flow cytometry and observed near-complete inhibition of infection with anti-CD4 Ab, but 25-OHC had no effect on infection (percentage of cells expressing GFP) (Fig. 5B). However, we detected a nearly 50% reduction of LTR-GFP expression (GFP MFI) in infected CD4+ T cells in the presence of 25-OHC (Fig. 5C). These results confirmed that 25-OHC inhibited replication at a postentry step of the viral life cycle.
Fig. 4.
T cell activation and HIV-1 infection of CD4+ T cells induce TFII-I expression. Flow cytometry analysis of intracellular Gag and TFII-I expression in HIV-1 NL4-3-infected primary naïve CD4+ T cells. Cells were first mock infected (top two rows) or infected with HIV-1 (bottom two rows) and 24 h later were stimulated with PHA (5 μg/ml) and IL-2 (20U/ml) or plate-bound anti-CD3 (2.5 μg/ml) and anti-CD28 (2.5 μg/ml) for 72 h in the presence (second and fourth row) or absence (first and third row) of 0.5 μg/ml of 25-OHC. Infected cells were detected by staining with a FITC-conjugated Ab against HIV-1 gag. TFII-I expression was assessed by indirect staining using an anti-TFII-I MAb and goat Alexa 647-conjugated anti-mouse antibody. HIV-1-infected cells are represented by the gated populations, and the percentage of Gag-positive cells within each gate is indicated. The MFI values of TFII-I for the total cell population (black) and gated infected cell populations (green) are indicated.
Fig. 5.
Inhibition of SREBP-2 activity with 25-OHC suppresses HIV-1 transcription. (A) Cells were first infected with NL4-3 and 24 h later were stimulated with plate-bound anti-CD3 Ab (2.5 mg/ml), anti-CD28 Ab (1.0 mg/ml), and 0.5 mg/ml of 25-OHC for 72 h, as indicated. Real-time PCR analysis showing that treatment of HIV-1-infected primary T cells with 25-OHC reduces the level of HIV-1 transcripts. Expression of GAPDH mRNA was used to normalize data. (B) Activated primary CD4+ T cells were pretreated for 2 h with 0.5 mg/ml of 25-OHC or 1 mg/ml anti-CD4 antibody before infection with HIV-1 GFP reporter virus. Levels of infection were assessed by flow cytometry after 48 h. (C) Cells were treated as described in the legend to panel B, and the GFP MFI of the GFP+ populations was determined by flow cytometry 72 h after infection to calculate relative expression. Error bars are the standard deviations of the triplicate experiments. Statistically significant differences are denoted by a single asterisk (*, P < 0.05) and were calculated using Student's t test.
TFII-I potentiates Tat-dependent HIV-1 LTR transcription.
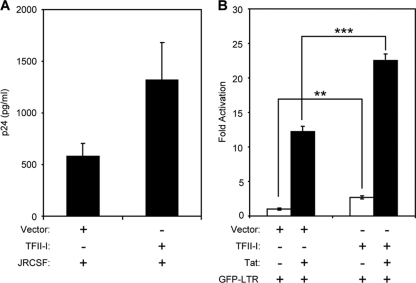
T cell activation increases SREBP2 activity, TFII-I expression levels, and HIV-1 LTR transcription. To determine if increasing the level of TFII-I is sufficient to enhance HIV-1 production, we cotransfected an HIV-1 provirus construct with a TFII-I expression vector into Jurkat T cells. Overexpression of TFII-I increased virus production by more than 2.5-fold, as assessed by p24 ELISA of cell culture supernatants (Fig. 6A).
Fig. 6.
TFII-I potentiates Tat-dependent HIV-1 LTR transcription. (A) Jurkat T cells were nucleofected with 1.5 μg of control empty vector (pCIS2) or TFII-I expression constructs along with 1.5 μg of provirus construct, pYK-JRCSF. Amounts of HIV-1 in supernatant were assessed by p24 ELISA 36 h after nucleofection. Error bars are the standard deviations of results of the triplicate experiments. (B) Primary CD4+ T cells were transfected with HIV-LTR GFP reporter plasmid (1.5 μg) and expression plasmids for Tat (0.5 μg) and/or TFII-I (3 μg). Cells were harvested for flow cytometry 16 h after transfection. The total amount of DNA in transfections was balanced by using empty vector. Values are means obtained from triplicate experiments, as measured by flow cytometry in units of mean fluorescence intensity (MFI) of GFP+ cells normalized to samples transfected with HIV-LTR GFP and empty vector. Error bars are the standard deviations of results of the triplicate experiments. Statistically significant differences are denoted by two (**, P < 0.01) or three (***, P < 0.001) asterisks and were calculated using Student's t test. The experiment shown is representative of experiments performed with cells obtained from three different donors.
TFII-I has been reported to bind and transactivate the HIV-1 LTR (14). To confirm that TFII-I enhances HIV-1 replication at the level of transcription, resting primary CD4+ T cells were cotransfected with a TFII-I expression vector and reporter construct containing the HIV-1 LTR driving expression of enhanced green fluorescent protein (EGFP) (Fig. 6B). As expected, TFII-I overexpression significantly increased LTR promoter activity (∼3-fold) in resting T cells. This showed that TFII-I expression levels could influence early virus transcription in resting T cells (28, 33). Furthermore, induction of TFII-I during T cell activation may enhance HIV-1 expression (32). Tat is the only HIV-1 protein directly involved in its own gene transcription and is essential for efficient transcriptional elongation of viral transcripts (10). To examine whether TFII-I can also influence Tat-activated LTR transcription, we cotransfected suboptimal levels of Tat (0.5 μg) with TFII-I expression vector and HIV-1 LTR GFP reporter construct. Tat stimulated HIV-1 LTR expression more than 12-fold, but when it was cotransfected with TFII-I, transcription was increased by more than 22-fold, showing that TFII-I potentiates Tat-dependent transcription. These results indicate that TFII-I may play a role in the dramatic enhancement of HIV-1 transcription mediated by Tat in activated T cells.
SREBP2/TFII-I pathway is critical for HIV-1 replication in primary CD4+ T cells.
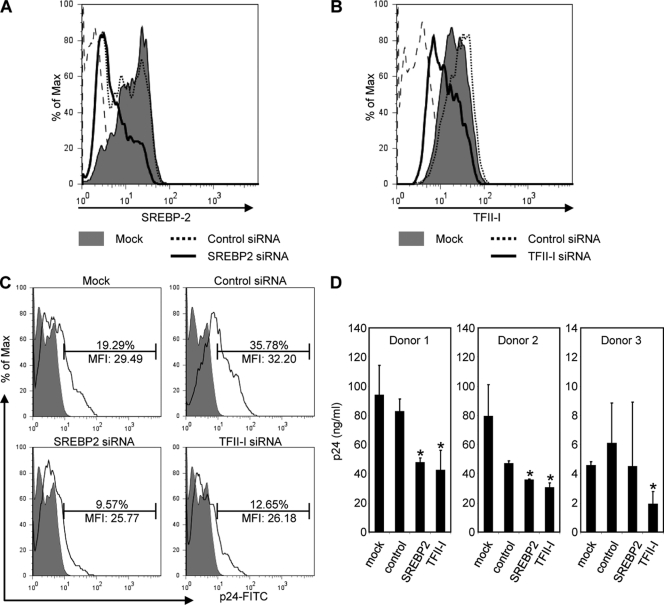
To demonstrate that SREBP2 and TFII-I are involved in HIV-1 replication in primary T cells, we knocked down expression of SREBP2 and TFII-I in primary CD4+ T cells using siRNA-mediated silencing. After siRNA transfection, CD4+ T cells were first infected with HIV-1 and then stimulated with anti-CD3/anti-CD28 Ab for 60 h to induce virus replication. As seen in Fig. 7A and B, transfection with siRNAs for SREBP2 and TFII-I resulted in a significant reduction in the corresponding proteins compared to that for mock- or control siRfNA-transfected cells, as assessed by flow cytometry. Using a Live/Dead fixable dead cell stain kit (Molecular Probes) for viability staining, we found that cells transfected with SREBP2 and TFII-I siRNA had relative cell viabilities of 98.1% and 98.6%, respectively, compared to control siRNA-transfected cells (data not shown). We found that SREBP2 and TFII-I siRNA-transfected cultures had significantly reduced levels of HIV-1 infection, as determined by intracellular p24 staining, compared to mock- or control siRNA-transfected cultures (9.57 and 12.65% compared to 19.29 and 35.78%, respectively) (Fig. 7C).
Fig. 7.
SREBP2-TFII-I pathway is essential for HIV-1 replication. (A and B) Histograms represent flow cytometry analysis of SREBP2 and TFII-I expression in primary CD4+ T cells mock transfected (gray filled) or transfected with nonsilencing (dotted line), SREBP2 (black thick line) (A), or TFII-I (black thick line) (B) siRNAs. Histogram plots indicate that cells transfected with TFII-I or SREBP2 siRNAs express lower levels of TFII-I than those that were transfected with nonsilencing siRNA or mock transfected. (C) Cells from panels A and B were analyzed by flow cytometry to assess levels of infection by staining with FITC-conjugated anti-gag Ab. The percentage of infected cells and the MFI of FITC anti-gag are indicted for each gated population. (D) The HIV-1 produced by cells in panel C was measured by p24 ELISA. Error bars are the standard deviations of results of triplicate experiments. Statistically significant differences are denoted by a single asterisk (*, P < 0.05) and were calculated using Student's t test. The experiments shown were performed with cells obtained from three different donors.
To correlate the flow cytometry data to HIV-1 production, we assessed HIV-1 in the culture supernatants of siRNA-treated cells by p24 ELISA (Fig. 7D). SREBP2 and TFII-I siRNA-transfected cells showed up to a 42% and 49% reduction in virus production, respectively, compared to control siRNA-transfected cells of three different donors. These results are consistent with the effect of 25-OHC inhibition of SREBP2 activity on HIV-1 replication in CD4+ T cells. The data confirm that the SREBP2-TFII-I pathway is critical for HIV-1 replication and that SREBP2 is the controlling factor for induction of TFII-I-dependent virus replication.
DISCUSSION
TFII-I is a widely expressed transcription factor that plays an important role in cellular calcium homeostasis and the transcriptional regulation of genes involved in cell cycle progression, endoplasmic reticulum stress response, angiogenesis, and embryonic development (9, 25). TFII-I is also an important component of a transcription factor complex that regulates LTR-driven transcription of HIV genes (26). The potential importance of the SREBP2/TFII-I pathway in HIV transcription is reflected by the fact that the binding site for TFII-I in the HIV LTR is one of the most highly conserved regions in the HIV genome (14). It is well established that HIV infection results in the induction of the cholesterol biosynthetic pathway via SREBP2-mediated activation of sterol response genes. Our results reveal for the first time that TFII-I is a sterol response gene controlled, at least in part, by SREBP2. Induction of this pathway by HIV-1 also increases the production of this critical transcription factor, thereby driving expression of viral proteins.
HIV-infected individuals develop a series of metabolic disorders, including lipodystrophy and dyslipidemia (12). Microarray and proteomic studies have revealed that HIV-1 infection of CD4+ T cells results in enhanced transcription and production of gene products involved in cholesterol metabolism and transport. These changes appear to result from HIV-1 activation of SREBP2, the master regulator of this cholesterol-dependent transcription. The clinical implications of our findings are highlighted by the recent observation that HIV RNA levels in antiretroviral-naïve patients correlate with serum cholesterol levels (6). Specifically, serum LDL cholesterol levels were inversely correlated with viral loads. These data fit a model in which HIV-1 transcription is modulated by LDL cholesterol, since uptake of LDL cholesterol results in feedback inhibition of SREBP-dependent transcription and lower levels of SREBP2-dependent proteins such as TFII-I. In agreement with this, Negredo et al. found that HIV-infected patients with low levels of PBMC-associated cholesterol had increased viral loads when treated with a statin (atorvastatin) during highly active antiretroviral therapy (HAART) interruption (21). Under conditions in which cholesterol levels are low and cholesterol biosynthesis is blocked, SREBP2 will be activated, resulting in enhanced expression of SREBP2-dependent proteins, including TFII-I. This in turn would result in enhanced HIV-1 transcription and HIV-1 replication. Hence, our observations provide a molecular explanation for the impact of cholesterol status on HIV-1 replication in vivo.
We have shown that pharmacological or genetic inhibition of SREBP2 inhibits both TFII-I expression and HIV-1 replication. We have also demonstrated that activation of the SREBP2/TFII-I pathway provides a link between T cell activation and HIV-1 transcription. The SREBP2/TFII-I pathway also provides a direct link between cellular cholesterol homeostasis and HIV-1 transcription. Recently, Robinzon et al. reported that measles virus infection of a neuronal cell line inhibits cholesterol biosynthesis and transcription of genes that are SREBP2 dependent (24). Interestingly, a prospective study of HIV-infected pediatric patients demonstrated a significant reduction of plasma HIV RNA during acute measles infection (19). It would be of interest to determine if measles virus infection modulates cholesterol metabolism of CD4+ T cells, since our finding that TFII-I expression is controlled by SREBP2 suggests that coinfection with measles virus may result in suppression of HIV-1 replication by inhibition of TFII-I-dependent HIV-1 transcription.
The results of recent studies on the pathogenesis of AIDS suggest that our findings may have important implications beyond HIV transcription. Sedaghat et al. have reported that activated CD4+ T cells from untreated HIV-positive individuals have dysregulated transcriptional programs enriched in type I interferon-responsive genes (27). The IRF-7 transcription factor is essential for type I interferon-dependent transcription (8), and a recent study has provided evidence that TFII-I may control IRF-7 expression (1). Our data show that T cell activation and HIV-1 infection induce high expression of TFII-I. Thus, our findings and those of the previous studies suggest that TFII-I may be involved in the aberrant interferon response in activated HIV-1-infected CD4+ T cells. Divergent type I interferon signaling mediated via IRF-7 is thought to distinguish pathogenic and nonpathogenic AIDS virus infections (15). It will be of interest to determine if TFII-I expression levels in activated CD4+ T cells from HIV-1-infected patients correlate with IRF-7 expression. Such a finding would suggest that the SREBP/TFII-I pathway may play a role in HIV-1 pathogenesis.
In this paper, we demonstrate for the first time that the TFII-I gene contains a functional sterol response element and is a sterol-responsive gene. We further demonstrate that expression of TFII-I, a factor critical in the regulation of HIV transcription, is regulated by SREBP2, thereby linking HIV-1 transcription to cellular cholesterol homeostasis. Our results have important implications for understanding HIV pathogenesis and may be relevant to the wide person-to-person variation seen in HIV-1 replication in vivo. Further insights into the role of the SREBP2/TFII-I pathway in HIV replication may provide new therapeutic approaches for inhibiting the virus.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NIH: U54 RR019192 (J.E.K.H., principal investigator), R01 HD040772 (J.E.K.H., principal investigator), and G12 RR003032, and Vanderbilt Institute for Clinical and Translational Research Scholar Award 5 KL2 RR024977-03 (H.E.T.).
We thank Jared D. Elzey for editorial assistance and members of the Center for AIDS Health Disparities Research for helpful suggestions. We also thank members of the Kalams Laboratory (Vanderbilt) for technical help with flow cytometry. Flow cytometry acquisition and analysis were performed at the NIH-funded Vanderbilt-Meharry CFAR Immunopathogenesis Core Facility (P30-AI-54999).
Author contributions: H.E.T. conceived the study, performed the flow cytometry experiments and molecular and biochemical studies, and drafted the manuscript. M.E.L. assisted in flow cytometry experiments. A.K.K. and W.P. performed real-time PCR experiments. J.E.K.H. supervised the project and together with W.P. was involved in writing the manuscript. All authors approved the final copy.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Ashworth T., Roy A. L. 2009. Phase specific functions of the transcription factor TFII-I during cell cycle. Cell Cycle 8:596–605 [DOI] [PubMed] [Google Scholar]
- 2. Bensinger S. J., et al. 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134:97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Billheimer J. T., Chamoun D., Esfahani M. 1987. Defective 3-ketosteroid reductase activity in a human monocyte-like cell line. J. Lipid Res. 28:704–709 [PubMed] [Google Scholar]
- 4. Bosque A., Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. del Real G., et al. 2004. Statins inhibit HIV-1 infection by down-regulating Rho activity. J. Exp. Med. 200:541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a. Dignam J. D., Lebovitz R. M., Roeder R. G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El-Sadr W. M., et al. 2005. Effects of HIV disease on lipid, glucose and insulin levels: results from a large antiretroviral-naive cohort. HIV Med. 6:114–121 [DOI] [PubMed] [Google Scholar]
- 7. Folks T., et al. 1986. Susceptibility of normal human lymphocytes to infection with HTLV-III/LAV. J. Immunol. 136:4049–4053 [PubMed] [Google Scholar]
- 8. Honda K., et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 9. Jackson T. A., Taylor H. E., Sharma D., Desiderio S., Danoff S. K. 2005. Vascular endothelial growth factor receptor-2: counter-regulation by the transcription factors, TFII-I and TFII-IRD1. J. Biol. Chem. 280:29856–29863 [DOI] [PubMed] [Google Scholar]
- 10. Jones K. A., Peterlin B. M. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717–743 [DOI] [PubMed] [Google Scholar]
- 11. Khatua A. K., Taylor H. E., Hildreth J. E., Popik W. 2009. Exosomes packaging APOBEC3G confer human immunodeficiency virus resistance to recipient cells. J. Virol. 83:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koutkia P., Grinspoon S. 2004. HIV-associated lipodystrophy: pathogenesis, prognosis, treatment, and controversies. Annu. Rev. Med. 55:303–317 [DOI] [PubMed] [Google Scholar]
- 13. Liao Z., Cimakasky L. M., Hampton R., Nguyen D. H., Hildreth J. E. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retroviruses 17:1009–1019 [DOI] [PubMed] [Google Scholar]
- 14. Malcolm T., Kam J., Pour P. S., Sadowski I. 2008. Specific interaction of TFII-I with an upstream element on the HIV-1 LTR regulates induction of latent provirus. FEBS Lett. 582:3903–3908 [DOI] [PubMed] [Google Scholar]
- 15. Mandl J. N., et al. 2008. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 14:1077–1087 [DOI] [PubMed] [Google Scholar]
- 16. Massey J. B., Pownall H. J. 2006. Structures of biologically active oxysterols determine their differential effects on phospholipid membranes. Biochemistry 45:10747–10758 [DOI] [PubMed] [Google Scholar]
- 17. Moog C., Aubertin A. M., Kirn A., Luu B. 1998. Oxysterols, but not cholesterol, inhibit human immunodeficiency virus replication in vitro. Antivir. Chem. Chemother. 9:491–496 [DOI] [PubMed] [Google Scholar]
- 18. Morrow M. P., et al. 2010. Stimulation of the liver X receptor pathway inhibits HIV-1 replication via induction of ATP-binding cassette transporter A1. Mol. Pharmacol. 78:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moss W. J., et al. 2002. Suppression of human immunodeficiency virus replication during acute measles. J. Infect. Dis. 185:1035–1042 [DOI] [PubMed] [Google Scholar]
- 20. Mujawar Z., et al. 2006. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 4:e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Negredo E., et al. 2006. The effect of atorvastatin treatment on HIV-1-infected patients interrupting antiretroviral therapy. AIDS 20:619–621 [DOI] [PubMed] [Google Scholar]
- 22. Nguyen D. H., Hildreth J. E. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U. S. A. 104:6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinzon S., et al. 2009. Impaired cholesterol biosynthesis in a neuronal cell line persistently infected with measles virus. J. Virol. 83:5495–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy A. L. 2007. Signal-induced functions of the transcription factor TFII-I. Biochim. Biophys. Acta 1769:613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sadowski I., Mitchell D. A. 2005. TFII-I and USF (RBF-2) regulate Ras/MAPK-responsive HIV-1 transcription in T cells. Eur. J. Cancer 41:2528–2536 [DOI] [PubMed] [Google Scholar]
- 27. Sedaghat A. R., et al. 2008. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J. Virol. 82:1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stevenson M., Stanwick T. L., Dempsey M. P., Lamonica C. A. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Treguier M., et al. 2004. Transcription factor sterol regulatory element binding protein 2 regulates scavenger receptor Cla-1 gene expression. Arterioscler. Thromb. Vasc. Biol. 24:2358–2364 [DOI] [PubMed] [Google Scholar]
- 30. van 't Wout A. B., et al. 2005. Nef induces multiple genes involved in cholesterol synthesis and uptake in human immunodeficiency virus type 1-infected T cells. J. Virol. 79:10053–10058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waheed A. A., Freed E. O. 2010. The role of lipids in retrovirus replication. Viruses 2:1146–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinberger L. S., Burnett J. C., Toettcher J. E., Arkin A. P., Schaffer D. V. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182 [DOI] [PubMed] [Google Scholar]
- 33. Wu Y., Marsh J. W. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506 [DOI] [PubMed] [Google Scholar]
- 34. Zheng Y. H., Plemenitas A., Fielding C. J., Peterlin B. M. 2003. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc. Natl. Acad. Sci. U. S. A. 100:8460–8465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Y., Zhang H., Siliciano J. D., Siliciano R. F. 2005. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J. Virol. 79:2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.