Abstract
Hepadnaviruses are the only known viruses that replicate by protein-primed reverse transcription. Beyond the conserved reverse transcriptase (RT) and RNase H domains, their polymerases (P proteins) carry a unique terminal protein (TP) domain that provides a specific Tyr residue, Tyr96 in duck hepatitis B virus (DHBV), to which the first nucleotide of minus-strand DNA is autocatalytically attached and extended by three more nucleotides. In vitro reconstitution of this priming reaction with DHBV P protein and cellular chaperones had revealed strict requirements for the Dε RNA stem-loop as a template and for catalytic activity of the RT domain plus RNA-binding competence of the TP domain. Chaperone dependence can be obviated by using a truncated P protein (miniP). Here, we found that miniP with a tobacco etch virus (TEV) protease cleavage site between TP and RT (miniPTEV) displayed authentic priming activity when supplied with α-32P-labeled deoxynucleoside triphosphates; however, protease cleavage revealed, surprisingly, that the RT domain was also labeled. RT labeling had identical requirements as priming at Tyr96 and originated from dNMP transfer to a unique Tyr residue identified as Tyr561 in the presumed RT primer grip motif. Mutating Tyr561 did not affect Tyr96 priming in vitro and only modestly reduced replication competence of an intact DHBV genome; hence, deoxynucleotidylated Tyr561 is not an obligate intermediate in TP priming. However, as a first alternative substrate for the exquisitely complex protein-priming reaction, dNMP transfer to Tyr561 is a novel tool to further clarify the mechanism of hepadnaviral replication initiation and suggests that specific priming inhibitors can be found.
INTRODUCTION
Hepadnaviruses, with human hepatitis B virus (HBV) as their prototypic member and a most important pathogen, are small hepatotropic DNA viruses that replicate their ∼3-kb relaxed circular DNA (rcDNA) genome through protein-primed reverse transcription. Because of numerous experimental restrictions with HBV, the related duck HBV (DHBV) is widely used as a model (39). Crucial for protein priming is the terminal protein (TP) domain which P proteins carry in addition to the reverse transcriptase (RT) and RNase H (RH) domains that are conserved in all RTs (Fig. 1 A). TP provides the specific Tyr residue (Tyr96 in DHBV P and Tyr63 in HBV P) to whose phenolic OH-group the first DNA nucleotide of minus-strand DNA is covalently attached (24, 55, 62).
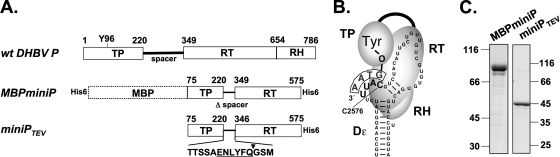
Fig. 1.
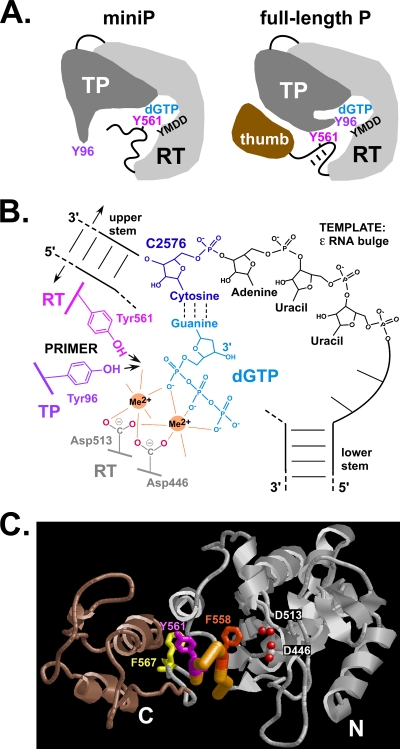
Structural aspects of DHBV protein priming and recombinant P proteins used. (A) Domain structures of wt DHBV P protein and miniP proteins. The P protein consists of conserved RT and RH domains plus, linked by a dispensable spacer, an N-terminal TP domain. Tyr96 (Y96) in TP serves as an acceptor for the 5′ nucleotide of minus-strand DNA. The numbers represent amino acid positions. MBPminiP is a fusion of a truncated P protein lacking the indicated terminal sequences and carrying an SPG linker instead of the spacer fused to MBP. miniPTEV contains a recognition site for tobacco etch virus (TEV) protease (underlined); the cleavage site is denoted by an arrow. (B) Schematic representation of primed P protein. Binding of P to Dε RNA, for full-length P requiring chaperones (not shown), induces a conformational change that disrupts base pairing in the upper Dε stem and appropriately positions Tyr96 over the initiation template in Dε (C2576) so as to allow the RT domain to copy the DNA primer (boxed). (C) Recombinant miniP proteins. MBPminiP (88 kDa) and miniPTEV (46 kDa) were expressed in E. coli, purified as described in Materials and Methods, and analyzed by SDS-PAGE and Coomassie blue staining.
Studies in HBV- or DHBV-transfected cells have provided a general outline of the hepadnaviral replication cycle (reviewed in references 8 and 27). Like core protein, P protein is translated from the pregenomic RNA (pgRNA) and then binds to the 5′ proximal ε (Dε for HBV) stem-loop on pgRNA, triggering RNA encapsidation and replication initiation. In this priming reaction a bulged region within ε templates the synthesis of a 3- or 4-nucleotide-long, TP-linked DNA oligonucleotide. Its translocation to a 3′-proximal RNA element, DR1*, and extension from there yields minus-strand DNA. Concomitant degradation of the RNA template by the RNase H activity of P spares some 15 to 18 nucleotides from the pgRNA's 5′ end (18, 26) which serve as a conventional RNA primer for plus-strand DNA synthesis, eventually yielding rcDNA. The covalent linkage of the minus-strand DNA 5′ end to TP remains intact throughout the process.
More mechanistic details on hepadnaviral protein-priming were revealed by cell-free systems in which recombinant P protein from DHBV, though not from HBV (20), perpetuates the initial replication steps in vitro (6, 9, 21, 52). Provision of a proper ε RNA template and α-32P-labeled deoxynucleoside triphosphates ([α-32P]dNTPs) results in covalent radiolabeling of the protein that can sensitively be monitored. The original system based on DHBV P protein in vitro translated in rabbit reticulocyte lysate (52) showed that priming requires catalytic activity of the RT domain and involves structural rearrangements in both the RNA (7) and P protein (45, 46). Consistent with data from DHBV-transfected cells (22, 47, 51), the sequence of the in vitro-synthesized oligonucleotide primer is 5′GTAA, complementary to the sequence 5′UUAC in the bulge of Dε (Fig. 1B), with C2576 representing the initiation template. An analogous mechanism applies to HBV (29). Replacement of Tyr96 in TP prevented detectable labeling (55, 62), confirming this residue as the major acceptor site for the first DNA nucleotide. More recently, DHBV P protein produced in E. coli as fusion with solubility-enhancing partners such as NusA (6, 40) or glutathione S-transferase (GST) (21) has successfully been reconstituted into initiation complexes with authentic priming activity. These studies demonstrated a strict dependence of priming activity on cellular chaperones and ATP (6, 21, 41) and revealed that chaperone activation involves the exposure of a buried RNA binding site in TP (1, 40); TP mutations preventing RNA binding also abolished priming. An even simpler in vitro system is provided by DHBV P protein truncated from both ends, miniP (Fig. 1A), which displays ε-dependent, yet chaperone- and ATP-independent, priming activity (6, 54). However, doubts have arisen as to whether this activity has the same strict template specificity as that of the full-length protein (25).
Protein priming rather than nucleic acid priming is used by some DNA viruses (36) as one strategy to ensure the lossless replication of 5′ ends in the face of an absolute requirement of all DNA polymerases for a primer (43). Although some RNA-dependent RNA polymerases can initiate replication de novo, protein priming is widespread among positive-strand RNA viruses (reviewed in reference 48), most prominently picornaviruses (reviewed in reference 56). Their 5′ terminally linked VPg (viral protein genome-linked) peptides may also block innate immune responses that are triggered by RNAs with a 5′ triphosphate (38, 44). For some viruses, direct structural information pertaining to the protein-priming mechanism is available, and yet indications of hepadnaviral protein priming are rare. Hepadnaviruses are the only reverse-transcribing elements that use this mechanism, possibly except for some mitochondrial retroplasmids from a filamentous fungus (17). Meaningful comparisons with protein priming in DNA and RNA viruses are obscured by the generally high sequence diversity among primer proteins, and even more so by the lack of hepadnavirus TP homologs in the database which restricts even structure modeling to the conserved RT and RH domains (see Discussion).
In this situation, limited proteolysis had proven an appropriate method to derive at least some information on the structural changes that accompany chaperone activation of nearly full-length P protein (40). To improve resolution, we intended to introduce a recognition site for the highly specific tobacco etch virus (TEV) protease between the TP and RT/RH domains and combine this with the chaperone independence of the truncated miniP protein. The modified miniP, miniPTEV, indeed displayed ε-dependent in vitro priming activity. Most surprisingly, however, TEV cleavage after priming revealed substantial 32P labeling not only in the TP domain but also in the RT domain. Similar results were independently obtained in J. Hu's lab and are reported in the accompanying study (10a). As shown below, we mapped the RT labeling site to a unique Tyr residue, Tyr561 in box E, one of seven sequence motifs (boxes A to E and boxes 1/G and 2/F; see Fig. S1 in the supplemental material) that are conserved in all RTs. We demonstrate that deoxynucleotidylation of this residue shares the essential features of the authentic priming reaction at Tyr96 but is not essential for viral replication.
MATERIALS AND METHODS
Plasmid constructs.
Vectors for P protein expression in Escherichia coli were based on pET28a(+) (Novagen), and viral sequences were from DHBV16 (GenBank accession no. K01834). pET28-MBPminiP encodes MBP fused to DHBV P amino acids (aa) 75 to 575, with the spacer (aa 221 to 348) replaced by Ser-Pro-Gly (6). pET28-miniPTEV encodes the same DHBV P sequence without the maltose-binding protein (MBP) part and with the tripeptide linker replaced by the peptide Thr-Thr-Ser-Ser-Ala-Glu-Asn-Leu-Tyr-Phe-Gln-Gly-Ser-Met, which includes a consensus motif for TEV protease (underlined). For in vitro translation in RRL, the complete P open reading frame (ORF) in pT7AMVpol16H6 (4) was replaced by the miniPTEV ORF from pET28-miniPTEV. Into the resulting plasmid, pT7AMV-miniPTEV, appropriate restriction fragments were inserted to restore the complete P protein C terminus (to aa 786; pT7AMV-P75TEV) or in addition the N terminus (pT7AMV-P1TEV). Schematic representations of the parental proteins are shown in Fig. 1A; additional variants, obtained by standard PCR mutagenesis, are listed in Table 1. For the expression of wild-type (wt) DHBV in LMH cells, plasmid pCD16, containing a 1.1×DHBV16 genome under the control of the cytomegalovirus immediate-early promoter (32), was used. In plasmids pCD16-Y561A and pCD16-Y561F, the codon for Tyr561 in P was changed to Ala and Phe via point mutations. For in vitro transcription of Dε and HBV ε RNAs, the T7 RNA polymerase promoter containing plasmids pDε1 (6) and PCR-derived mutants with nucleotide C2576 replaced by A, G, or T and pCHG-3068-T7 (5), respectively, served as templates. All plasmid constructs were verified by DNA sequencing.
Table 1.
miniPTEV terminal deletion and substitution variants
| Variant | Positiona |
aa substitution(s) | |
|---|---|---|---|
| N terminus | C terminus | ||
| miniPTEV | 75 | 575* | |
| N-80 | 80 | 575* | |
| N-90 | 90 | 575* | |
| N-120 | 120 | 575* | |
| C-566 | 75 | 566 | |
| YMHD | 75 | 575* | D513H |
| T3mut | 75 | 575* | Y175G, Y181S |
| 96F | 90 | 575* | Y96F |
| 96F566 | 90 | 566 | Y96F |
| Cysmin | 90 | 575* | Y96F, C518S, C533S |
| 424F | 90 | 575* | Y424F |
| 452F | 90 | 575* | Y452F |
| 473/4FF | 90 | 575* | Y473F, Y474F |
| 511F | 90 | 575* | Y511F, M512V, Y96F |
| 561A | 90 | 575* | Y561A |
| 561F | 75 | 566 | Y561F |
| 558Y | 75 | 566 | F558Y, Y561F |
| 567Y | 75 | 567 | F567Y, Y561F |
The N- and C-terminal amino acid positions are indicated (numbers refer to DHBV16 P). *, Constructs terminating at position 575 contain a C-terminal hexahistidine tag.
Recombinant miniP expression and purification.
miniP proteins were expressed in E. coli BL21 Codonplus RIL cells (Stratagene). MBPminiP was purified by immobilized nickel affinity chromatography (IMAC) under native conditions as described previously (6) with the following modifications. Cells were lysed in buffer A (50 mM sodium phosphate [pH 6.8], 200 mM NaCl, 5 mM MgCl2, 0.1% octyl-tetraoxyethylene, 25 mM imidazole, 10 mM β-mercaptoethanol) supplemented with lysozyme (0.5 mg/ml), Benzonase (2.5 U/ml; Novagen), and Complete EDTA-free protease inhibitor (20 μl/ml; Roche Applied Science). After sonication, the lysate was cleared by ultracentrifugation (100,000 × g, 90 min, 4°C), and IMAC was performed in buffer A, followed by gel filtration on a HiLoad 16/60 Superdex 200-pg column (GE Healthcare) in buffer B (50 mM sodium phosphate [pH 6.8], 200 mM NaCl, 5 mM MgCl2, 0.1% octyl-tetraoxyethylene, 1 mM dithiothreitol [DTT]). Peak fractions (0.5 to 1.0 mg/ml) were pooled and stored at −80°C. miniPTEV and its variants were refolded from inclusion bodies (IBs) (11). Bacterial pellets were resuspended in buffer C (50 mM Tris-HCl, 50 mM NaCl, 5 mM DTT, 0.5 mM EDTA, 5% glycerol [pH 8.0]) supplemented with lysozyme (final concentration, 0.5 mg/ml) and Benzonase (final concentration, 2.5 U/ml). After incubation for 30 min at 23°C, the lysate was sonicated and supplemented with Triton X-100 (final concentration, 1% [vol/vol]), and miniP IBs were pelleted by centrifugation (8,000 × g, 15 min). IBs were washed twice in buffer C plus 1% Triton X-100 and once in detergent-free buffer C. IBs were solubilized in buffer D (7 M guanidine-HCl, 50 mM Tris-HCl, 0.5 mM EDTA, 10 mM DTT [pH 7.5]). The solution was cleared by ultracentrifugation (100,000 × g, 45 min), and miniP was refolded by rapid dilution into buffer E [50 mM Tris-HCl, 50 mM NaCl, 1 M 3-(1-pyridino)-1-propane sulfonate (pH 7.5)] and subsequent incubation for 2 h at 23°C. Malfolded proteins were removed by ultracentrifugation (100,000 × g, 45 min).
RNA synthesis.
Uncapped RNAs were generated by T7 RNA polymerase mediated in vitro transcription (AmpliScribe T7 high-yield transcription kit;, Epicentre) as described previously (6).
In vitro priming assays.
MBPminiP/Dε RNA initiation complexes were reconstituted by mixing an aliquot of purified MBPminiP (see above) with an equal volume of buffer E (50 mM Tris-HCl [pH 8.0], 5 mM DTT, 2 μM Dε RNA) and subsequent incubation for 1 h at 37°C. miniPTEV/Dε RNA complexes were reconstituted by adding to refolded miniPTEV Dε RNA to a final concentration of 1 μM and incubation for 1 h at room temperature. Subsequently, a 0.5 volume of priming buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 6 mM MnCl2, 2 μCi of [α-32P]dNTP [3,000 Ci/mmol]) was added, and the samples were incubated at 37°C for 1 h. MnCl2 was omitted from the priming buffer in Mn2+-free samples. Reactions were stopped by adding 1 volume of sodium dodecyl sulfate (SDS) sample buffer and boiling the mixtures at 100°C for 5 min. Aliquots of 10 μl were analyzed by SDS-PAGE. Gels were vacuum dried, and primed miniP was detected by phosphorimaging (Typhoon FLA 7000; GE Healthcare). 32P signals were quantified by using ImageQuant TL software (GE Healthcare).
TEV cleavage of primed miniPTEV.
To [α-32P]dNTP-primed miniPTEV, prepared as described above, EDTA was added to a final concentration of 5 mM. Aliquots of 20 μl were mixed with 1 μl of a 1-mg/ml stock of home-made recombinant TEV protease (50), followed by incubation at 37°C for 2 h. The reaction was stopped by adding 10 μl of SDS sample buffer and boiling at 100°C for 5 min. Samples were analyzed by SDS-PAGE and phosphorimaging.
Cysteine-specific chemical cleavage.
[α-32P]dGTP-primed miniPTEV was treated with 2-nitro-5-thiocyanobenzoate (NTCB) as described previously (14). In brief, primed miniP was denatured in 1% SDS, and potential disulfide bridges were reduced by 1 mM DTT prior to treatment with NTCB. Cleavage was monitored by SDS-PAGE, followed by autoradiography and phosphorimaging.
Acid and base stability assay.
[α-32P]dGTP-primed miniPTEVY96F was cleaved with TEV protease as described above. Then, 30-μl aliquots were treated in parallel with 3 μl of 1 M NaOH, 3 μl of 1 M HCl, or 3 μl of H2O, followed by incubation for 1 h at 37°C. Basic and acidic samples were neutralized with 3 μl of 1 M HCl and 3 μl of 1 M NaOH, respectively. The samples were analyzed by SDS-PAGE and phosphorimaging.
Priming assays and TEV cleavage of in vitro-translated P proteins.
P protein variants were expressed in rabbit reticulocyte lysate in the presence of in vitro-transcribed Dε RNA and subjected to [α-32P]dGTP priming or were expressed in the presence of [35S]methionine, without priming, essentially as previously described (9). Subsequent TEV protease digestion and analysis by SDS-PAGE and phosphorimaging were conducted as described above.
Transient-transfection experiments.
Chicken LMH hepatoma cells were cultured and transfected with pCD16, pCD16-Y561A, and pCD16-Y561F as previously described (41). At 4 days posttransfection, cytoplasmic lysates were prepared, and viral DNA from cytoplasmic nucleocapsids was isolated and detected by Southern blotting using a 32P-labeled full-length DHBV DNA probe (10). 32P signals were quantified by phosphorimaging. Capsids from cytoplasmic lysates were separated by native agarose gel electrophoresis and analyzed by immunoblotting as described previously (49) using a polyclonal rabbit anti-DHBV core protein antiserum and a peroxidase-conjugated secondary antibody, followed by detection with ECL Plus (GE Healthcare). Chemiluminescent signals were recorded on X-ray film or on a Fuji LAS 3000 instrument and quantified using AIDA software (Raytest).
RESULTS
Authentic in vitro priming activity of recombinant miniP protein.
miniP was initially expressed in E. coli as a fusion to an N terminally His6-tagged maltose-binding protein (MBPminiP; Fig. 1A). MBP is smaller than the previously used NusA (6, 41) and yet provided for a similar solubility enhancement, such that sufficient amounts for the subsequent priming assays could be purified under native conditions (Fig. 1C, left panel). Priming competence was assessed by incubation with in vitro-transcribed Dε RNA in the presence of Mg2+, yet without chaperones, and the subsequent addition of a mix of [α-32P]dATP plus unlabeled dGTP and dTTP (the constituent nucleotides of the authentic DNA primer GTAA; see Fig. 1B) in Mn2+-containing buffer (see below), followed by SDS-PAGE and autoradiography (Fig. 2 A). A strong 32P signal at the position expected for MBPminiP was observed with Dε RNA (lane 1) but not with HBV ε RNA (lane 4) or a 30-fold-higher concentration of tRNA (lane 3); the presence of excess tRNA did not detectably reduce Dε RNA-mediated priming (lane 2). Hence, MBPminiP displayed overall the same strict RNA template specificity as previously seen for nearly full-length P protein (6); however, firm conclusions on the exact initiation site were not yet possible.
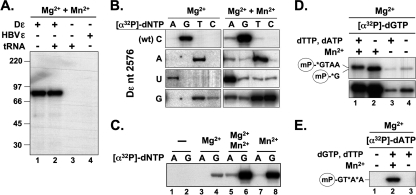
Fig. 2.
MBPminiP displays authentic priming activity. (A) The cognate Dε RNA is required for MBPminiP priming. MBPminiP was incubated with the indicated RNAs at final concentrations of 1.5 μM for Dε and HBV ε and 50 μM for yeast tRNA. Priming was tested in the presence of unlabeled dTTP and dGTP (at 10 μM each) plus 2 μCi of [α-32P]dATP. Samples were analyzed by SDS-PAGE and autoradiography. (B) MBPminiP initiates primer synthesis in a Dε templated manner at the authentic site. Priming assays were performed with wt Dε RNA or its variants C2576U, C2576A, and C2576G, plus the indicated individual [α-32P]dNTPs. Reactions contained either 1.7 mM Mg2+ or 1.7 mM Mg2+ plus 2.0 mM Mn2+. Note that for proper visualization, the exposure times for the Mg2+-only reactions were ∼5-fold (wt Dε and variant C2576A) and ∼10-fold (Dε variants C2576U and C2576G) longer than for the Mg2+ + Mn2+ reactions. (C) Mn2+ as the only bivalent metal ion does not impair template specificity. For the removal of Mg2+, MBPminiP was dialyzed against buffer without MgCl2. Priming assays with wt Dε RNA and [α-32P]dATP or [α-32P]dGTP were performed without (−) or with prior back-addition of the indicated bivalent cations. (D and E) MBPminiP requires Mn2+ to extend the first minus-strand DNA nucleotide. Priming assays were performed with [α-32P]dGTP (D) or [α-32P]dATP (E). Reactions were supplemented or not with unlabeled dNTPs and Mn2+ as indicated. The two panels in panel C show two different exposures of the same gel. The encircled mP denotes miniP, the asterisks the 32P-labeled nucleotides. An upward shift (D) and incorporation of dA at the third and fourth primer positions (E) in the presence of unlabeled dNTPs was only seen upon Mn2+ supplementation.
In the authentic priming reaction the 3′-terminal bulge nucleotide C2576 templates the first nucleotide of the DNA primer, resulting in the covalent attachment of a complementary dG residue to Tyr96 in TP. To test whether this also holds true for MBPminiP, we supplemented the priming reactions with only one α-32P-labeled dNTP; because Mn2+ is known to enhance the priming activity but was also reported to reduce dNTP specificity (25), we performed these assays in the presence of either Mg2+ alone (Fig. 2B, left panel) or Mg2+ plus Mn2+ (Fig. 2B, right panel). Finally, we included variant Dε RNAs in which C2576 was replaced by A, U, or G. With Mg2+ as the only divalent cation, almost exclusively the deoxynucleotide complementary to position 2576 was detectably incorporated, except for the Dε variant C2576G, where both the complementary dC and dT gave relatively intense signals. The additional presence of Mn2+ enhanced the signals by ∼20-fold, allowing us to estimate that with wt Dε RNA as a template the dG was incorporated ∼20-fold more efficiently than dA and ∼100-fold better than dT; dCTP produced no signal, indicating an at least 1,000-fold discrimination. Thus, the overall nucleotide preference remained the same as with Mg2+ alone, including the acceptance of both dC and dT with the C2576G Dε variant as a template. To further evaluate a potential influence of Mn2+ on the specificity of first nucleotide incorporation, we removed all divalent cations by dialysis and then added wt Dε RNA plus either dGTP or dATP, and either no divalent cation, Mg2+, Mg2+ plus Mn2+, or Mn2+ only (Fig. 2C). The lack of signal without exogenously added divalent cation (lanes 1 and 2) confirmed their efficient removal by dialysis. Back-addition of Mg2+ allowed the incorporation of dG but not dA, the addition of Mg2+ plus Mn2+ strongly enhanced dG incorporation and led to a faint signal with dA; importantly, the same pattern was observed with Mn2+ only. These data are fully consistent with MBPminiP using the authentic C2576 as an initiation template and with Mn2+ enhancing the efficiency but not substantially reducing the specificity of the priming reaction. Hence, MBPminiP recapitulates all crucial aspects of first nucleotide incorporation seen in the chaperone-assisted reaction with full-length P protein. A possible rationale for the relaxed dC versus dT with the C2576G RNA variant is given in the Discussion.
To investigate the ability of MBPminiP to synthesize the full-length GTA(A) primer, we performed priming assays using [α-32P]dGTP, with or without adding a mix of unlabeled dTTP plus dATP. Primer elongation from one nucleotide to three or four nucleotides is expected to cause a slight retardation of primed MBPminiP during SDS-PAGE. Such an upward shift was indeed reproducibly seen in the presence of Mg2+ plus Mn2+, whereas with only Mg2+ the mobility of the primed MBPminiP was not different from that obtained with dGTP as the only dNTP (Fig. 2D). Furthermore, in the absence of Mn2+ MBPminiP could not be labeled by [α-32P]dATP even in the presence of unlabeled dGTP plus dTTP (Fig. 2E, lane 3). Hence, MBPminiP is capable of full-length primer synthesis but, in contrast to full-length P, does so in an Mn2+-dependent fashion. While these latter data are consistent with those reported by others (25), the reasons for the apparent discrepancies regarding maintained versus relaxed template specificities remain to be determined.
Specifically cleavable miniPTEV reveals covalent 32P labeling of the RT domain during in vitro priming with [α-32P]dNTPs.
Although MBPminiP displayed largely authentic priming activity, it might be argued that the still bulky MBP domain affects its properties. We noticed that miniP was highly expressed in E. coli even without MBP and yet was virtually insoluble. As an alternative, we developed a protocol to renature the unfused miniP from IBs and applied this to miniPTEV, which contains an engineered TEV protease recognition site between TP and RT (Fig. 1A and C, right panel).
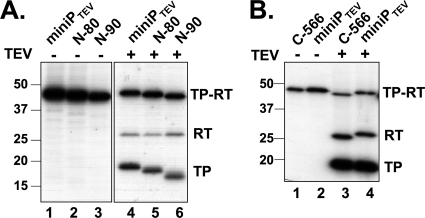
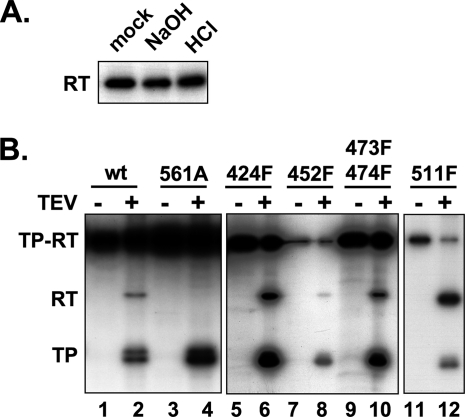
Enzymatic activity of the refolded miniPTEV was demonstrated by strong 32P labeling upon in vitro priming using wt Dε RNA and [α-32P]dGTP (Fig. 3 A, lane 1). Subsequent treatment with TEV protease generated the expected 32P-labeled TP fragment and yet, unexpectedly, also a fragment with an apparent molecular mass matching that of the clipped-off RT domain (Fig. 3A, lane 4). To confirm this interpretation, we analyzed a set of terminal miniPTEV deletion variants (see Table 1) in the same way. Deletions in TP shifted the TP band but not the putative RT band (Fig. 3A, lanes 4 to 6), and the reverse was seen for a deletion in the RT domain (Fig. 3B, lanes 3 to 4). Hence, beyond the established linkage to TP, the priming reaction had led to the covalent transfer to the RT domain of at least that part of dGTP that contains the former [α-32P]phosphate. Treatment with lambda protein phosphatase that dephosphorylates phosphoserine, phosphothreonine, and phosphotyrosine residues did not reduce RT labeling, whereas [γ-32P]ATP failed to produce any labeling (see Fig. S2 in the supplemental material); the modification therefore most likely involved dGMP transfer (deoxyguanylation) rather than phosphorylation.
Fig. 3.
TEV protease cleavage reveals 32P labeling of the RT domain during α-32P priming. Priming assays were performed with miniPTEV and N-terminal (A) or C-terminal (B) miniPTEV truncation variants. Aliquots of each sample were treated with TEV protease and analyzed as described in Fig. 2. Undigested miniPTEV and cleavage products are denoted by TP-RT, RT, and TP, respectively. Evidence against RT labeling being due to phosphorylation is presented in Fig. S2 in the supplemental material.
RT modification requires RT activity and occurs independently from primer synthesis at Tyr96.
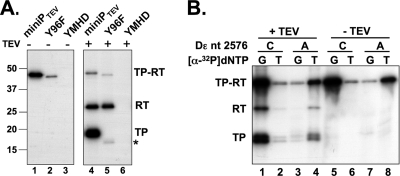
Authentic initiation of negative-strand DNA synthesis requires Tyr96 in TP as an acceptor site and a catalytically active RT domain. To investigate whether this was also true for RT labeling, either function was knocked out in miniPTEV, and the catalytically inactive YMHD and Tyr96-deficient 96F variants (see Table 1) were analyzed by in vitro priming and TEV protease cleavage (Fig. 4 A). The YMHD mutation abolished 32P labeling of both domains, indicating that RT labeling is as well catalyzed by the RT activity of the RT domain itself. As expected, the Y96F mutation strongly reduced the TP signal, though not completely, suggesting a low level of noncanonical labeling at TP sites different from Tyr96. However, RT labeling was not significantly affected compared to wt miniPTEV (Fig. 4A, lane 4 versus lane 5). Besides confirming that RT labeling involves deoxynucleotidyl rather than phosphoryl transfer, the dependence on polymerase activity strongly suggests that RT labeling is mechanistically similar to, yet independent from, Tyr96-linked protein priming.
Fig. 4.
RT labeling shares essential features with priming at Tyr96. (A) RT labeling depends on catalytic activity of miniP but not on Tyr96. Priming assays with Dε RNA plus [α-32P]dGTP were performed with miniPTEV or variants with disabled RT activity (YMHD) or with Tyr96 replaced by Phe (96F). Aliquots from each reaction were analyzed as described in Fig. 2. RT labeling was abolished by the YMHD mutation but not the 96F mutation. Note that miniPTEV 96F starts with TP amino acid 90 rather than amino acid 75 of TP (see Table 1), causing a 1.7-kDa lower molecular mass. The residual labeling of TP (asterisk) despite the replacement of Tyr96 suggests the presence in TP of additional acceptor sites; this labeling was also strictly dependent on an active RT domain (no signal with the YMHD variant). The dependence of RT labeling on the cognate Dε RNA and the RNA binding ability of TP is shown in Fig. S3 in the supplemental material. (B) RT labeling depends on dNTP complementarity to the authentic initiation template. Priming assays were performed with wt Dε RNA or its variant C2576A and either [α-32P]dGTP or [α-32P]dTTP as indicated. Aliquots from each reaction were analyzed as described in Fig. 2. Note the nearly constant ratio of TP versus RT labeling in all reactions.
RT modification is strictly dependent on the cognate Dε RNA and on an RNA-binding-competent TP domain.
Next, we investigated the role of RNA in RT labeling. miniPTEV was incubated with various RNAs and supplemented with [α-32P]dGTP. As with MBPminiP (Fig. 2A), 32P labeling of miniPTEV was exclusively observed with Dε RNA (see Fig. S3A in the supplemental material). The lack of signal with HBV ε RNA or tRNA indicated that, as for TP priming, the cognate ε RNA is essential for RT labeling.
TP contributes crucially to ε RNA binding; deletion of 120 or more residues from the N terminus residues prevents RNA binding (53), and so do various mutations in and around a highly conserved C proximal TP motif (aa 176 to 183) termed T3 (12). In full-length P protein its RNA-binding activity needs to be exposed by chaperones (1, 40). Introducing two such TP mutations into miniPTEV (N-120 and T3mut; see Table 1) resulted in a complete loss of detectable priming (see Fig. S3B in the supplemental material), demonstrating that the RNA-binding activity of TP is essential for RT labeling. Thus, the RT modification shares two more essential features of authentic TP priming.
Like TP priming, RT modification is dependent on base complementarity between the dNTP substrate and the Dε bulge nucleotide 2576.
To address whether RT labeling, like TP priming, was dependent on complementarity between incoming dNTP and the authentic initiation template in Dε RNA, we incubated miniPTEV with wt Dε RNA (C2576) or its variant C2576A and with either [α-32P]dGTP or [α-32P]dTTP. Consistent with the MBPminiP results (Fig. 2B and C), with wt Dε RNA as a template miniPTEV incorporated dG much more efficiently than dT, and the reverse was seen for the variant C2576A RNA (Fig. 4B, panel −TEV). TEV cleavage (panel +TEV) revealed an approximately proportional distribution of the label to the TP and RT fragments for all RNA-dNTP combinations, indicating that the efficiency of both TP and RT labeling is largely determined by complementarity between the dNTP substrate and the RNA base at position 2576. Hence, priming at Tyr96 and RT modification use a virtually identical mechanism.
We also assessed whether further nucleotides could be added to the dNMP linked to Tyr561. The results obtained using a similar mobility shift assay as shown in Fig. 2D were not fully conclusive (see Fig. S4 in the supplemental material). As with MBPminiP, the addition of all three primer-constituent dNTPs caused a visible upward shift of the uncleaved protein and, after TEV cleavage, of the TP fragment; a much less pronounced, though reproducible, shift was seen for the RT fragment. Hence, the extension reaction on RT was either inefficient, or it had only a limited impact on the electrophoretic mobility of the RT fragment.
The RT nucleotidylation site is located downstream of Cys518.
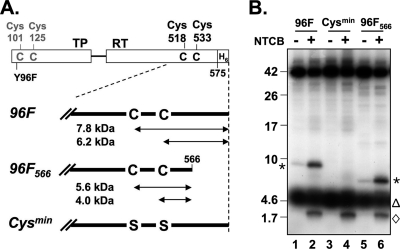
To simplify mapping of the RT nucleotidylation site(s), we suppressed TP labeling by using the miniPTEV variant Y96F, ending with aa 575, for priming with wt Dε RNA and [α-32P]dGTP and treated the primed protein with the cysteine-specific cleavage agent NTCB. miniPTEV contains two cysteines, at positions 518 and 533, in the RT domain and two more close to the truncated N terminus of TP (Fig. 5 A). NTCB produced two specific small products with mobilities corresponding to about 9 and 2 kDa (Fig. 5B, lane 2). Given the substantial variability in the mobility of small peptides during SDS-PAGE (see, for example, reference 34) and the unknown influence of the covalent modification, this was compatible with the label being present in a C-terminal 6.2-kDa fragment generated by cleavage at Cys533 or in a 7.8-kDa fragment generated by a partial cleavage at Cys518; the 2-kDa fragment could have arisen from cleavage between the cysteines. However, other interpretations were not excluded. For clarification, we truncated miniPTEV slightly further to end after amino acid 566 (miniPTEV Y96F566). This led to a complete loss of the 9-kDa band in favor of a new product migrating at ∼7 kDa; the mobility of the ∼2-kDa band was not affected. Since very faint bands at the 9- and 7-kDa positions, respectively, were also seen in the mock-incubated reactions, we sought to verify Cys specificity by replacing both Cys518 and Cys533 with Ser (miniPTEV-Cysmin). This modified protein produced no signal at the 9-kDa position. The 2-kDa band was again unaffected, suggesting that it may originate from residual TP priming at a site different from Tyr96. Together, these data demonstrated that the RT deoxynucleotidylation site was located between Cys518 or Cys533 and the C-terminal residue 566.
Fig. 5.
Cys-specific chemical cleavage maps the RT labeling site to the sequence between positions 518 or 533 and 566. (A) A Cys map of miniPTEV. miniP contains four Cys residues at the indicated positions. Calculated masses of cleavage products from the RT domain are given in kilodaltons. Variant 96F566 has a slightly further truncated C terminus; in variant Cysmin, the two Cys residues in RT were replaced by Ser, in addition to the Y96F mutation. (B) RT is labeled in its C-terminal region. Priming assays were performed with wt Dε RNA plus [α-32P]dGTP and the indicated miniPTEV variants. Samples were treated with NTCB and analyzed by SDS-PAGE on 16% Tris-Tricine gels before autoradiography. Asterisks denote RT derived C-terminal peptides. The 2-kDa band (⋄) was not dependent on RT Cys residues and thus may originate from residual TP labeling close to Cys101/Cys125. The diffuse 32P signals at ∼5 kDa (▵) occurred in all reactions and probably do not represent protein-bound label.
The RT domain is specifically modified at Tyr561.
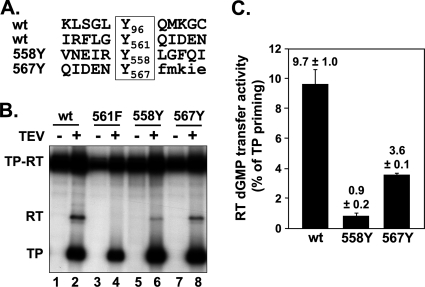
The most plausible explanation for the above-described results was that in a reaction very similar to authentic priming, a dGMP moiety was covalently attached to one or few C proximal residues in the RT domain. In principle, dGMP could be linked by a phosphodiester bond to Tyr, Ser, or Thr or by a phosphoamide bond to His, Lys, or Arg. The latter bonds are acid labile; seryl and threonyl but not tyrosyl phosphodiesters are alkali labile (13). The RT modification product was resistant to treatment with both 0.1 N HCl and 0.1 M NaOH (Fig. 6 A), strongly arguing for a tyrosyl linkage. To confirm this hypothesis and to obtain independent evidence for the NTCB mapping data, we mutated all seven Tyr residues in the miniPTEV RT domain except Y375. Tyr511 is part of the YMDD active-site motif, and in HIV-1 RT a Tyr→Phe exchange at this site reduces enzymatic activity; however, full activity is restored by replacing the following Met residue by Val (19). Since RT labeling depends on RT activity (see above), we combined the Y511F mutation with an M512V exchange. Furthermore, the 511F variant but not the others contained the Y96F mutation to suppress TP priming (Table 1). Monitoring RT labeling by [α-32P]dGTP priming and TEV cleavage revealed substantial variations in overall priming efficiency, and yet all variants but one still showed detectable RT labeling (Fig. 6B). The exception was variant Y561A, which produced strong labeling of TP but no signal at the RT position. This phenotype was not dependent on the specific Y561A exchange; it was also observed with variant Y561F (see below). Hence, in line with the mapping data shown above, RT is selectively modified at Tyr561.
Fig. 6.
The RT labeling site is located at Tyr561. (A) RT labeling is resistant against acid and base. miniPTEV variant 96F was subjected to priming and TEV protease cleavage and then treated or not (mock) for 1 h at 37°C with 0.1 M NaOH or 0.1 M HCl. Products were analyzed as described in Fig. 2. Only the part of the autoradiogram containing the RT related products is shown. (B) Mutation of Tyr561 but not of other Tyr residues in RT impedes RT labeling. miniPTEV variants bearing the indicated Tyr mutations in RT (in variant 511F combined with Y96F; see Table 1) were subjected to priming and TEV protease cleavage and analyzed as described in Fig. 3. RT domain labeling was detectable in all variants except for 561A.
RT deoxynucleotidylation requires RT DNA polymerase activity in cis.
It is generally assumed that the P-ε interaction leads to copackaging of one copy of P protein plus one copy of pgRNA per newly forming nucleocapsid (2, 60). In contrast, during in vitro priming the miniPTEV molecules are not physically separated from each other; hence, RT labeling may either occur within one molecule, i.e., with Tyr561 replacing Tyr96 in the primer binding site in cis, or by one molecule causing RT deoxynucleotidylation of another, i.e., in trans. A specific concern was that improperly folded molecules in the recombinant P protein preparation might act as such trans-substrates. We therefore sought to determine whether the lack of Tyr561 deoxynucleotidylation in the catalytically inactive miniPTEV YMHD variant could be rescued by co-refolding with the catalytically competent, yet RT modification-deficient miniPTEV Y561A variant. As shown in Fig. S5 in the supplemental material, the Y561A mutant failed to trans-complement the YMHD mutant. Hence, RT deoxynucleotidylation occurs in cis, indicating that both the TP residue Tyr96 and the RT residue Tyr561 of the same polypeptide can occupy the primer binding site of the RT catalytic center.
Artificially introduced Tyr residues in the vicinity of position 561 can serve as dNMP acceptor sites.
Based on the data reported above, Tyr561, but none of the other RT Tyr residues, was able to be loaded into the primer position of the catalytic center in a fashion comparable to that of Tyr96, indicating that some sequence or structural feature distinguishes Tyr561 from the other Tyr residues in RT. What such features might be for Tyr96 is unknown, and yet the sequence contexts of Tyr96 and Tyr561 bear a limited similarity (Fig. 7 A). We therefore replaced the two close-by Phe residues at positions 558 and 567 by Tyr, combined with the Y561F exchange, and sought to determine whether these artificially introduced Tyr residues with their differing neighboring sequences could act as substrates for deoxynucleotidylation. In addition to wt-like TP priming signals, both mutant proteins became detectably labeled in the RT domain (Fig. 7B), albeit to an ∼3-fold (F567Y) and ∼10-fold (F558Y) lesser extent compared to Tyr561 (Fig. 7C). Hence, Tyr residues at three different locations in the vicinity of position 561 can enter the primer position in the RT active site. Although the primary sequence context may contribute to this property, this suggests that structural flexibility in the C-terminal miniPTEV region is at least as important.
Fig. 7.
Tyr residues in the vicinity of Tyr561 can act as dNMP acceptors. (A) Primary sequence contexts of Tyr96 and Tyr561 and of variants 558Y and 567Y in which the genuine Phe residues were replaced by Tyr residues in a Y561F background. (B) Autoradiography after priming with wt Dε RNA plus [α-32P]dGTP. The indicated miniPTEV variants were tested for RT modification as described in Fig. 3. (C) Quantification of RT labeling. Efficiency of RT modification was determined by quantification of the 32P-labeling intensity of the RT band normalized to that of the TP band within the same lane. Numbers represent mean values ± the standard deviation from three experiments.
Restoring the C-terminal P protein sequences suppresses RT deoxynucleotidylation.
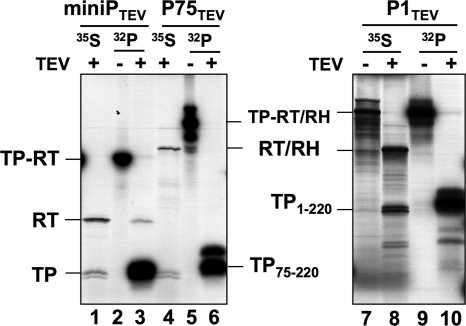
Deletion of the terminal regions in miniPTEV may promote the suspected structural flexibility around Tyr561. We therefore analyzed the effects on RT deoxynucleotidylation caused by reintroducing the deleted regions. P75TEV contains the truncated TP domain plus the entire RT and RH domains, and P1TEV comprises the entire TP, RT, and RH domains. Since these larger proteins are difficult to produce in E. coli and because of their chaperone dependence, the variant proteins, including miniPTEV itself, were in vitro translated in RL in the presence of Dε RNA and primed with [α-32P]dGTP, followed by TEV cleavage (Fig. 8); cleavage products of unprimed [35S]methionine-labeled proteins served as markers. Primed miniPTEV generated a 32P-labeled RT band, demonstrating that RT modification is not restricted to E. coli expressed miniP. In contrast, 32P-labeled RT-bands were not detectable with P1TEV and P75TEV, although TP priming and TEV cleavage occurred efficiently. Thus, the C-terminal parts of the RT domain and/or the RH domain suppress RT deoxynucleotidylation, likely by restricting conformational freedom of the P protein region around aa 561.
Fig. 8.
RT deoxynucleotidylation occurs with in vitro-translated miniPTEV but is suppressed by restoring the deleted C-terminal P protein sequences. miniPTEV, the C terminally extended variant P75TEV and the N plus C terminally extended variant P1TEV were expressed by in vitro translation in the presence of wt Dε RNA, primed with [α-32P]dGTP, and cleaved with TEV protease (lanes 32P). [35S]methionine-labeled and TEV protease digested in vitro translation products served as size markers for the TP and RT fragments (lanes 35S). For miniPTEV, a 32P-labeled band comigrating with the 35S-labeled RT domain was detectable, albeit weakly (lane 3 versus lane 1); no 32P-labeled products were seen at the positions of the clipped-off RT/RH domains for P75TEV (lane 6 versus lane 4) and P1TEV (lane 10 versus lane 8).
Tyr561 deoxynucleotidylation is not essential for DHBV replication.
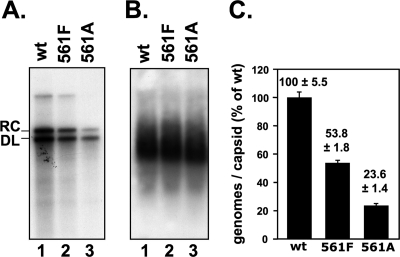
To test for a role of RT deoxynucleotidylation in DHBV replication, we introduced the Y561A and Y561F mutations into replication-competent DHBV vectors and transfected them into LMH cells, in parallel with an analogous vector encoding wt DHBV16 as a reference. At 4 days posttransfection, replicative DNA intermediates were analyzed by Southern blotting of DNA extracted from cytoplasmic lysates (Fig. 9). Genomes carrying either mutation reproducibly supported replication to ca. 50% (Y561F) and 20% (Y561A), respectively, of wt DHBV levels. Hence, deoxynucleotidylation at Y561 is not essential for DHBV replication.
Fig. 9.
Tyr561 deoxynucleotidylation is not essential for DHBV replication. LMH cells were transfected with plasmids encoding wt DHBV or mutant genomes with Tyr561 in P replaced by Phe (561F) or Ala (561A) and analyzed for replication competence and capsid formation. (A) Replicative DNA intermediates. DNA from cytoplasmic nucleocapsids was analyzed by Southern blotting. (B) Capsid formation. Aliquots from cytoplasmic lysates were subjected to native agarose gel electrophoresis and DHBV capsids were detected by immunoblotting against DHBV core protein. (C) Quantification of replicative intermediates. DNA signals from panel A were quantitated by phosphorimaging, and the sum of signals at the rcDNA and dlDNA positions was normalized to the capsid signal from the same transfection. Numbers represent mean values ± the standard deviation from two independent experiments.
DISCUSSION
Using a site specifically cleavable DHBV P protein derivative, miniPTEV, we identified Tyr561 as a hitherto-unknown (“cryptic”) dNMP acceptor site in the RT domain of the protein. Tyr561 is located in the highly conserved box E element that in all RTs of known structure constitutes the “primer grip” motif close to enzymes' active site. In all aspects tested, the reaction at Tyr561 mimicked the well-established priming reaction at Tyr96 in the TP domain. Although not essential for virus replication, Tyr561 deoxynucleotidylation provides the first example of an alternative substrate to Tyr96 that meets the exquisitely complex requirements for forming a catalytically active initiation complex. Below we discuss the validity of our data, the specificity of Tyr561 deoxynucleotidylation, its potential physiological relevance for the virus and the host, and its implications for structural aspects of hepadnaviral protein priming.
miniP faithfully recapitulates authentic priming at Tyr96.
Our results were largely obtained in an in vitro reconstitution system using a recombinant, truncated DHBV P protein that does not require chaperone assistance for enzymatic activity (6, 54). Although we had previously shown that this simplified system recapitulates important aspects of the authentic protein priming reaction, it has recently been reported that a similar truncated P protein, termed miniRT2, can incorporate only a single dNMP residue (54) and displays relaxed specificity for the cognate Dε RNA, especially in the presence of Mn2+ (25). Because Mn2 has long been known to stimulate activity of various polymerases, including protein-priming viral RNA polymerases (30, 31), all our previous priming assays were performed in its presence. We therefore reevaluated the properties of our miniP construct, including bivalent metal ion dependence. MBPminiP was able to synthesize primers longer than one nucleotide but, in line with data from other researchers (25), only in the presence of Mn2+ (Fig. 2D and E). However, we did not see a relaxed template specificity. A detectable priming signal was obtained only with Dε RNA but not HBV ε RNA or excess tRNA; neither was tRNA able to compete with the cognate RNA. A set of Dε RNA variants with mutations at the genuine initiation template, C2576, revealed a strong preference for incorporation of the dNMP that was complementary to this residue, in full support of an authentically templated reaction. The weak signals obtained with noncomplementary dNTPs in the Mn2+-containing reactions may simply reflect the much higher overall priming efficiency, such that proportionally weaker signals in the Mg2+-only reactions would not have been detected. The only exception was the Dε variant C2576G, which supported the incorporation of both dC and dT regardless of the presence or absence of Mn2+. Possibly, in this mutant G2576 can pair with U2604 opposite the bulge (Fig. 1B) such that A2575 now represents the 3′ terminus of the bulge and may become useable as an initiation template. Altogether, these data validate the use of miniP as a simplified mimic of full-length P protein. Hence, we are confident that the data discussed below reflect intrinsic properties of the DHBV P protein and are not artifacts caused by peculiarities of the reconstitution system.
Specificity of RT deoxynucleotidylation for residue Tyr561.
The function of TP residues Tyr96 in DHBV and Tyr63 in HBV P protein as acceptor sites for the first DNA nucleotide is firmly established (24, 55, 62). However, since most previous studies used longer contiguous P protein chains, dNMP incorporation into other sites would have gone unnoticed. Our TEV protease data combined with mapping by chemical cleavage, stability toward acid and alkali, and the abolition of RT labeling by the replacement of Tyr561 with Ala or Phe established this Tyr residue in the RT domain as the major alternative site (Fig. 6B). Tyr561 labeling was not caused by a major folding defect since both the Ala and the Phe variants supported TP in vitro priming (Fig. 6B and 7), as well as viral replication (Fig. 9).
Of note, we reproducibly observed that mutating Tyr96 to Phe did not completely abrogate a signal at the TP position after protease cleavage (e.g., Fig. 4A and 6B). Since Phe lacks modifiable functional groups, alternative acceptor sites must exist also in TP (unless there is misincorporation of Tyr for Phe during overexpression in E. coli). However, the labeling efficiency of these TP sites was routinely 5- to10-fold weaker than that of the RT domain, which therefore is the dominant alternative acceptor.
Mechanism of RT deoxynucleotidylation at Tyr561.
The hallmarks of Tyr96 mediated priming are its dependence on catalytic activity of the RT domain and on Dε RNA, which in turn requires contributions to RNA binding from other TP regions (1, 40). The very same was observed for Tyr561 deoxynucleotidylation; no reaction occurred with noncognate RNA or when RT activity was disabled by the YMHD mutation. The requirement for ε RNA to promote Tyr561 deoxynucleotidylation could simply have reflected the RNA's function as a structural cofactor to bring P protein into an enzymatically active state (46). However, in strict analogy to Tyr96 priming, dNMP transfer to Tyr561 was governed by complementarity to the authentic Dε RNA initiation template at position 2576 (Fig. 4B). This also rules out, for either reaction, that P protein has an intrinsic, template-independent preference for dGTP. Furthermore, mutations in P protein, unless at Tyr96 or Tyr561, had nearly identical impacts on the overall efficiency of both TP and RT labeling (e.g., Fig. 6B). Thus, both Tyr96 and Tyr561 are able to productively occupy the RT's active site such that the phenolic OH group, like the 3′ OH of a nucleic acid primer, is properly lined up so as to attack the α-phosphate of the first dNTP that is base paired with the initiation template in Dε (C2576 in wt Dε RNA), as displayed in Fig. 10 B. Direct structural evidence that the priming amino acid side chain occupies the identical site as the 3′ end of a conventional nucleic acid primer exists, for instance, for the VPg of foot-and-mouth disease virus (16).
Fig. 10.
Mechanistic features of TP priming at Tyr96 and RT Tyr561 deoxynucleotidylation. (A) Schematic comparison of the TP-RT domains in DHBV miniP and full-length P. The RT domain is depicted with palm and fingers containing the dNTP pocket (dGTP) and the active site (YMDD). In full-length P they are connected to the thumb via the primer grip (box E) hairpin which harbors Tyr561. Intramolecular interactions (which may include the RH domain [not shown]) stabilize the hairpin and prevent Tyr561 but not Tyr96 from being maneuvered into the active site. In miniP, the lack of downstream sequences destabilizes the hairpin such that Tyr561 (and Tyr residues in its vicinity) can replace Tyr96. (B) Model of templated dNMP transfer to alternative Tyr acceptor sites. The model illustrates the complex arrangement required for ε-templated protein priming. C2576 in the bulge (only the template region is explicitly depicted) must be positioned so as to specify the identity of the incoming dGTP by base pairing (dotted lines). The triphosphate moiety of dGTP is held in place via interactions with the two bivalent metal ions (Me2+) required for catalysis by all polymerases; in DHBV P they are coordinated by Asp446 (from box A) and Asp513 (the first D in the YMDD motif). Coordination with a free site on the second metal ion brings the phenolic OH group of Tyr96 (red) or Tyr561 (brown) into a proper position for nucleophilic attack on the dGTP α-phosphate, generating a Tyr-dGMP phosphodiester and pyrophosphate (not shown). Successful adaptation of both Tyr96 and Tyr561 into such an arrangement implies that both are in a sufficiently flexible environment. For clarity, the link between the upper and lower stem opposite the bulge is depicted with interrupted dashed lines. (C) Predicted 3D structure of the RT domain in DHBV P protein and positions of RT-intrinsic dNMP acceptor sites. The graphic representation is based on the highest ranking model predicted by the LOMETS metaserver (58). The thumb subdomain deleted in miniP is shown in brown, and the carboxylate oxygens of Asp446 and Asp513 are shown as red dots. The backbone trace of the primer grip hairpin is highlighted in orange; residues Phe558, Tyr561, and Phe567 are shown with their side chains. The ability of all three to act as dNMP substrates in miniP is difficult to envisage without destruction of the hairpin structure. A stereo view of the same model, rotated by 90°, is provided in Fig. S6 in the supplemental material.
Some evidence suggests that the dNMP linked to Tyr561 may be extendable to a limited extent (see Fig. S4 in the supplemental material); however, our current data do not firmly establish such an extension or that the additional nucleotides are templated by the ε bulge. Thus, a potential role of Tyr561 deoxynucleotidylation in virus replication, if any (see below), would probably be confined to the addition of the very first nucleotide.
A structurally flexible environment appears critical for Tyr561 deoxynucleotidylation and probably also for authentic priming at Tyr96.
Despite the presence of six more Tyr residues, RT deoxynucleotidylation occurred exclusively at Tyr561 or at Tyr residues artificially introduced in its immediate vicinity. The simultaneous requirement for catalytic activity in cis suggests spatial proximity of Tyr561 to a properly folded catalytic center as one decisive feature. No direct structural data exist for any hepadnaviral P protein, but the presence of universally conserved motifs (boxes 1/G and 2/F and boxes A to E; see Fig. S1 in the supplemental material) has allowed researchers to generate plausible, HIV-1 RT-based models for the HBV P protein RT domain (3, 15). These models conform to the right-hand architecture consisting of fingers, palm, and thumb domains found in many polymerases (42). For DHBV P protein, our previous mutational analysis had strongly suggested a similar structure of the dNTP pocket as in retroviral RTs (10). That the entire domain adopts a typical RT fold is supported by predictions from the LOMETS metaserver (58). It combines the results from various prediction algorithms into 10 most likely models, the highest ranking of which forms the basis of the structure shown in Fig. 10C (see also Fig. S6 in the supplemental material).
Tyr561 is part of box E (residues 556 to 563), which in HIV-1 RT ternary primer-template complexes contacts the primer close to its 3′ end (37). This primer grip forms a β-hairpin that neighbors the YMDD active site, and a similar structure is predicted for the DHBV P protein box E sequence, as implicated schematically in Fig. 10A and in detail in Fig. 10C. The side chains of Asp446 (from box A) and Asp513 (from the YMDD motif, box C) coordinating the two bivalent metal ions that are essential for catalysis in all polymerases (42) are highlighted as reference points. The α-carbon of Tyr561 would not be far, and yet its side chain would not be suitably oriented to reach into the active site without substantial distortion, if not destruction, of the hairpin structure. The same holds for the side chains of residues 558 and 567. Hence, the ability of all three sites to act as dNMP acceptor (Fig. 7) is most plausibly explained by the entire primer grip motif adopting a flexible state in miniP. The decreased efficiency of RT labeling upon back-addition of the terminal regions deleted from miniPTEV suggests that the lack of stabilizing interactions with the deleted thumb region, and possibly the RH domain, contribute importantly to this enhanced flexibility.
Our initial suspicion that the remotely similar sequence context to Tyr96 was responsible for selective Tyr561 deoxynucleotidylation was not supported by the results obtained with the Tyr558 and Tyr567 mutants, whose sequence context lacks such similarity (Fig. 7A). We also replaced the sequence around Tyr561 by that surrounding Tyr96; while RT deoxynucleotidylation did occur, it appeared to be slightly less efficient than that of the original RT sequence (data not shown). Testing a more comprehensive set of tyrosine-containing sequences for their dNMP acceptor qualities should clarify a potential contribution of primary sequence context. At present, however, we consider proximity to the active site and structural flexibility to be the most important criteria.
The same may then hold true for Tyr96 as the authentic priming substrate (Fig. 10B). Several positive-strand RNA virus VPgs (33) are proposed to be intrinsically disordered (see reference 57 for a recent general review); direct evidence for disorder around the dNMP acceptor site (Ser232) is available for the three-domain TP of the double-stranded DNA bacteriophage phi29 (23). In the priming domain (residues 173 to 266) as such, flexibility is crucial to allow for the diverse structural alterations that accompany its gradual displacement by the growing DNA chain. For hepadnaviral TP, intrinsic disorder is supported by its ability to provide Tyr96 as an acceptor even upon massive N-terminal deletions (with two residues upstream of Tyr96 being sufficient; J. Beck, unpublished data). Clearly, however, both Tyr96 and Tyr561 can access the polymerase active site only in the absence of a rigid, bulky environment that would cause steric clashes with the catalytically active, folded RT structure. Whether deoxynucleotidylation can simultaneously occur at Tyr96 and Tyr561 within the same P protein molecule is currently not known. However, modification of one Tyr residue is not a prerequisite for modification of the other.
Is Tyr561 deoxynucleotidylation physiologically relevant?
RT deoxynucleotidylation could be important for virus replication but also for the virus's relationship with the host. An intriguing possibility, supported by the identical template dependencies of dNMP transfer to Tyr96 and Tyr561 and the strict conservation of a Tyr residue at the Tyr561 equivalent position in all hepadnavirus P proteins, was that Tyr561 with bound dNMP was a covalent intermediate in Tyr96 priming. Such a two-step mechanism had been suspected for the self-uridylylation of poliovirus 3D polymerase, with subsequent transfer to VPg (3B) as a final target, but was experimentally made unlikely (35). Our results are likewise not compatible with a ping-pong mechanism in hepadnaviral protein priming. Mutation of Tyr561 did not abolish in vitro priming at Tyr96, and genomes with substitutions of Tyr561 were replication competent (Fig. 9), albeit with reduced efficiency. For the Y561A mutant this correlates with an increased aggregation tendency of the variant P protein (Y. Wang and M. Nassal, unpublished results), indicating a partial folding problem rather than a specific functional defect. Conversely, dNMP transfer to Tyr561 did not require Tyr96; hence, the two reactions are not directly coupled.
Another option was that Tyr561 acts as alternative true protein priming site, analogously to a recent proposal for a protein-priming potyvirus (33). For hepadnaviruses, however, this is highly unlikely because a Y96F mutant DHBV genome still containing Tyr561 was shown not to support the formation of replicative protein-linked DNA (51, 55). This also excludes a potential role in the replication of the suspected cryptic priming sites in TP. Deoxynucleotidylation of cellular proteins could affect the course of infection; ribonucleotidyl transfer is indeed an important regulatory mechanism. However, DHBV P protein strongly discriminates against using NTPs instead of dNTPs (10). Deoxynucleotidylation appears to be unlikely as well. Even though unpackaged P protein molecules are found in cells (59) and might fortuitously be processed to truncated forms comparable to miniP, dNMP transfer activity would require them to be bound to ε RNA and to be catalytically active. Furthermore, we obtained no evidence for any trans-substrate activity in vitro (see Fig. S5 in the supplemental material).
Conclusions.
In summary, our data revealed Tyr561 in the RT domain of DHBV P protein as a specific alternative dNMP acceptor to Tyr96 in TP. Although not crucial for viral replication, the striking mechanistic similarities with the authentic reaction at Tyr96 suggest deoxynucleotidylation of Tyr561 as a most useful model to promote further understanding of hepadnavirus protein priming, in particular its structural and primary sequence requirements. On the applied side, there is a great need for improved anti-HBV therapeutics that have other targets than the current nucleos(t)ide analogs (28), which exclusively inhibit the polymerase activity of P protein and are prone to resistance development (61). The multifactorial requirements of the priming reaction suggest that it as an exquisitely specific target. The ability of P protein to deoxynucleotidylate an alternative Tyr residue in the presence of the authentic Tyr96 substrate implies that small molecule substrate analogs can be found that efficiently compete with the analogous reaction at Tyr63 in HBV P protein.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grants from the Deutsche Forschungsgemeinschaft (Na 154/4-4 and Na 154/7-3).
We thank Christine Rösler for excellent technical help with the cell culture experiments and Yongxiang Wang for the Y561A mutant pCD16 plasmid, as well as for stimulating discussions of the primer grip motif.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Badtke M. P., Khan I., Cao F., Hu J., Tavis J. E. 2009. An interdomain RNA binding site on the hepadnaviral polymerase that is essential for reverse transcription. Virology 390:130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartenschlager R., Schaller H. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartholomeusz A., Tehan B. G., Chalmers D. K. 2004. Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir. Ther. 9:149–160 [PubMed] [Google Scholar]
- 4. Beck J., Nassal M. 1996. A sensitive procedure for mapping the boundaries of RNA elements binding in vitro translated proteins defines a minimal hepatitis B virus encapsidation signal. Nucleic Acids Res. 24:4364–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck J., Nassal M. 1995. Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res. 23:4954–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beck J., Nassal M. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128–36138 [DOI] [PubMed] [Google Scholar]
- 7. Beck J., Nassal M. 1998. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 18:6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beck J., Nassal M. 2007. Hepatitis B virus replication. World J. Gastroenterol. 13:48–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beck J., Nassal M. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beck J., Vogel M., Nassal M. 2002. dNTP versus NTP discrimination by phenylalanine 451 in duck hepatitis B virus P protein indicates a common structure of the dNTP-binding pocket with other reverse transcriptases. Nucleic Acids Res. 30:1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a. Boregowda R. K., Lin L., Zhu Q., Tian F., Hu J. 2011. Cryptic protein priming sites in two different domains of duck hepatitis B virus reverse transcriptase for initiating DNA synthesis in vitro. J. Virol. 85:7754–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgess R. R. 2009. Refolding solubilized inclusion body proteins. Methods Enzymol. 463:259–282 [DOI] [PubMed] [Google Scholar]
- 12. Cao F., et al. 2005. Identification of an essential molecular contact point on the duck hepatitis B virus reverse transcriptase. J. Virol. 79:10164–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooper J. A., Hunter T. 1981. Changes in protein phosphorylation in Rous sarcoma virus-transformed chicken embryo cells. Mol. Cell. Biol. 1:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crimmins D. L., Mische S. M., Denslow N. D. 2005. Chemical cleavage of proteins in solution. Curr. Protoc. Protein Sci. Chapter11:Unit 11–4 [DOI] [PubMed] [Google Scholar]
- 15. Das K., et al. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrer-Orta C., Agudo R., Domingo E., Verdaguer N. 2009. Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase. Curr. Opin. Struct. Biol. 19:752–758 [DOI] [PubMed] [Google Scholar]
- 17. Galligan J. T., Marchetti S. E., Kennell J. C. 2011. Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed. Mob DNA 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haines K. M., Loeb D. D. 2007. The sequence of the RNA primer and the DNA template influence the initiation of plus-strand DNA synthesis in hepatitis B virus. J. Mol. Biol. 370:471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris D., Yadav P. N., Pandey V. N. 1998. Loss of polymerase activity due to Tyr-to-Phe substitution in the YMDD motif of human immunodeficiency virus type-1 reverse transcriptase is compensated by Met-to-Val substitution within the same motif. Biochemistry 37:9630–9640 [DOI] [PubMed] [Google Scholar]
- 20. Hu J., Boyer M. 2006. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80:2141–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu J., Toft D., Anselmo D., Wang X. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang H., Loeb D. D. 1997. Insertions within epsilon affect synthesis of minus-strand DNA before the template switch for duck hepatitis B virus. J. Virol. 71:5345–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamtekar S., et al. 2006. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 25:1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lanford R. E., Notvall L., Lee H., Beames B. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin L., Wan F., Hu J. 2008. Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions. J. Virol. 82:5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loeb D. D., Hirsch R. C., Ganem D. 1991. Sequence-independent RNA cleavages generate the primers for plus-strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 10:3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nassal M. 2008. Hepatitis B viruses: reverse transcription a different way. Virus Res. 134:235–249 [DOI] [PubMed] [Google Scholar]
- 28. Nassal M. 2009. New insights into HBV replication: new opportunities for improved therapies. Fut. Virol. 4:55–70 [Google Scholar]
- 29. Nassal M., Rieger A. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paul A. V., et al. 2003. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J. Virol. 77:891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poranen M. M., et al. 2008. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 36:6633–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Protzer U., Nassal M., Chiang P. W., Kirschfink M., Schaller H. 1999. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc. Natl. Acad. Sci. U. S. A. 96:10818–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rantalainen K. I., Eskelin K., Tompa P., Makinen K. 2011. Structural flexibility allows the functional diversity of potyvirus genome-linked protein VPg. J. Virol. 85:2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Receveur-Brechot V., Bourhis J. M., Uversky V. N., Canard B., Longhi S. 2006. Assessing protein disorder and induced folding. Proteins 62:24–45 [DOI] [PubMed] [Google Scholar]
- 35. Richards O. C., et al. 2006. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J. Virol. 80:7405–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salas M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39–71 [DOI] [PubMed] [Google Scholar]
- 37. Sarafianos S. G., et al. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidt A., Endres S., Rothenfusser S. 2011. Pattern recognition of viral nucleic acids by RIG-I-like helicases. J. Mol. Med. 89:5–12 [DOI] [PubMed] [Google Scholar]
- 39. Schultz U., Grgacic E., Nassal M. 2004. Duck hepatitis B virus: an invaluable model system for HBV infection. Adv. Virus Res. 63:1–70 [DOI] [PubMed] [Google Scholar]
- 40. Stahl M., Beck J., Nassal M. 2007. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J. Virol. 81:13354–13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stahl M., Retzlaff M., Nassal M., Beck J. 2007. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. 35:6124–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steitz T. A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395–17398 [DOI] [PubMed] [Google Scholar]
- 43. Steitz T. A. 2006. Visualizing polynucleotide polymerase machines at work. EMBO J. 25:3458–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takeuchi O., Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tavis J. E., Ganem D. 1996. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tavis J. E., Massey B., Gong Y. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 72:5789–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tavis J. E., Perri S., Ganem D. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Dijk A. A., Makeyev E. V., Bamford D. H. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85:1077–1093 [DOI] [PubMed] [Google Scholar]
- 49. Vorreiter J., et al. 2007. Monoclonal antibodies providing topological information on the duck hepatitis B virus core protein and avihepadnaviral nucleocapsid structure. J. Virol. 81:13230–13234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walker A., Skamel C., Vorreiter J., Nassal M. 2008. Internal core protein cleavage leaves the hepatitis B virus capsid intact and enhances its capacity for surface display of heterologous whole chain proteins. J. Biol. Chem. 283:33508–33515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang G. H., Seeger C. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang G. H., Seeger C. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663–670 [DOI] [PubMed] [Google Scholar]
- 53. Wang G. H., Zoulim F., Leber E. H., Kitson J., Seeger C. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68:8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X., Qian X., Guo H. C., Hu J. 2003. Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase. J. Virol. 77:4471–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weber M., et al. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wimmer E., Paul A. V. 2010. The making of a picornavirus genome, p. 33–55 In Ehrenfeld E., Domingo E., Roos R. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 57. Wright P. E., Dyson H. J. 2009. Linking folding and binding. Curr. Opin. Struct. Biol. 19:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu S., Zhang Y. 2007. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35:3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yao E., Tavis J. E. 2003. Kinetics of synthesis and turnover of the duck hepatitis B virus reverse transcriptase. J. Biol. Chem. 278:1201–1205 [DOI] [PubMed] [Google Scholar]
- 60. Zhang Z., Tavis J. E. 2006. The duck hepatitis B virus reverse transcriptase functions as a full-length monomer. J. Biol. Chem. 281:35794–35801 [DOI] [PubMed] [Google Scholar]
- 61. Zoulim F., Locarnini S. 2009. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 137(e1-2):1593–1608 [DOI] [PubMed] [Google Scholar]
- 62. Zoulim F., Seeger C. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.