Abstract
The packaging of the adenovirus (Ad) genome into a capsid displays serotype specificity. This specificity has been attributed to viral packaging proteins, the IVa2 protein and the L1-52/55K protein. We previously found that the Ad17 L1-52/55K protein was not able to complement the growth of an Ad5 L1-52/55K mutant virus, whereas two other Ad17 packaging proteins, IVa2 and L4-22K, could complement the growth of Ad5 viruses with mutations in the respective genes. In this report, we investigated why the Ad17 L1-52/55K protein was not able to complement the Ad5 L1-52/55K mutant virus. We demonstrate that the Ad17 L1-52/55K protein binds to the Ad5 IVa2 protein in vitro and the Ad5 packaging domain in vivo, activities previously associated with packaging function. The Ad17 L1-52/55K protein also associates with empty Ad5 capsids. Interestingly, we find that the Ad17 L1-52/55K protein is able to complement the growth of an Ad5 L1-52/55K mutant virus in conjunction with the Ad17 structural protein IIIa. The same result was found with the L1-52/55K and IIIa proteins of several other Ad serotypes, including Ad3 and Ad4. The Ad17 IIIa protein associates with empty Ad5 capsids. Consistent with the complementation results, we find that the IIIa protein interacts with the L1-52/55K protein in vitro and associates with the viral packaging domain in vivo. These results underscore the complex nature of virus assembly and genome encapsidation and provide a new model for how the viral genome may tether to the empty capsid during the encapsidation process.
INTRODUCTION
The adenovirus (Ad) capsid is an icosahedron that contains at least 12 different viral proteins (reviewed in references 14 and 27). The three major proteins, hexon, penton base, and fiber, constitute the facets, vertices, and projections of the particle, respectively. Three viral core proteins (V, VII, and mu) interact with and condense the viral DNA and mediate interactions between the core and the capsid. A series of minor capsid components, IIIa, VI, VIII, and IX, contribute to capsid structure and stability. The viral genome is covalently linked to the terminal protein. The packaging of the Ad genome into the capsid is believed to follow the paradigm of bacteriophage assembly, with the insertion of viral DNA into a preassembled empty capsid (17). After DNA encapsidation, the Ad protease within the virus particle is activated by a peptide from pVI and viral DNA to cleave precursor capsid proteins into the mature forms, an important aspect of Ad maturation and the formation of fully infectious virus particles (12).
Ad viral DNA packaging requires the packaging domain located at the left end of the viral genome between nucleotides 200 and 380 (17). This region contains seven functional packaging elements, known as A repeats, with the consensus sequence 5′-TTTGN8CG-3′ (6, 25). Three Ad proteins, IVa2, L1-52/55K, and L4-22K, are known to associate with the packaging domain in vitro and/or in vivo, and each is required for viral DNA packaging (5, 7, 9, 16, 19–21, 26, 29–32). The IVa2 protein binds to the CG motif and the L4-22K protein binds the TTTG motif of the packaging A repeats in vitro and in vivo (5, 19, 26, 29). The L1-52/55K protein has only been found to be associated with the packaging domain in vivo, and the L1-52/55K protein associates with IVa2 in vitro (8, 19, 21). The IVa2 and L1-52/55K proteins are required for packaging of Ad DNA into the capsid (7, 9, 32). IVa2 mutant viruses produce empty virus particles devoid of viral DNA (15). Depending on the mutant, L1-52/55K mutant viruses produce empty capsids devoid of viral DNA (7) or containing left-end Ad DNA sequences (9). Interestingly, a temperature-sensitive mutant in the IIIa capsid protein (ts112) has a similar phenotype when grown at the restrictive temperature (3). An L4-22K mutant virus displays a complex phenotype with defects in viral late gene expression, as well as the production of infectious virus particles, consistent with a role for the L4-22K protein in the packaging of viral DNA (13, 16). While these cis-acting DNA sequences and trans-acting viral proteins involved in Ad DNA packaging have been identified, the mechanisms of how the Ad particle is assembled and how the viral DNA is packaged are poorly understood.
More than 50 human Ad serotypes have been identified. Based on the ability to agglutinate red blood cells, serological cross-reactivity, and viral genome homology, these serotypes are further classified into six subgroups, A to F (1). The pseudopackaging of the genome of one Ad serotype into the capsid of another Ad serotype is generally inefficient except with highly related viruses (18). The Ad IVa2 and L1-52/55K proteins have been implicated in the serotype specificity of Ad DNA packaging (28, 32). In an earlier study, we found that the Ad17 L1-52/55K protein was not able to substitute for the Ad5 L1-52/55K protein in an infection with an Ad5 L1-52/55K mutant virus (28). In contrast, both the Ad17 IVa2 and L4-22K proteins were able to functionally substitute for their Ad5 counterparts (28). These results suggested the possibility that the serotype specificity is determined by the interaction between the L1-52/55K protein and the Ad packaging domain and/or capsid components during virus assembly.
In this report, we investigated why the Ad17 L1-52/55K protein was not able to complement the Ad5 L1-52/55K mutant virus. We demonstrate that the Ad17 L1-52/55K protein binds to the Ad5 IVa2 protein in vitro and the Ad5 packaging domain in vivo, activities previously associated with packaging function. The Ad17 L1-52/55K protein also associates with empty Ad5 capsids. Interestingly, we find that the Ad17 L1-52/55K protein is able to complement the growth of an Ad5 L1-52/55K mutant virus in conjunction with the Ad17 structural protein IIIa. The same result was found with the L1-52/55K and IIIa proteins of several other Ad serotypes, including Ad3 and Ad4. Consistent with the complementation results, we find that the IIIa protein interacts with the L1-52/55K protein in vitro and associates with the viral packaging domain in vivo. These results underscore the complex nature of virus assembly and genome encapsidation and provide a new model for how the viral genome may tether to the empty capsid during the encapsidation process.
MATERIALS AND METHODS
Cells and viruses.
293 cells were grown in Dulbecco's modified Eagle's minimal essential medium (DMEM) (Gibco) supplemented with 10% bovine calf serum (HyClone). 293 cells stably expressing the Ad5 L1-52/55K protein were grown in DMEM supplemented with 10% fetal bovine serum in the presence of Geneticin at 500 μg/ml (7). A new 293 cell line that stably expresses the Ad17 L1-52/55K protein was isolated following selection with Geneticin as described for 293-L1. The mutant virus pm8001 was propagated in 293-L1 cells (7). The temperature-sensitive mutant ts147 (10) was propagated at the permissive temperature 32°C. Virus particles were isolated by cesium chloride equilibrium gradient centrifugation and quantified as previously described (4).
Expression vectors, antibodies, protein coprecipitation, and chromatin immunoprecipitation (ChIP).
Ad5 IVa2 protein was expressed using vector pcDNA3 (Invitrogen). Ad5 and Ad17 L1-52/55K proteins were expressed using vector pcDNA3 and contained a C-terminal Strep tag (WSHPNFEL). Ad5 and Ad17 IIIa genes were amplified by PCR and cloned into pEF-1 (Invitrogen) to introduce a C-terminal V5 epitope tag. The Ad3, Ad4, and Ad12 L1-52/55K and IIIa genes were introduced into the same expression vectors described above. The chimeric Ad5/17 and Ad17/5 IIIa constructs were generated by PCR with breakpoints for the chimeras at amino acid 410 in Ad5 IIIa and at the corresponding position in the Ad17 IIIa protein.
Ad5 protein IIIa amino acids 1 to 438 were expressed in Escherichia coli Rosetta strain, and purified IIIa protein was used to immunize rabbits (Lampire Biologicals, PA) to generate a polyclonal anti-IIIa antibody. Rabbit polyclonal antibodies to the Ad5 L1-52/55K and IVa2 proteins were previously described (19). Other antibodies used in these studies were from commercial sources: anti-V5 tag antibody (Invitrogen) and anti-Strep tag antibody (IBA Tagnology).
Transfections were performed using Fugene 6 transfection reagent (Roche) according to the manufacturer's instructions. For L1-52/55K-IVa2 interaction assays, 293 cells were transfected with a IVa2 expression vector alone or cotransfected with IVa2 and L1-52/55K expression vectors. Two days after transfection, cells were harvested and lysed in F lysis buffer (10 mM Tris, pH 7.4, 50 mM NaCl, 10% glycerol, 0.5% Triton X-100). Ad5 and Ad17 Strep-tagged L1-52/55K proteins were precipitated using Strep-Tactin beads, and the beads were washed using F lysis buffer. The final pellet was resuspended in SDS sample buffer and boiled. IVa2 protein was detected using anti-IVa2 polyclonal rabbit antibody, and the L1-52/55K proteins were detected using anti-Strep antibody. For L1-52/55K-IIIa interaction assays, 293 cells were transfected with a V5-tagged IIIa expression vector alone or cotransfected with Strep-tagged L1-52/55K expression vectors. The extracts were processed as described above using Strep-Tactin beads for precipitation or using anti-V5 tag antibody and protein A-Sepharose beads for immunoprecipitation. Protein IIIa was detected by Western blotting using anti-IIIa antibody or anti-V5 tag antibody, and the L1-52/55K protein was detected using anti-Strep antibody or anti-L1-52/55K antibody, as indicated in the figures.
Chromatin immunoprecipitation experiments were performed as previously described (19). 293 cells that expressed the Ad5 or Ad17 L1-52/55K protein were infected with pm8001 virus at 100 particles/cell, or 293 cells were infected with ts147 virus at 100 particles/cell. Eighteen to 24 h postinfection (h.p.i.), cells were processed for ChIP experiments. The serum-containing medium was removed from the cell monolayer, and serum-free medium was added. Formaldehyde was added to a final concentration of 1% and incubated for 10 min at 37°C, followed by the addition of glycine to 125 mM at room temperature for 5 min. Cells were washed twice with an ice-cold phosphate-buffered saline solution, and the cells were detached by scraping into phosphate-buffered saline and then pelleted at 1,000 × g for 4 min at 4°C. The cell pellet was resuspended in SDS lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS). Chromatin was sheared by sonication (Branson Sonifier 450) with 3 sets of 20- to 30-s pulses, output 7, on ice. Samples were centrifuged at 16,000 × g for 10 min at 4°C. Amounts of 50 to 100 μg of sheared chromatin were immunoprecipitated using anti-L1-52/55K, anti-IVa2, or anti-IIIa polyclonal rabbit antibodies. Immunoprecipitates were washed as described previously (19). Samples were analyzed by quantitative PCR using Ad5 packaging domain-specific primers (19), as well as internal primer pairs far removed from the packaging domain (approximately at Ad5 nucleotides 9000 and 36000) with a Fitzyme quantitative PCR (qPCR) kit (Fitzyme). The binding of proteins to packaging domain sequences was normalized to binding to the internal or right-end Ad5 sequences, and the data are represented as the fold enrichment of protein binding to packaging domain sequences.
Mutant virus complementation assay.
293 cells were transfected with vectors expressing Ad5 or Ad17 L1-52/55K proteins alone or in combination with expression vectors containing the Ad5 or Ad17 protein IIIa genes. Following transfection, cells were infected with the L1-52/55K mutant virus pm8001. Three days after infection, cell lysates were prepared and infectious virus yields were determined by plaque assay using 293-L1 cells.
RESULTS
The Ad17 L1-52/55K protein interacts with the Ad5 IVa2 protein in vitro and the Ad5 packaging domain in vivo.
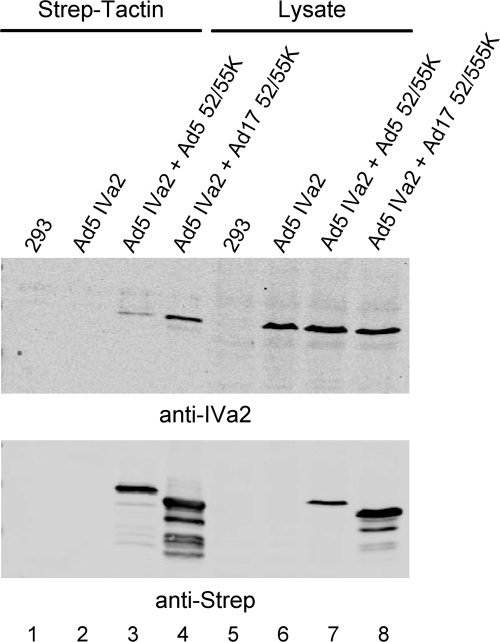
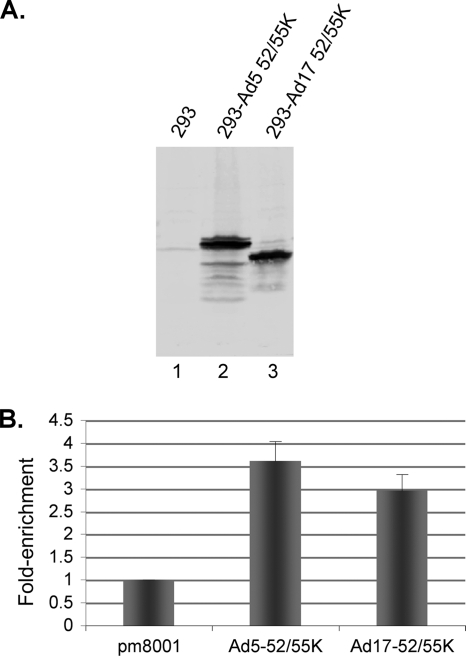
The Ad5 (Ad subgroup C) L1-52/55K and IVa2 proteins interact when analyzed by coimmunoprecipitation in vitro (8). To determine if the inability of the Ad17 (Ad subgroup D) L1-52/55K protein to complement the growth of the Ad5 L1-52/55K mutant virus pm8001 reflected the inability of the Ad17 L1-52/55K protein to bind the Ad5 IVa2 protein, the interaction of these two proteins was examined by coprecipitation. 293 cells were cotransfected with vectors that expressed the Ad5 IVa2 protein and Strep-tagged versions of the Ad5 or Ad17 L1-52/55K protein. Cell lysates were incubated with Strep-Tactin beads to bind the tagged L1-52/55K proteins, the beads were washed, and coprecipitation of Ad5 IVa2 was examined by Western blot analysis (Fig. 1). The Ad17 L1-52/55K protein was found to bind the Ad5 IVa2 protein to a similar extent as the Ad5 L1-52/55K protein when adjusted for L1-52/55K protein expression levels (Fig. 1, top and bottom, lanes 3, 4, 7, and 8). The Ad5 L1-52/55K protein binds to the packaging domain in vivo as analyzed using ChIP assays (19, 21). To investigate whether the Ad17 L1-52/55K protein binds to the Ad5 packaging domain, we developed a new 293 cell line that stably expresses the Ad17 L1-52/55K protein. Comparable levels of the Ad5 and Ad17 L1-52/55K proteins were expressed in the previously described 293-Ad5-L1-52/55K cell line (7) and the new 293-Ad17-L1-52/55K cell line (Fig. 2A). 293-Ad5-L1 and 293-Ad17-L1 cells were infected with the L1-52/55K mutant virus pm8001 (7), and at 18 h.p.i., ChIP assays were performed using anti-L1-52/55K antiserum. Samples were analyzed by quantitative PCR using Ad5 packaging domain-specific primers (19), as well as internal primer pairs far removed from the packaging domain (approximately at Ad5 nucleotides 9000 and 36000). The binding of proteins to packaging domain sequences was normalized to binding to the internal or right-end Ad5 sequences, and the data are represented as the fold enrichment of protein binding to packaging domain sequences. These results (Fig. 2B) demonstrated that the Ad17 L1-52/55K protein was able to bind the Ad5 packaging domain in vivo in pm8001-infected cells. These results indicate that the inability of the Ad17 L1-52/55K protein to complement the Ad5 L1-52/55K mutant virus pm8001 is not due to the inability of the Ad17 L1-52/55K protein to bind the Ad5 IVa2 protein or the Ad5 packaging domain but, rather, that some other interaction may be missing.
Fig. 1.
The Ad17 L1-52/55K protein binds to the Ad5 IVa2 protein in vitro. 293 cells were transfected with a vector that expressed the Ad5 IVa2 protein (lanes 2 and 6) or cotransfected with vectors that expressed IVa2 and Strep-tagged versions of the Ad5 or Ad17 L1-52/55K protein (lanes 3, 4, 7, and 8). Lanes 1 and 5 correspond to untransfected 293 cells. Cell lysates were incubated with Strep-Tactin beads to bind the tagged L1-52/55K proteins, the beads were washed, and coprecipitation of Ad5 IVa2 was examined by Western blot analysis using antibodies against IVa2 (top) and the Strep tag (bottom). Lanes 1 to 4 show the precipitated proteins. Lanes 5 to 8 show protein expression levels with equal concentrations of cell extract.
Fig. 2.
The Ad17 protein binds to the Ad5 packaging domain in vivo. (A) Western blot analysis shows L1-52/55K protein expression levels with equal concentrations of cell extract from 293-Ad5-L1 (lane 2) and 293-Ad17-L1 (lane 3) cells. (B) 293-Ad5-L1 and 293-Ad17-L1 cells were infected with the L1-52/55K mutant virus pm8001 (7), and ChIP assays were performed using anti-L1 antiserum. Immunoprecipitation of the Ad5 packaging domain was quantified by qPCR. The quantification of viral DNA captured by ChIP using 293 cells infected with pm8001 but without L1-52/55K protein expression was set at 1. Binding of proteins to packaging domain sequences was normalized to binding to the internal or right-end Ad5 sequences, and the data are represented as the fold enrichment of protein binding to packaging domain sequences. Error bars show standard deviations.
The Ad17 L1-52/55K and IIIa proteins complement the growth of Ad5 L1-52/55K mutant virus pm8001.
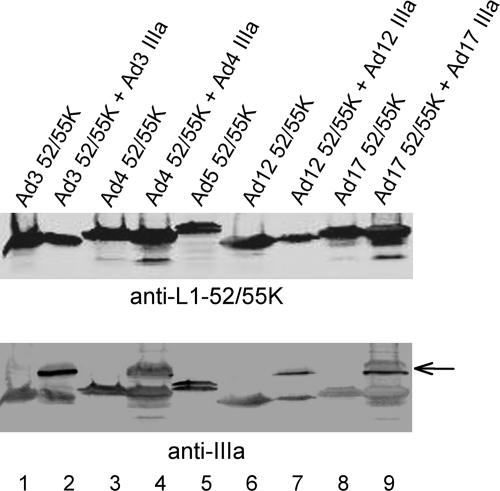
The Ad IIIa protein is a well-known structural protein in the virus capsid that is located underneath the vertices of the penton base and connects to the peripentonal hexons (11, 22–24). Interestingly, the growth of a protein IIIa temperature-sensitive mutant virus (ts112) at the nonpermissive temperature results in the formation of light intermediate virus particles (3), the same result found with L1-52/55K mutant viruses (7, 9). This prompted us to examine the effect of expressing the Ad17 IIIa protein in conjunction with the Ad17 L1-52/55K protein in a complementation assay with Ad5 L1-52/55K mutant virus pm8001 (Table 1). Interestingly, the coexpression of the Ad17 IIIa and Ad17 L1-52/55K proteins rescued the growth of pm8001, whereas the expression of the Ad17 L1-52/55K protein alone or the coexpression of the Ad17 L1-52/55K protein with the Ad5 IIIa protein did not. The latter result was anticipated since the Ad5 IIIa protein was already expressed in infections with the Ad5 mutant virus pm8001. We note that the level of complementation using the Ad17 L1-52/55K and IIIa proteins is only ∼25% of that observed using the corresponding Ad5 proteins, possibly because the heterologous IIIa protein is in competition with the endogenous Ad5 IIIa protein. To begin to investigate which sequences in the Ad17 IIIa protein mediate the complementation of mutant virus pm8001 in conjunction with the Ad17 L1-52/55K protein, we generated two Ad5 and Ad17 IIIa chimeric proteins that contained the N terminus of Ad5 protein IIIa fused to the C terminus of Ad17 protein IIIa (Ad5/17 IIIa) and the N terminus of Ad17 protein IIIa fused to the C terminus of Ad5 protein IIIa (Ad17/5 IIIa). The Ad17/5 chimeric IIIa protein was able to complement the growth of mutant virus pm8001, whereas the Ad5/17 IIIa protein was not (Table 1). Next, we examined whether the same result could be achieved with other Ad serotypes. The L1-52/55K proteins of Ad3 (Ad subgroup B) and Ad4 (Ad subgroup E) alone could not rescue the growth of Ad5 mutant virus pm8001, whereas in both cases, virus rescue was achieved when the respective IIIa proteins were coexpressed in the assays (Table 1). The L1-52/55K and IIIa proteins of Ad12 (Ad subgroup A), however, were not able to rescue the growth of mutant virus pm8001 in this assay (Table 1). Similar levels of the Ad5, Ad3, and Ad4 L1-52/55K and IIIa proteins were expressed in these assays (Fig. 3), although we cannot exclude the possibility that the anti-Ad5 polyclonal antisera used in these experiments may have different affinities for the L1-52/55K and IIIa proteins of the different Ad serotypes. The Ad12 IIIa protein did not express as well as the IIIa proteins of the other Ad serotypes in transfected cells, which makes the interpretation of the complementation result difficult.
Table 1.
L1-52/55K and IIIa proteins of different Ad serotypes complement the growth of the Ad5 L1-52/55K mutant virus pm8001
| Protein(s) expresseda | Virus yield |
|---|---|
| None | 0 |
| Ad5 52/55K | 13,267 |
| Ad5 52/55K + Ad5 IIIa | 8,633 |
| Ad17 52/55K | 0 |
| Ad17 52/55K + Ad5 IIIa | 0 |
| Ad17 52/55K + Ad17 IIIa | 2,267 |
| Ad17 52/55K + Ad5/17 IIIa | 0 |
| Ad17 52/55K + Ad17/5 IIIa | 1,500 |
| Ad3 52/55K | 0 |
| Ad3 52/55K + Ad3 IIIa | 3,533 |
| Ad4 52/55K | 0 |
| Ad4 52/55K + Ad4 IIIa | 7,433 |
| Ad12 52/55K | 0 |
| Ad12 52/55K + Ad12 IIIa | 0 |
293 cells were transfected overnight with vectors that expressed the L1-52/55K proteins of different Ad serotypes alone or in combination with vectors that expressed the IIIa proteins of the respective serotypes or Ad5/17 IIIa chimeric proteins. The next day, transfected cells were infected with mutant virus pm8001. Cell lysates were prepared 72 h.p.i., and infectious virus yields were quantified by plaque assay on the pm8001 complementing cell line 293-L1. The results represent the total infectious virus yields.
Fig. 3.
L1-52/55K and IIIa protein expression levels in transfected cells. 293 cells were transfected with vectors that expressed the Ad L1-52/55K proteins of different Ad serotypes without (lanes 1, 3, 5, 6, and 8) or with (lanes 2, 4, 7, and 9) cotransfection of vectors to express the corresponding IIIa proteins of the respective serotypes. The top blot was probed with anti-L1-52/55K polyclonal rabbit antiserum. The same blot was reprobed with anti-IIIa polyclonal rabbit antibody (bottom). The IIIa proteins are indicated by an arrow.
The Ad5 L1-52/55K and IIIa proteins interact in vitro and the IIIa protein binds to the packaging domain in vivo.
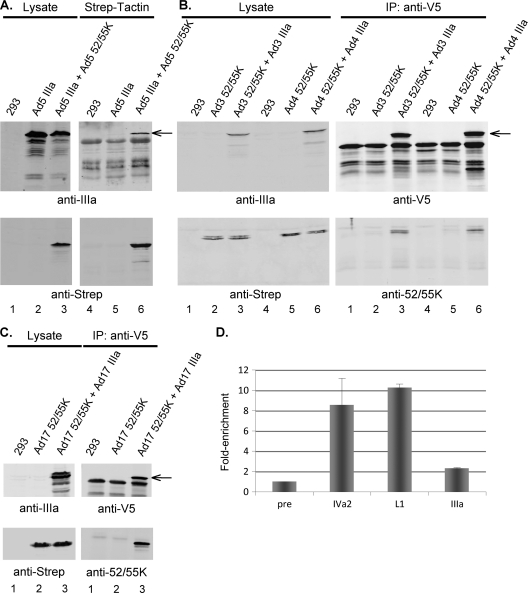
We examined whether the Ad5 L1-52/55K and IIIa proteins interact with each other by coprecipitation analysis. 293 cells were transfected with a vector to express V5-tagged IIIa protein without or with cotransfection with a vector to express the Strep-tagged L1-52/55K protein. L1-52/55K protein was precipitated from cell lysates using Strep-Tactin beads, the beads were washed, and precipitation of the IIIa protein was examined by Western blot analysis (Fig. 4A). These results demonstrated that the Ad5 IIIa and L1-52/55K proteins interact in vitro. We also analyzed whether the IIIa and L1-52/55K proteins of other Ad serotypes interact. 293 cells were transfected with a vector to express V5-tagged IIIa proteins of Ad3, Ad4, or Ad17 without or with cotransfection with a vector to express the corresponding Strep-tagged L1-52/55K protein. IIIa proteins were immunoprecipitated from cell lysates using anti-V5 tag antibody and protein A-Sepharose beads, the beads were washed, and coprecipitation of the L1-52/55K proteins was examined by Western blot analysis (Fig. 4B and C). These results demonstrated that the IIIa and L1-52/55K proteins of different Ad serotypes interact in vitro.
Fig. 4.
The IIIa and L1-52/55K proteins of different Ad serotypes interact in vitro, and the Ad5 IIIa protein binds the packaging domain in vivo. (A) 293 cells were transfected with a vector to express Ad5 V5-tagged IIIa protein (lanes 2, 3, 5, and 6) without (lanes 2 and 5) or with cotransfection with a vector to express the Strep-tagged Ad5 L1-52/55K protein (lanes 3 and 6). Cell lysates were precipitated using Strep-Tactin beads, the beads were washed, and coprecipitation of protein IIIa with the L1-52/55K protein was examined by Western blot analysis using antibodies against IIIa (top) and the Strep tag (bottom). Lanes 1 to 3 show protein expression levels with equal concentrations of cell extract. Lanes 4 to 6 show the precipitated proteins. The arrow indicates the IIIa protein (top). (B) 293 cells were transfected with vectors to express Ad3 or Ad4 Strep-tagged L1-52/55K proteins (lanes 2, 3, 5, and 6) without (lanes 2 and 5) or with cotransfection with a vector to express the V5-tagged IIIa protein of the corresponding Ad serotype (lanes 3 and 6). IIIa proteins were immunoprecipitated (IP) from cell lysates using anti-V5 tag antibody and protein A-Sepharose beads, the beads were washed, and coprecipitation of the L1-52/55K protein with protein IIIa was examined by Western blot analysis using antibodies against the IIIa protein or the V5 tag (top) and the Strep tag or the L1-52/55K protein (bottom). The left panels show protein expression levels with equal concentrations of cell extract. The right panels show the precipitated proteins. The arrow indicates the IIIa protein (top). (C) Coimmunoprecipitation of the Ad17 IIIa and L1-52/55K proteins was examined as described for panel B. (D) 293 cells were infected with the mutant virus ts147 at the restrictive temperature. Infected cells were processed for ChIP using preimmune rabbit serum or polyclonal rabbit antibodies directed against the IVa2, L1-52/55K, and IIIa proteins. The quantification of viral DNA captured by ChIP using preimmune serum was set at 1. Binding of proteins to packaging domain sequences was normalized to binding to the internal or right-end Ad5 sequences, and the data are represented as fold enrichment of protein binding to packaging domain sequences. Error bars show standard deviations.
We next examined, using ChIP, whether the Ad5 IIIa protein binds to the packaging domain. The temperature-sensitive mutant ts147 was used for this analysis. ts147 contains a mutation in the hexon gene that blocks capsid assembly at the restrictive temperature (10). This mutant was used to attempt to enrich for unpackaged viral genomes with bound proteins. We previously verified that the results obtained using mutant ts147 were also observed using wild-type Ad5 (18). 293 cells were infected with ts147 and incubated at the restrictive temperature (40°C). At 18 h.p.i., the cells were processed for ChIP. Preimmune serum and antibodies directed against the Ad5 IVa2, L1-52/55K, and IIIa proteins were used for immunoprecipitation. Immunoprecipitation of the Ad5 packaging domain was quantified by qPCR as described above. These results (Fig. 4D) demonstrated that the IIIa protein bound to the packaging domain in vivo; we note that while the binding was modest, it was statistically significant (P value = 0.033). We do not know if these results reflect a lower level of IIIa protein binding to packaging domain sequences in vivo in comparison to the binding of IVa2 and L1-52/55K proteins or a limitation of the anti-IIIa antibody used in the analysis.
The Ad17 L1-52/55K and Ad17 IIIa proteins enter empty virus particles independently.
The Ad5 L1-52/55K protein is found in empty virus particles that lack viral DNA (7), showing that while this protein does bind to the packaging sequences, a pool of the L1-52/55K protein behaves in an independent manner. We examined whether the Ad17 L1-52/55K protein or Ad17 IIIa protein could enter empty Ad5 virus particles independently of each other. Mutant virus pm8001 was used to infect 293-Ad5-L1 or 293-Ad17-L1 cells that express the Ad5 or Ad17 L1-52/55K proteins, respectively, or used to infect 293 cells that were transfected with a vector to express V5-tagged Ad17 IIIa protein. Empty virus particles were purified on CsCl equilibrium gradients and analyzed by Western blotting (Fig. 5). The Ad5 and Ad17 L1-52/55K proteins were found in pm8001 empty virus particles (lanes 1 and 2). The Ad17 IIIa protein also was found in pm8001 empty virus particles in the absence of the L1-52/55K protein (lane 4). From these results, we conclude that the Ad17 L1-52/55K protein is not required for the Ad17 IIIa protein to enter into an empty virus particle, nor is the Ad17 IIIa protein required for the Ad17 L1-52/55K protein to enter empty virus particles.
Fig. 5.
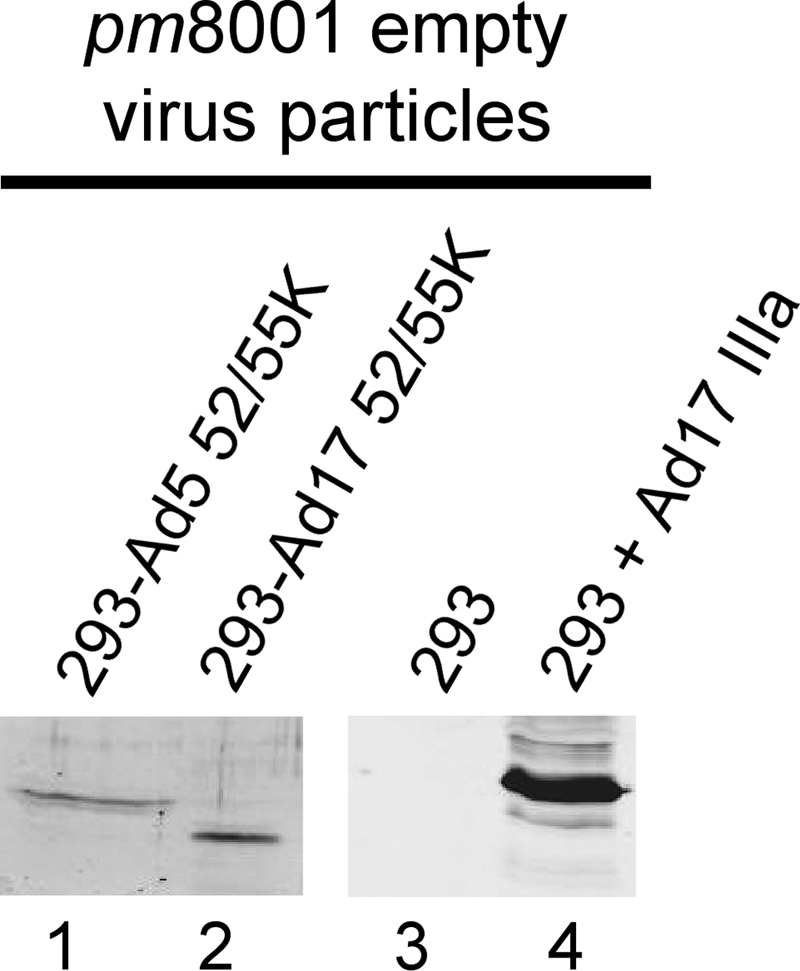
The Ad17 L1-52/55K and Ad17 IIIa proteins are found in empty Ad5 virus particles. 293 cells were transfected with vectors to express the Ad5 (lane 1) or Ad17 (lane 2) L1-52/55K proteins or with a vector to express V5 epitope-tagged Ad17 IIIa protein (lane 4). Lane 3 corresponds to untransfected 293 cells. Cells were infected with mutant virus pm8001, and empty virus particles were subsequently purified by CsCl equilibrium centrifugation. Purified virus particles were analyzed by Western blotting using polyclonal rabbit antibody directed against the Ad5 L1-52/55K protein (lanes 1 and 2) or anti-V5 tag monoclonal mouse antibody (lanes 3 and 4). Empty virus particles were quantified by Western blotting for hexon (lanes 1 and 2) or by using a protein determination assay (lanes 3 and 4). The particle load was within 2-fold for each sample.
DISCUSSION
We previously demonstrated that the L1-52/55K protein is involved in the serotype specificity of Ad genome packaging (28). In this analysis, we assayed for the ability of the packaging proteins of Ad17 to complement the growth of Ad5 viral mutants with mutations in the respective genes. In another study, using a chimeric virus that contained the inverted terminal repeats and packaging domain of Ad5 fused with the rest of the Ad7 genome, a role for the IVa2 protein in serotype-specific viral genome packaging was shown (32). We believe that the different conclusions of these reports reflect the very different ways in which the assays were conducted. In the current report, we suggest an even more complex picture of how Ad DNA is packaged, with the demonstration of a role for the capsid structural protein IIIa in the serotype specificity of Ad genome packaging.
We probed the reason(s) why the Ad17 L1-52/55K protein was not able to complement the growth of the Ad5 L1-52/55K mutant virus pm8001. We conclude from these analyses that the Ad17 L1-52/55K protein is able to bind to the Ad5 IVa2 protein in vitro, as well as to the Ad5 packaging domain in vivo (Fig. 1 and 2). How the L1-52/55K protein is recruited to the packaging domain is not known at present, although the interaction with the IVa2 protein is not required (21). The Ad17 L1-52/55K protein could enter empty Ad5 virus particles apparently as efficiently as the Ad5 L1-52/55K protein (Fig. 5). These results suggested the possibility of a serotype-specific missing component that may bridge the viral genome with the empty capsid. Our results suggest that this component is the viral structural protein IIIa. The IIIa proteins of Ad3 (Ad subgroup B), Ad4 (Ad subgroup E), and Ad17 (Ad subgroup D) could complement the growth of the L1-52/55K mutant virus pm8001 (Ad subgroup C) in conjunction with the L1-52/55K protein of the corresponding serotype (Table 1). The Ad12 (Ad subgroup A) L1-52/55K and IIIa proteins were not able to complement pm8001, which may reflect reduced Ad12 IIIa protein expression in the transfection assays. Alternatively, Bailey and Mautner compiled a phylogenetic tree between different Ad serotypes and showed that among these, subgroups A (Ad12) and F are the most distinct from the other viruses (1). This may represent the basis for our observation.
These results predicted that the L1-52/55K and IIIa proteins may interact, and the L1-52/55K and IIIa proteins of different Ad serotypes were found to coprecipitate with one another in vitro (Fig. 4A to C). We also found, using ChIP assays, that the Ad5 IIIa protein was bound to the Ad5 packaging domain in vivo (Fig. 4D). Whether or not this requires the L1-52/55K protein is under investigation. We envision two scenarios that may explain the observations that the L1-52/55K, IVa2, and L4-22K packaging proteins, as well as the IIIa structural protein, are found both in empty capsids devoid of viral DNA (7, 9, 15, 31) and bound to viral packaging sequences (19, 21) (Fig. 4D), showing both DNA-independent and DNA-dependent localization. First, there may be two separate pools of these proteins where capsid-associated components may be poised as a portal vertex, for example, ready to receive viral DNA for encapsidation. A second pool of these proteins may be bound to packaging sequences and promote an interaction between the viral DNA and capsid components. Alternatively, these viral proteins may be associated with empty capsids and interact with viral packaging sequences when viral DNA docks with the empty capsid. The latter idea is an attractive model, but it is not suggested by the results of experiments that we previously reported (19). With prior ChIP analyses that demonstrated the binding of the L1-52/55K and IVa2 proteins to the packaging domain in vivo, we utilized the viral mutant ts147 in some of these assays. This temperature-sensitive mutant produces no virus capsids at the restrictive temperature (10), and yet, we found the L1-52/55K and IVa2 proteins bound to the packaging domain under these conditions. This supports the idea that there may be separate pools of these packaging and structural proteins that help to bring together viral DNA with the empty capsid.
Elegant structural studies have demonstrated that the structural protein IIIa within the virus particle is located underneath the vertices of the penton base and connects to the peripentonal hexons (11, 22–24). Recently, it was reported that the IVa2 protein is located at a unique capsid vertex that may represent a portal for genome encapsidation (2). In such a situation, part of the IIIa protein could be situated at a portal vertex and available for protein-protein interaction with components outside the capsid. We speculate that such an interaction may explain the roles of the IIIa protein as a structural component of the capsid and in viral DNA packaging. Our data would suggest an interaction with the L1-52/55K protein at this position of the IIIa protein. The orientation of the IIIa protein within the virus particle is such that the N terminus is situated below the penton base at the vertices. This is consistent with the pm8001 complementation data with the Ad5/17 chimeric IIIa proteins where the N terminus but not the C terminus of the Ad17 IIIa protein is required for pm8001 complementation in conjunction with the Ad17 L1-52/55K protein (Table 1). The N terminus of IIIa is the region that may be accessible to components outside the capsid at a portal vertex.
Finally, the region in the L1-52/55K protein that interacts with the IVa2 protein is located within the N-terminal 173 amino acids (20). A comparison between these sequences of the Ad5 and Ad17 L1-52/55K proteins shows that the major differences are located within the N-terminal 60 amino acids of Ad5, which are truncated to 35 amino acids in Ad17. Since the Ad17 L1-52/55K protein interacts with the Ad5 IVa2 protein, this suggests that the region of interaction is between Ad5 L1-52/55K amino acids 61 and 173.
ACKNOWLEDGMENTS
We thank Michael Imperiale (University of Michigan) for the 293-L1 cell line and mutant virus pm8001. We thank members of our laboratory for informed discussions and Ilana Shoshani for excellent technical help.
This work was supported by NIH grant AI041636.
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Bailey A., Mautner V. 1994. Phylogenetic relationships among adenovirus serotypes. Virology 205:438–452 [DOI] [PubMed] [Google Scholar]
- 2. Christensen J. B., et al. 2008. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 82:9086–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Halluin J. C., Milleville M., Boulanger P. A., Martin G. R. 1978. Temperature-sensitive mutant of adenovirus type 2 blocked in virion assembly: accumulation of light intermediate particles. J. Virol. 26:344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans J. D., Hearing P. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ewing S. G., Byrd S. A., Christensen J. B., Tyler R. E., Imperiale M. J. 2007. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 81:12450–12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grable M., Hearing P. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gustin K. E., Imperiale M. J. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gustin K. E., Lutz P., Imperiale M. J. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasson T. B., Soloway P. D., Ornelles D. A., Doerfler W., Shenk T. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kauffman R. S., Ginsberg H. S. 1976. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol. 19:643–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H., et al. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mangel W. F., Toledo D. L., Ding J., Sweet R. M., McGrath W. J. 1997. Temporal and spatial control of the adenovirus proteinase by both a peptide and the viral DNA. Trends Biochem. Sci. 22:393–398 [DOI] [PubMed] [Google Scholar]
- 13. Morris S. J., Leppard K. N. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nemerow G. R., Pache L., Reddy V., Stewart P. L. 2009. Insights into adenovirus host cell interactions from structural studies. Virology 384:380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ostapchuk P., Almond M., Hearing P. 2011. Characterization of empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 85:5524–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ostapchuk P., Anderson M. E., Chandrasekhar S., Hearing P. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ostapchuk P., Hearing P. 2005. Control of adenovirus packaging. J. Cell Biochem. 96:25–35 [DOI] [PubMed] [Google Scholar]
- 18. Ostapchuk P., Hearing P. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ostapchuk P., Yang J., Auffarth E., Hearing P. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perez-Romero P., Gustin K. E., Imperiale M. J. 2006. Dependence of the encapsidation function of the adenovirus L1 52/55-kilodalton protein on its ability to bind the packaging sequence. J. Virol. 80:1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perez-Romero P., Tyler R. E., Abend J. R., Dus M., Imperiale M. J. 2005. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 79:2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reddy V. S., Natchiar S. K., Stewart P. L., Nemerow G. R. 2010. Crystal structure of human adenovirus at 3.5 A resolution. Science 329:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saban S. D., Silvestry M., Nemerow G. R., Stewart P. L. 2006. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 80:12049–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. San Martin C., et al. 2008. Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. J. Mol. Biol. 383:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmid S. I., Hearing P. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyler R. E., Ewing S. G., Imperiale M. J. 2007. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 81:3447–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vellinga J., Van der Heijdt S., Hoeben R. C. 2005. The adenovirus capsid: major progress in minor proteins. J. Gen. Virol. 86:1581–1588 [DOI] [PubMed] [Google Scholar]
- 28. Wohl B. P., Hearing P. 2008. Role for the L1-52/55K protein in the serotype specificity of adenovirus DNA packaging. J. Virol. 82:5089–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang T. C., Yang Q., Maluf N. K. 2009. Interaction of the adenoviral IVa2 protein with a truncated viral DNA packaging sequence. Biophys. Chem. 140:78–90 [DOI] [PubMed] [Google Scholar]
- 30. Zhang W., Imperiale M. J. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang W., Imperiale M. J. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang W., Low J. A., Christensen J. B., Imperiale M. J. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446–10454 [DOI] [PMC free article] [PubMed] [Google Scholar]