Abstract
Respiratory syncytial virus (RSV) invades host cells via a type I fusion (F) glycoprotein that undergoes dramatic structural rearrangements during the fusion process. Neutralizing monoclonal antibodies, such as 101F, palivizumab, and motavizumab, target two major antigenic sites on the RSV F glycoprotein. The structures of these sites as peptide complexes with motavizumab and 101F have been previously determined, but a structure for the trimeric RSV F glycoprotein ectodomain has remained elusive. To address this issue, we undertook structural and biophysical studies on stable ectodomain constructs. Here, we present the 2.8-Å crystal structure of the trimeric RSV F ectodomain in its postfusion conformation. The structure revealed that the 101F and motavizumab epitopes are present in the postfusion state and that their conformations are similar to those observed in the antibody-bound peptide structures. Both antibodies bound the postfusion F glycoprotein with high affinity in surface plasmon resonance experiments. Modeling of the antibodies bound to the F glycoprotein predicts that the 101F epitope is larger than the linear peptide and restricted to a single protomer in the trimer, whereas motavizumab likely contacts residues on two protomers, indicating a quaternary epitope. Mechanistically, these results suggest that 101F and motavizumab can bind to multiple conformations of the fusion glycoprotein and can neutralize late in the entry process. The structural preservation of neutralizing epitopes in the postfusion state suggests that this conformation can elicit neutralizing antibodies and serve as a useful vaccine antigen.
INTRODUCTION
Respiratory syncytial virus (RSV) belongs to the family Paramyxoviridae of RNA viruses. RSV, human metapneumovirus, pneumonia virus of mice (PVM), and avian pneumoviruses form the subfamily Pneumovirinae. RSV is a leading cause of pneumonia and bronchiolitis in infants and the elderly and is estimated to cause 30 million lower respiratory tract infections and more than 60,000 deaths worldwide each year (32). This global health burden could be ameliorated by an effective vaccine, but one does not currently exist. A series of RSV vaccine trials in the 1960s that evaluated an alum-precipitated formalin-inactivated whole virus did not prevent infection and caused enhanced disease severity upon natural RSV infection (11, 20, 22, 23), in part due to poor elicitation of fusion-inhibiting and neutralizing antibodies (17, 30, 31). Severe disease caused by RSV can be reduced by monthly injections of the monoclonal antibody palivizumab (Synagis) (21), but the high cost of treatment restricts its use. Thus, the creation of a safe and effective RSV vaccine is still urgently needed.
RSV-neutralizing antibodies target either the attachment (G) glycoprotein or the fusion (F) glycoprotein (40). The F glycoprotein is a type I viral fusion protein, a class of entry machines that includes influenza virus hemagglutinin and HIV-1 envelope. Much of our knowledge regarding the structural rearrangements of type I fusion proteins derives from crystal structures of the influenza virus hemagglutinin (7, 9, 41) and paramyxovirus F (10, 39, 43, 44) glycoproteins, which have been determined in the pre- and postfusion states. Type I fusion proteins are synthesized as inactive, single-chain polypeptides that assemble into trimers. The proteins become active after cleavage by host proteases, which liberate a hydrophobic stretch of amino acids called the fusion peptide. After binding to the target cell and subsequent activation, the metastable prefusion protein undergoes a series of dramatic structural rearrangements that result in the insertion of the fusion peptide into the target cell membrane, followed by the formation of a stable helical bundle that forms as the viral and cell membranes are apposed (reviewed in reference 12).
Unlike most type I viral fusion proteins, the RSV F precursor (F0) is cleaved by a furin-like protease at two sites, which generates three fragments. The shorter, N-terminal fragment (F2) is covalently attached to the larger, C-terminal fragment (F1) by two disulfide bonds. F2 is predicted to have two N-glycosylation sites at Asn27 and Asn70, and F1 is predicted to have one N-glycosylation site at Asn500. The intervening fragment of 27 amino acids is predicted to have 2 or 3 N-glycosylation sites depending on the RSV subtype, but this fragment dissociates after cleavage and is not found in the mature protein (6). Neutralizing antibodies, such as palivizumab and 101F, target epitopes that have been mapped to linear regions in the F1 subunit, referred to as antigenic site II and site IV, respectively (3, 5). The structures of the epitope peptides bound to 101F and motavizumab, a potent derivative of palivizumab, have been determined (28, 29), and both structures suggest that the epitopes are larger than the linear peptides. Modeling of these epitopes on the RSV F glycoprotein has been performed using structures of soluble F ectodomains from paramyxoviruses in the subfamily Paramyxovirinae (43, 44), but low sequence homology decreased modeling accuracy and limited conclusions that could be drawn.
To obtain structural information on the RSV F glycoprotein ectodomain, we created a soluble, furin-cleaved ectodomain construct and determined its structure. Here, we present the 2.8-Å crystal structure of the RSV F glycoprotein in the postfusion state. The structure reveals that the 101F and motavizumab epitopes exist in the F glycoprotein in conformations that are similar to the antibody-bound peptide structures. Binding experiments demonstrate that the postfusion state can bind 101F and palivizumab with nanomolar affinity and can bind motavizumab with picomolar affinity. Modeling predicts the full extent of the epitopes and reveals that 101F interactions are contained within a single protomer, whereas motavizumab recognizes residues on two protomers in the trimer. These results are discussed in the context of antibody-mediated RSV neutralization and vaccine design.
MATERIALS AND METHODS
RSV F glycoprotein expression and purification.
F glycoprotein constructs were derived from the A2 strain (accession no. P03420) with three naturally occurring substitutions (P102A, I379V, and M447V) to enhance expression. A mammalian codon-optimized gene encoding RSV F ΔFP (RSV F residues 1 to 513, with fusion peptide residues 137 to 146 deleted [ΔFP]) with a C-terminal human rhinovirus (HRV) 3C site, 8×His tag, and StreptagII was synthesized by GeneArt (Regensburg, Germany) and subcloned into a mammalian expression vector derived from pLEXm (4). Protein was expressed by transient transfection of HEK293F cells in suspension at 37°C for 4 to 5 days, (Invitrogen, Carlsbad, CA) and initially purified via Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen, Valencia, CA) using an elution buffer consisting of 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 250 mM imidazole, pH 8.0. The protein was further purified over StrepTactin resin according to the manufacturer's instructions (Novagen, Darmstadt, Germany). After incubation with HRV 3C protease (Novagen), the protein was passed back over Ni2+-NTA to remove uncleaved protein and affinity tags. The protein was further purified on a Superdex 200 gel filtration column (GE Healthcare) with a running buffer of 2 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.02% NaN3, and the eluted protein was concentrated to ∼6 mg/ml. Similar procedures were used to express and purify the complete RSV F ectodomain (residues 1 to 513) with a C-terminal Factor Xa site and a 6×His tag.
Crystallization and data collection.
Crystallization conditions were screened using a Cartesian Honeybee crystallization robot, and initial crystals were grown by the vapor diffusion method in sitting drops at 20°C by mixing 0.2 μl of RSV F ΔFP with 0.2 μl of reservoir solution (20% [wt/vol] polyethylene glycol [PEG] 3000, 0.1 M sodium citrate, pH 5.5). These crystals were manually reproduced in hanging drops over a range of PEG 3000 concentrations, and large single crystals were obtained by streak seeding at 14 to 16% PEG 3000. The crystals were flash frozen in liquid nitrogen in 25% (wt/vol) PEG 3000, 15% (vol/vol) (2R,3R)-butanediol, and 0.1 M citrate, pH 5.6. Anisotropic data to 2.8 Å were collected at a wavelength of 1.00 Å at the SER-CAT beamline ID-22 (Advanced Photon Source, Argonne National Laboratory).
Crystals were also obtained in 23% (wt/vol) PEG 3000, 0.1 M acetate, pH 4.5, using a 2:1 protein/reservoir ratio. These crystals were not single, but after being flash frozen in 26% PEG 3000, 0.1 M acetate, pH 4.5, and 15% (2R,3R)-butanediol, they diffracted X rays to 3.2 Å. Data were collected at a wavelength of 1.00 Å at the SER-CAT beamline ID-22.
Structure determination, model building, and refinement.
Diffraction data were integrated and scaled with the HKL2000 suite (34) and then processed by the diffraction anisotropy server (38). A molecular replacement solution for the P21 data set using the structure of the RSV F six-helix bundle (Protein Data Bank [PDB] ID 1G2C) (45) as a search model was obtained using PHASER (27). The initial phases were improved dramatically by PARROT (15) due to the 6-fold noncrystallographic symmetry, and the resulting phases were sufficient for automatic model building by BUCCANEER (14). Manual model building was carried out using COOT (18), and refinement of individual sites, translation/libration/screw (TLS) parameters, and group b factors was performed in PHENIX (1). Final data collection and refinement statistics are presented in Table 1. All structural images were created using PyMol (The PyMol Molecular Graphics System, version 1.1; Schrödinger, LLC).
Table 1.
X-ray crystallographic data collection and refinement statistics
| Parameter | Valuea |
|
|---|---|---|
| RSV F ΔFP | RSV F ΔFP | |
| PDB ID | 3RRR | 3RRT |
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 113.2, 131.5, 164.3 | 71.0, 81.9, 272.0 |
| β (°) | 103.2 | |
| Resolution (Å) | 50–2.80b | 50–3.20 |
| Rmerge | 15.9 (35.3) | 20.8 (47.0) |
| I /σI | 7.5 (1.4) | 6.0 (1.7) |
| Completeness (%) | 66.6c (16.7) | 93.9 (82.3) |
| Redundancy | 3.2 (1.5) | 3.1 (2.7) |
| Refinement | ||
| Resolution (Å) | 2.82 | 3.15 |
| No. of reflections | 71,850 | 25,927 |
| Rwork/Rfree (%) | 22.1/26.2 | 25.3/28.2 |
| No. of atoms | ||
| Protein | 19,947 | 10,064 |
| Ligand (NAG) | 168 | 0 |
| B factors | ||
| Protein | 47.4 | 75.9 |
| Ligand (NAG) | 97.0 | |
| RMS deviations | ||
| Bond length (Å) | 0.005 | 0.008 |
| Bond angle (°) | 0.86 | 1.09 |
Values in parentheses are for the highest-resolution shell.
The high-resolution cutoffs along the reciprocal axes a*, b*, and c* are 2.8, 3.4, and 3.2 Å, respectively.
The data are >95% complete to 3.80 Å, and the first shell with <50% completeness is 3.32 to 3.15 Å.
Surface plasmon resonance.
All experiments were carried out on a Biacore 3000 instrument (GE Healthcare). RSV F ΔFP was covalently coupled to a CM5 chip at 330 response units (RU), and a blank surface with no antigen was created under identical coupling conditions for use as a reference. Antigen-binding fragments were serially diluted 2-fold, starting at 25 nM, into 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% polysorbate 20 (HBS-EP) and injected over the immobilized RSV F ΔFP and reference cell at 40 μl/min. The 3.1 nM concentration was performed in duplicate. The data were processed with SCRUBBER-2 and double referenced by subtraction of the blank surface and a blank injection (no analyte). Binding curves were globally fit to a 1:1 binding model.
Protein structure accession numbers.
The atomic coordinates and structure factors for RSV F ΔFP have been deposited in the Protein Data Bank with accession numbers 3RRR and 3RRT.
RESULTS
Crystallization of RSV F in the postfusion state.
To obtain a postfusion state of RSV F, we initially expressed the soluble ectodomain, residues 1 to 513. This protein expressed well in HEK293F cells and was efficiently cleaved into F2 and F1 subunits but eluted as a broad, high-molecular-weight peak from a gel filtration column. This was consistent with previous reports that demonstrated that cleaved, soluble F glycoproteins in the postfusion state aggregate into rosettes of trimers held together by exposed fusion peptides (6, 13). To prevent rosette formation, we deleted the first 9 amino acids of the fusion peptide (residues 137 to 146), which was previously shown to produce monodispersed trimers in the postfusion state as judged by electron microscopy (36). This deletion prevented aggregation by our new construct, called RSV F ΔFP, which eluted from a gel filtration column as a symmetrical, monodispersed peak with a retention volume consistent with a glycosylated trimer.
Glycosylated RSV F ΔFP crystallized under several conditions, and under one of these conditions (20% PEG 3000, 0.1 M citrate, pH 5.5), crystals with at least three different space groups and unit cell dimensions were obtained. A single crystal in space group P21, with cell constants a, 113.2; b, 131.5; c, 164.3 Å; and β, 103.2° diffracted X-rays along one axis to 2.8 Å. The diffraction was anisotropic, with X-rays extending to only 3.2 and 3.5 Å along the other two axes, as judged by the Diffraction Anisotropy Server (38). A partial molecular replacement solution was obtained using the structure of the RSV six-helix bundle as a search model (45). The asymmetric unit contained two six-helix bundles, and the phases were greatly improved by 6-fold noncrystallographic symmetry averaging using PARROT, with the figure of merit improving from 0.181 to 0.677 (15). After automatic and manual model building, the structure was refined to an Rcrys/Rfree of 22.1%/26.2% and had good geometry. The two trimers in the asymmetric unit were similar, with a Cα root mean square deviation (RMSD) of 0.9 Å. This structure was used to solve a second crystal form of RSV F ΔFP that crystallized under similar conditions in space group P212121 with unit cell dimensions a, 71.0; b, 81.9; and c, 272.0 Å. Data for this crystal extended to 3.2 Å, and the asymmetric unit contained only a single trimer. This structure was refined to an Rcrys/Rfree of 25.3%/28.2%, and data collection and refinement statistics for both structures are presented in Table 1. The structures of the F glycoprotein in the two different crystal forms are similar (Cα RMSD, 0.9 Å), and the first three chains of the higher-resolution structure are presented and analyzed below.
Structure of postfusion RSV F.
The structure of trimeric RSV F ΔFP has a cone shape that had previously been observed by electron microscopy (8) (Fig. 1). The presence of the characteristic six-helix bundle (45), whose formation coincides with membrane fusion, confirms that the structure is in the postfusion state. All of the residues in F2 had good electron density and were modeled, with the exception of the C-terminal 11 amino acids that precede the first furin cleavage site. All of the residues in F1 were modeled with the exception of 9 residues that form a loop between Cys322 and Cys333. This loop resides at the top of the molecule and is disordered in all six chains in the asymmetric unit, suggesting that the loop is flexible. The truncated fusion peptide has been modeled in one of the three protomers in each trimer, and it forms an unstructured coil that is held in place by a lattice contact. Electron density is also observed for the proximal N-acetyl-d-glucosamine moiety attached to Asn70 and Asn500, which are two of the three N-linked glycan sites on the mature protein. The electron density around Asn27 is weak, however, likely due to its proximity to the N terminus of F2 (Gln26), and thus a third N-acetyl-d-glucosamine moiety was not modeled. The mature F glycoprotein contains 7 disulfide bonds, 4 of which are highly conserved in all F glycoproteins from the family Paramyxoviridae. For the remaining three disulfide bonds that are conserved in the subfamily Pneumovirinae but not the subfamily Paramyxovirinae F glycoproteins, the RSV F ΔFP structure reveals disulfide bonds formed between Cys37-Cys439, Cys313-Cys343, and Cys322-Cys333, confirming the predicted arrangement (16).
Fig. 1.
Structure of RSV F ΔFP in the postfusion state. The RSV F ectodomain with a truncated fusion peptide adopts the postfusion state, as evidenced by the characteristic postfusion six-helix bundle of N- and C-terminal helices. (Left) Ribbon representation of an F2/F1 monomer, colored from blue to red for the N terminus (term) of F2 to the C terminus of F1, respectively. (Middle) Ribbon representation of an F2/F1 monomer colored tan and rotated 90° about the y axis. Stick representations of the N-acetyl-d-glucosamines (NAG) attached to Asn70 and Asn500 are shown, with oxygen atoms colored red and nitrogen atoms colored blue. (Right) Ribbon representation of the trimeric F glycoprotein, with each F2/F1 monomer a different color. The location of the fused membrane is represented by two solid lines.
The postfusion structure also revealed that the motavizumab and 101F epitopes (sites II and IV) are solvent exposed and in conformations that are similar to those observed in the antibody-bound peptide structures (28, 29) (Fig. 2). In addition, Pro389, whose mutation leads to loss of binding by antibodies targeted to antigenic site I (25), resides at the tip of a loop on the top of the F glycoprotein (Fig. 2). The locations, exposures, and conformations of these three antigenic sites suggested that the postfusion F conformation is capable of binding antibodies directed against these sites. To investigate, surface plasmon resonance experiments were performed by flowing antigen-binding fragments of 101F, palivizumab, motavizumab, and the site I-directed antibody 131-2a (2) over immobilized RSV F ΔFP. Both 101F and palivizumab bound the postfusion state with KD (equilibrium dissociation constant) values of 2.4 nM and 3.7 nM, respectively (Fig. 3). 131-2a bound with a KD of 3.1 nM, and motavizumab bound with a very low off rate that resulted in a KD of <0.1 nM (Fig. 3). The affinities of the antibodies for the postfusion F glycoprotein are similar to those measured for an F glycoprotein construct designed to be stabilized in the prefusion conformation (29) (see Fig. S1 in the supplemental material). Thus, the postfusion conformation of RSV F is able to bind antibodies that target three distinct neutralizing epitopes.
Fig. 2.
Neutralizing epitopes on RSV F ΔFP. Epitopes for motavizumab (antigenic site II), 101F (antigenic site IV), and 131-2a (antigenic site I) are solvent exposed and in conformations compatible with antibody binding. Residues 254 to 277 are colored red (antigenic site II), residues 429 to 437 are colored blue (antigenic site IV), and atoms in Pro389 are shown as spheres (antigenic site I).
Fig. 3.
Binding of neutralizing antibodies to RSV F ΔFP as measured by surface plasmon resonance. Antibodies 101F, palivizumab, motavizumab, and 131-2a bind tightly to the postfusion conformation of RSV F. For each antigen-binding fragment, five 2-fold dilutions starting at 25 nM were measured, with the 3.1 nM concentration measured in duplicate. The red lines represent the best fit of the kinetic data to a 1:1 binding model.
Conservation of the 101F epitope.
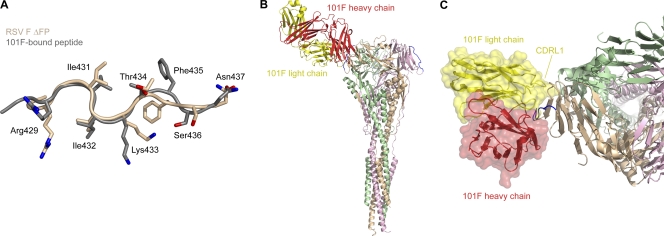
We previously determined the structure of 101F in complex with a peptide spanning RSV F residues 422 to 438, and it revealed that residues 427 to 437 contact the antibody (28). To determine how well the epitope in the context of the F glycoprotein matched the antibody-bound peptide epitope, a superposition was performed (Fig. 4A). The alignment revealed that the positions of the two N-terminal residues differed greatly, but residues 429 to 437 were in similar conformations in both structures, having a Cα RMSD of 1.0 Å. In this range, the position of Phe435 varied the most between the two structures, though it is unclear whether 101F binding requires a conformational change of Phe435, because its side chain points away from the antibody in the complex structure (28). It does, however, form two hydrogen bonds with the 101F heavy chain via its backbone nitrogen and oxygen atoms.
Fig. 4.
Structural conservation of the 101F epitope. The 101F epitope on the postfusion RSV F ΔFP glycoprotein is in a conformation similar to that of the 101F-bound peptide, and modeling suggests that CDRL1 contacts regions on the F glycoprotein outside the linear epitope. (A) Least-squares superposition of residues 429 to 437 from RSV F ΔFP (tan) and the 101F-bound peptide structure (gray) (28) (PDB ID 3O45), as viewed by the antibody. Side chains of residues that contact 101F in the peptide-bound structure are shown as sticks. Oxygen atoms are colored red; nitrogen atoms are colored blue. (B) Model of 101F bound to RSV F ΔFP based on the alignment in panel A. The 101F heavy chain is colored red, and the light chain is colored yellow. RSV F ΔFP residues 429 to 437 are colored blue. (C) Top view of 101F binding to RSV F ΔFP. A transparent molecular surface of 101F is shown over a ribbon representation of the heavy and light chains. The 101F CDRL1 is labeled.
We previously demonstrated that 101F binds to the epitope peptide approximately 16,000-fold more weakly than to the F glycoprotein (28), suggesting that the complete epitope contained residues outside the linear region. To investigate, we modeled the complex of 101F bound to the postfusion F glycoprotein using the alignment described above (Fig. 4B). The resulting model showed that 101F would likely contact additional residues in the cysteine-rich domain, with the first complementarity-determining region of the light chain (CDR L1) mediating the majority of the additional interactions (Fig. 4C). This agrees well with mutagenesis data that showed substitution of three residues on CDR L1 increased the binding affinity of a 101F derivative by over 30-fold (A. Del Vecchio, P. Tsui, P. J. Branigan, L. Conrad, N. Day, C. Liu, R. Sweet, S.-J. Wu, J. A. Melero, J. Luo, G. Canziani, M. Tornetta, G. Raghunathan, and V. Koka, U.S. patent application no. 11,261,356). Of the 3 residues, only Tyr27D (Kabat numbering (21b); D indicates that this is the 5th residue numbered 27) contacts the peptide in the complex structure (28), and it buries just 21 Å2 at the interface, suggesting that the increased affinity resulting from the substitution of these 3 residues is due to enhancement of interactions outside the linear epitope. Our model predicts that the additional residues on the F glycoprotein may include amino acids 418 to 423 and 451 to 456. Additional contacts with residues on the other two protomers in the trimer do not seem possible, and thus the 101F epitope is likely fully present in an RSV monomer.
Conservation of the motavizumab epitope.
We next compared the structure of the motavizumab epitope in the postfusion state to our previous structure of the peptide epitope in complex with motavizumab (29). The structures were strikingly similar, with a Cα RMSD of 0.4 Å for residues 255 to 276 (Fig. 5A). The side chains are also in similar positions, with an all-atom RMSD of 1.1 Å. This result and the unbound structure of palivizumab (21a) suggest that motavizumab binds the F glycoprotein with a lock-and-key fit and requires little conformational change. This may occur because many of the individual amino acid changes that were incorporated into motavizumab were selected because they increased the on-rate of motavizumab for the F glycoprotein (42), and on-rate enhancement is often associated with decreased conformational change upon binding.
Fig. 5.
Structural conservation of the motavizumab epitope. The motavizumab epitope on the postfusion RSV F ΔFP glycoprotein exists in a conformation that is very similar to that of the motavizumab-bound peptide. A model of motavizumab bound to RSV F ΔFP suggests that motavizumab contacts residues on two protomers in the trimer. (A) Least-squares superposition of residues 255 to 276 from RSV F ΔFP (tan) and the motavizumab-bound peptide structure (gray) (29) (PDB ID 3IXT) as viewed by the antibody. Side chains of residues that contact motavizumab in the peptide-bound structure are shown as sticks. Oxygen atoms are colored red; nitrogen atoms are blue. (B) Model of motavizumab bound to RSV F ΔFP based on the alignment in panel A. The motavizumab heavy chain is colored green, and the light chain is colored blue. RSV F ΔFP residues 254 to 277 are colored red. (C) Close-up of motavizumab binding to RSV F ΔFP. The helix-loop-helix epitope (red) resides in the tan protomer, whereas Asn454 and Lys465 are from the pink protomer.
It has been shown that motavizumab binds its epitope peptide approximately 6,000-fold more weakly than the F glycoprotein (42; G. I. Tous, M. A. Schenerman, J. Casas-Finet, Z. Wei, and D. S. Pfarr, U.S. patent application 11,230,593), suggesting that its epitope is larger than the helix-loop-helix. This was further supported by the structure of motavizumab in complex with the peptide epitope (29), which showed that Arg29 in the motavizumab light chain made no contact with the peptide yet provided a 4-fold increase in the on rate compared to the native serine found in palivizumab (42). To investigate the extent of this epitope, we modeled the complex of motavizumab bound to the postfusion F glycoprotein using the Cα alignment of residues 255 to 276 (Fig. 5B). Notably, this model showed that Arg29 in the motavizumab light chain was within hydrogen-bonding distance of Asn454 from an F protomer other than the one that contained the helix-loop-helix (Fig. 5C). An additional interaction may also be possible between Asp54 in the motavizumab heavy chain and Lys465 in the same F protomer that contains Asn454, though this would require rotation of Asp54 in the motavizumab heavy chain. Collectively, these results suggest that the motavizumab epitope is more complex than the helix-loop-helix and is quaternary in nature, involving at least two F protomers in the trimer.
Comparison to other paramyxovirus postfusion structures.
Alignment of the RSV F postfusion structure with the homologous parainfluenza virus 3 (PIV3) and Newcastle disease virus (NDV) F postfusion structures reveals that the overall folds of the proteins are similar (Fig. 6), despite sharing less than 15% sequence identity (37) (see Fig. S2 and S3 in the supplemental material). The motavizumab epitope, however, is one region that is in a different conformation in the RSV structure than in the PIV3 and NDV structures. In the related structures, the homologous region does not contain antiparallel alpha helices but rather perpendicular alpha helices (Fig. 6). Interestingly, in the prefusion PIV5 structure, this region exists in a helix-loop-helix conformation that resembles the motavizumab bound state (29). This suggests that in parainfluenza virus F glycoproteins, this epitope is distorted during the structural rearrangement from the pre- to the postfusion state, though it is difficult to be certain in the absence of either a postfusion PIV5 structure or a prefusion PIV3 or NDV structure.
Fig. 6.
Comparison of paramyxovirus F glycoproteins in the postfusion state. Superposition of RSV F ΔFP with uncleaved PIV3 and NDV F0 glycoproteins reveals that the structures are similar, though differences in the region containing the motavizumab epitope exist. RSV F ΔFP is colored tan, PIV3 F0 (43) (PDB ID 1ZTM) is colored magenta, NDV F0 (39) (PDB ID 3MAW) is colored blue, and all structures are shown as Cα ribbons. The boxed images magnify the regions containing the motavizumab epitope.
DISCUSSION
The structure of the RSV F glycoprotein ectodomain in the postfusion state provides a look at neutralizing epitopes in the context of the trimeric protein (Fig. 2). We also demonstrate that antibodies targeting three antigenic sites on the F glycoprotein bind tightly to this defined state (Fig. 3), which has implications for their mechanisms of neutralization. Though we demonstrate binding to the postfusion state, these antibodies must neutralize prior to membrane fusion and formation of the six-helix bundle, and therefore, we infer that they can bind intermediate states that at least partially resemble the postfusion conformation. Recently, Magro et al. showed that 101F and an antibody specific for antigenic site II bound virus before incubation with target cells, remained bound during ultracentrifugation, and neutralized the virus (26). This result is consistent with 101F and motavizumab being able to bind the prefusion conformation, as well. Thus, 101F and motavizumab can likely bind the fusion glycoprotein in the prefusion, postfusion, and intermediate states, which results from their epitopes residing in domains that are not expected to undergo large structural rearrangements during the fusion process (44). This is different from the modes of action of other RSV-neutralizing compounds, such as antigenic site I-directed antibodies, which appear not to bind the prefusion state (26), and some small molecules and heptad repeat B-derived peptides that target various intermediate states of the F glycoprotein (24, 35) (Fig. 7). Thus, we propose that 101F and motavizumab are compatible with multiple F glycoprotein conformations and would not block the structural rearrangements of a soluble F glycoprotein but, rather, exert their effect only when F is in the process of opposing two membranes.
Fig. 7.
Binding of RSV-neutralizing molecules to different F glycoprotein conformations. A schematic of the RSV F glycoprotein in the prefusion, intermediate, and postfusion states is shown, along with a table indicating whether various RSV-neutralizing molecules are expected to bind the different states. The green cylinders represent heptad repeat B (HRB), the red ovals represent domains I and II and the portion of domain III that appears to be conformationally static (as described by Yin et al. [44]), and the blue ovals represent the portion of domain III that does refold to form heptad repeat A (blue cylinders). TMC353121 is a small molecule that inhibits fusion by disrupting formation of the postfusion six-helix bundle (35).
The high degree of structural conservation of both the 101F and motavizumab epitopes in the postfusion state (Fig. 4 and 5) and the demonstration that both antibodies bind tightly to this protein (Fig. 3) suggest that the postfusion state is capable of eliciting neutralizing antibodies. This would be in contrast to some other viral fusion glycoproteins, such as HIV-1 gp41. It was recently demonstrated that, unlike HIV-1-neutralizing antibodies 2F5 and 4E10, which are thought to target a fusion intermediate of gp41, nonneutralizing antibodies bind to the postfusion six-helix bundle conformation (19, 33). Based on these results, the authors proposed that the postfusion form of gp41 thus acts as a decoy to assist HIV-1 immune evasion by inducing an ineffective immune response (19). Therefore, if the postfusion conformation of RSV F can induce neutralizing antibodies, which the structure predicts, it suggests that RSV and HIV-1 have evolved different mechanisms for evading neutralization. It also suggests that the stable postfusion conformation of RSV F may be a better vaccine antigen than the metastable prefusion state, though potentially deleterious T-cell epitopes are present in both conformations.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at SER-CAT (Southeast Regional Collaborative Access Team) for help with X-ray diffraction data collection; Marie Pancera, Mallika Sastry, and other members of the Structural Biology Section and Structural Bioinformatics Section for helpful comments on the manuscript; and Man Chen for technical assistance.
Support for this work was provided by the Intramural Research Program (National Institute of Allergy and Infectious Diseases). Use of sector 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
J.S.M. purified, crystallized, and solved the structure of RSV F ΔFP and performed the SPR experiments. Y.Y. expressed RSV F ΔFP and assisted in its purification. J.S.M., B.S.G., and P.D.K. designed experiments and analyzed data. J.S.M. wrote the initial draft of the paper, on which all of the authors commented.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Adams P. D., et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 2. Anderson L. J., Bingham P., Hierholzer J. C. 1988. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J. Virol. 62:4232–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arbiza J., et al. 1992. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 73:2225–2234 [DOI] [PubMed] [Google Scholar]
- 4. Aricescu A. R., Lu W., Jones E. Y. 2006. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62:1243–1250 [DOI] [PubMed] [Google Scholar]
- 5. Beeler J. A., van Wyke Coelingh K. 1989. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J. Virol. 63:2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Begoña Ruiz-Argüello M., et al. 2002. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology 298:317–326 [DOI] [PubMed] [Google Scholar]
- 7. Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43 [DOI] [PubMed] [Google Scholar]
- 8. Calder L. J., et al. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122–131 [DOI] [PubMed] [Google Scholar]
- 9. Chen J., et al. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417 [DOI] [PubMed] [Google Scholar]
- 10. Chen L., et al. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255–266 [DOI] [PubMed] [Google Scholar]
- 11. Chin J., Magoffin R. L., Shearer L. A., Schieble J. H., Lennette E. H. 1969. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 89:449–463 [DOI] [PubMed] [Google Scholar]
- 12. Colman P. M., Lawrence M. C. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309–319 [DOI] [PubMed] [Google Scholar]
- 13. Connolly S. A., Leser G. P., Yin H.-S., Jardetzky T. S., Lamb R. A. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol. Crystallogr. 62:1002–1011 [DOI] [PubMed] [Google Scholar]
- 15. Cowtan K. 2010. Recent developments in classical density modification. Acta Crystallogr D Biol. Crystallogr. 66:470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Day N., et al. 2006. Contribution of cysteine residues in the extracellular domain of the F protein of human respiratory syncytial virus to its function. Virol. J. 3:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delgado M. F., et al. 2009. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat. Med. 15:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Frey G., et al. 2010. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat. Struct. Mol. Biol. 17:1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fulginiti V. A., et al. 1969. Respiratory virus immunization. I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am. J. Epidemiol. 89:435–448 [DOI] [PubMed] [Google Scholar]
- 21. IMpact-RS. V. Study Group 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531–537 [PubMed] [Google Scholar]
- 21a. Johnson L. S., Bradford B. October 2005. Crystals and structure of Synagis Fab. U. S. patent 6,955,717
- 21b. Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S., Foeller C. 1991. Sequences of proteins of immunological interest, 5th ed. NIH publication no. 91-3242 U.S. Department of Health and Human Services, Public Health Service, National Institutes of Health, Bethesda, MD [Google Scholar]
- 22. Kapikian A. Z., Mitchell R. H., Chanock R. M., Shvedoff R. A., Stewart C. E. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89:405–421 [DOI] [PubMed] [Google Scholar]
- 23. Kim H. W., et al. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422–434 [DOI] [PubMed] [Google Scholar]
- 24. Lambert D. M., et al. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 93:2186–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lopez J. A., et al. 1998. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J. Virol. 72:6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magro M., Andreu D., Gomez-Puertas P., Melero J. A., Palomo C. 2010. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J. Virol. 84:7970–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLellan J. S., et al. 2010. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J. Virol. 84:12236–12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLellan J. S., et al. 2010. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 17:248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murphy B. R., et al. 1986. Dissociation between serum neutralizing and glycoprotein antibody responses of infants and children who received inactivated respiratory syncytial virus vaccine. J. Clin. Microbiol. 24:197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murphy B. R., Walsh E. E. 1988. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J. Clin. Microbiol. 26:1595–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nair H., et al. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicely N. I., et al. 2010. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat. Struct. Mol. Biol. 17:1492–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 35. Roymans D., et al. 2010. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc. Natl. Acad. Sci. U. S. A. 107:308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruiz-Arguello M. B., et al. 2004. Thermostability of the human respiratory syncytial virus fusion protein before and after activation: implications for the membrane-fusion mechanism. J. Gen. Virol. 85:3677–3687 [DOI] [PubMed] [Google Scholar]
- 37. Smith B. J., Lawrence M. C., Colman P. M. 2002. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15:365–371 [DOI] [PubMed] [Google Scholar]
- 38. Strong M., et al. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swanson K., et al. 2010. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology 402:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh E. E., Hruska J. 1983. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 47:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson I. A., Skehel J. J., Wiley D. C. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373 [DOI] [PubMed] [Google Scholar]
- 42. Wu H., et al. 2007. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 368:652–665 [DOI] [PubMed] [Google Scholar]
- 43. Yin H.-S., Paterson R. G., Wen X., Lamb R. A., Jardetzky T. S. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yin H. S., Wen X., Paterson R. G., Lamb R. A., Jardetzky T. S. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao X., Singh M., Malashkevich V. N., Kim P. S. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. U. S. A. 97:14172–14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.