Abstract
The HIV-1 envelope glycoprotein is a trimeric complex of heterodimers composed of a surface glycoprotein, gp120, and a transmembrane component, gp41. The association of this complex with CD4 stabilizes the coreceptor-binding site of gp120 and promotes the exposure of the gp41 helical region 1 (HR1). Here, we show that a 15-amino-acid peptide mimetic of the HIV-1 coreceptor CCR5 fused to a dimeric antibody Fc domain (CCR5mim-Ig) bound two gp120 molecules per envelope glycoprotein complex and by itself promoted HR1 exposure. CCR5mim-Ig also stabilized the association of a CD4-mimetic peptide with the envelope glycoprotein. A fusion of the CD4- and CCR5-mimetic peptides, DM1, bound gp120 and neutralized R5, R5X4, and X4 HIV-1 isolates comparably to CD4, and they did so markedly more efficiently than either peptide alone. Our data indicate that the potency of DM1-Ig derives from its avidity for the HIV-1 envelope glycoprotein trimer and from the bidirectional induction of its receptor-mimetic components. DM1 has significant advantages over other inhibitors that target both coreceptor and CD4-binding sites, and it may serve as a lead for a new class of HIV-1 inhibitor peptides.
INTRODUCTION
The entry of HIV-1 into a target cell requires the expression of the cellular receptor CD4 and a coreceptor, principally the chemokine receptor CCR5 or CXCR4 (5, 13, 20, 46). Virus association with CD4 triggers conformational changes in gp120 that promote high-affinity association with the coreceptor and expose helical region 1 (HR1) of gp41 (21, 37, 40, 45). The association of gp120 with CD4 and a coreceptor induces additional conformational changes in which gp41 helical regions 1 and 2 associate, placing viral and cellular membranes in close apposition and thereby promoting lipid mixing and viral fusion (2, 14, 42). HIV-1 variants that utilize CCR5 as a coreceptor, R5 viruses, mediate transmission and predominate through the asymptomatic stages of infection (7). In some cases, coinciding with a decline in immune function, variants emerge in infected individuals that also infect cells via CXCR4 (20). These include R5X4 viruses, which retain their use of CCR5, and X4 viruses, which cannot efficiently infect cells through CCR5. Viruses passaged extensively in cell lines also generally exhibit an X4 phenotype (11).
The ectodomains of CCR5 and CXCR4 are comprised of an amino terminus and three extracellular loops held in place by seven transmembrane helices (3). The entry of R5 viruses depends largely on the amino terminus and the second extracellular loop of CCR5 (16, 36). The CCR5 amino terminus includes a specific arrangement of sulfotyrosines and aspartic acids critical for HIV-1 entry (3, 17). Sulfotyrosines also can be found at the CXCR4 amino terminus (3, 15), in the heavy-chain CDR3 regions of the CD4-inducible (CD4i) HIV-1 neutralizing antibodies E51 and 412d (6) and of the recently described broadly neutralizing antibodies PG9 and PG16 (34, 41). All of these molecules associate with a region of gp120 that overlaps with its CCR5-binding site (6, 34, 35). The structure of gp120 complexed with 412d (22) localizes two sulfotyrosine-binding pockets to two gp120 regions critical for coreceptor association, at the conserved base of the third variable loop, and in the fourth conserved region (8, 35).
We previously described a tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of E51 (12), a CD4i antibody closely related to 412d and obtained from the same individual (6). Even more so than 412d, the pattern of E51 sulfotyrosines bears a striking resemblance to that found in CCR5. This E51-derived peptide, which we refer to here as CCR5mim, binds gp120 and neutralizes HIV-1 more efficiently than sulfopeptides directly based on the CCR5 amino terminus (12). The basis for this more efficient binding and neutralization is likely the greater flexibility and solubility of the E51-derived peptide. Although it resembles CCR5, this peptide also binds and neutralizes R5X4 and X4 isolates as well, which is consistent with the high level of the conservation of the gp120 sulfate-binding pockets across clade and coreceptor preference (9, 22, 35).
Here, we further explore the properties of CCR5mim by comparing it with CD4mim, a natural amino acid form of a previously described CD4-mimetic peptide (32). We observe that when each of these peptides is dimerized via the Fc region of human IgG1, CD4mim-Ig binds soluble gp120 with higher affinity but CCR5mim-Ig more avidly associates with the cell-expressed HIV-1 envelope glycoprotein trimer. Moreover, CCR5mim-Ig enhances the association of CD4mim-Ig with the trimer and, like soluble CD4, can promote the exposure of helical region 1 of gp41. Consistently with these properties, a fusion of both receptor-mimetic peptides binds gp120 and neutralizes HIV-1 comparably to CD4. We suggest that this double-mimetic peptide can be made more potent and difficult to escape than most neutralizing antibodies.
MATERIALS AND METHODS
Plasmids and cells.
Plasmids encoding CCR5mim-Ig (pΔE51-Ig), nC5aR-Ig, S1-Ig, CD4-Ig, and p17b-Ig have been described previously (12, 18, 44). nC5aR-Ig is a fusion peptide of human IgG1 with the tyrosine-sulfated amino terminus of the C5a receptor (18); S1-Ig is a similar fusion peptide with the S1 domain of the severe acute respiratory syndrome (SARS) coronavirus entry protein (44); CD4-Ig is a similar fusion with the first two immunoglobulin domains of CD4 (6); p17b-Ig is a similar fusion peptide with a peptide identical to the CDR3 region of the CD4i antibody 17b (12). Plasmids encoding CD4mim-Ig, DM1-Ig, and T20-Ig were created by generating sequences encoding these peptides using overlapping PCR and ligating the resulting fragments into the NheI and BamHI restriction sites of a previously described pcDM8-derived plasmid expressing the signal sequence of CD5 and the Fc domain of human IgG1. Plasmids encoding the control constructs CD4mim-F/A-Ig and YYYEE-Ig were generated by modifying plasmids encoding CD4mim-Ig and CCR5mim-Ig, respectively, using the QuikChange method (Agilent). Plasmids encoding monomeric Fc forms of CD4-Ig, CD4mim-Ig, and CCR5mim-Ig were generated by altering, using the QuikChange method, the human IgG1 Fc domain codons to encode the following changes: C219A, C226N, C229G, N297D, L368R, F405H, and Y407E (numbering according to the Kabat database). Plasmids expressing various clade B and clade C gp120 proteins were generated by introducing a stop codon at the gp120/gp41 junction from expression plasmids encoding the full-length HIV envelope glycoprotein that were described previously (ADA, YU2, HXB2, 89.6, BR20.4, SG3, SA32, ELI, and MCGP1 isolates [5, 12, 17]), obtained from the NIH AIDS Research and Reference Reagent Program (ConB, ConC, CF402.1, and Th966 [28]), or a generous gift from Cynthia Derdeyn (MBP40 and FBP10; Emory University). The plasmid encoding the NL4-3 HIV-1 genome in which the NL4-3 env gene has been replaced by that of the R5X4 isolate 89.6 (or has been deleted entirely), as well as the nef gene replaced by a gene encoding green fluorescent protein (GFP), was described previously (5, 12, 17). Human embryonic kidney 293T cells were obtained from the American Type Culture Collection. GHOST-CCR5 and GHOST-CXCR4 cells expressing GFP driven by a long terminal repeat promoter were obtained from the NIH AIDS Research and Reference Reagent Program.
Purification of peptide-Ig proteins.
293T cells were transfected with plasmids encoding CCR5mim-Ig, CD4mim-Ig, CD4-Ig, DM1-Ig, or variants thereof and a plasmid encoding tyrosyl-protein transferase 2 (TPST-2) as previously described (6). Six hours posttransfection, cells were washed and subsequently incubated in FreeStyle 293 expression medium (Invitrogen). Medium was harvested after 48 h and proteins precipitated with protein A-Sepharose at 4°C for 16 h. Beads were washed in phosphate-buffered saline (PBS) containing 0.5 M NaCl and eluted with 50 mM sodium citrate-50 mM glycine (pH 2.5) into the same volume of 1 M Tris, pH 7.4. Purified proteins were dialyzed in PBS and concentrated with Centricon filters (Amicon). Proteins were quantified by comparison to bovine serum albumin (BSA) standards in a Coomassie-stained SDS-PAGE gel and with a Micro-BCA protein assay (Thermo Scientific).
Immunoprecipitation of gp120.
293T cells transfected by the calcium phosphate method with plasmids expressing peptide-Ig variants and a plasmid expressing TPST-2 at a 10:1 ratio were metabolically labeled with 35S-Express (Perkin Elmer). Labeled supernatants were harvested 2 days posttransfection. In parallel, 293T cells were transfected with plasmids expressing the gp120 molecules of a range of HIV-1 isolates and metabolically labeled in the same manner. Fc fusion protein was quantified by SDS-PAGE and diluted to equivalent concentrations. Five hundred μl of gp120-containing supernatant then was incubated for 30 min at room temperature with 300 μl of diluted supernatants containing peptide-Ig protein. Protein A-Sepharose (Amersham) was added to the mixture, which subsequently was incubated overnight at 4°C. Protein A-Sepharose was washed three times with PBS and analyzed by SDS-PAGE.
Flow-cytometric analysis of envelope glycoprotein trimers.
Envelope glycoproteins were expressed on the cell surface by transfecting human embryonic kidney 293T cells with plasmids expressing the envelope glycoproteins lacking cytoplasmic residues 732 to 876 (HXB2 numbering). The surface expression of envelope glycoprotein was assessed 2 days later by flow cytometry using patient sera and anti-human IgG (Fc-specific) secondary antibody. The binding of various Fc fusion proteins was assessed by incubating aliquots of the same cells with purified protein at the indicated concentrations for 45 min on ice, followed by phycoerythrin (anti-human Fc)- or DyLight649 (anti-murine Fc)-conjugated secondary antibody (Jackson ImmunoResearch Laboratories).
Infection assays.
293T cells were transfected with a plasmid expressing an envelope glycoprotein from one of a number of HIV-1 isolates together with plasmid encoding the genome of the NL4-3 HIV-1 isolate lacking functional env and nef genes and expressing GFP. Supernatants of transfected cells were harvested, and reverse transcriptase activity was measured as previously described (5). These supernatants, diluted to 20,000 reverse transcriptase counts per ml, were added to GHOST-CCR5 or GHOST-CXCR4 cells in the presence of the indicated concentrations of p17b-Ig, DM1-Ig, CD4mim-Ig, or CD4-Ig. Media were changed the following day, and GFP expression in infected cells was measured 2 days postinfection by flow cytometry.
RESULTS
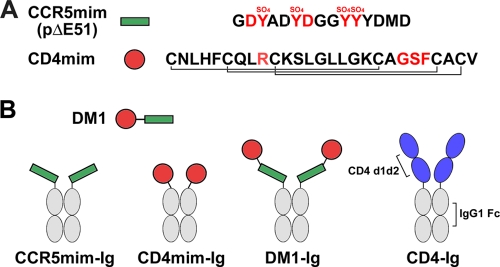
We have shown previously that a 15-amino-acid peptide derived from the CDR3 region of the tyrosine-sulfated HIV-1 neutralizing antibody E51 binds gp120 and inhibits HIV-1 entry (12). This association was localized to a conserved coreceptor-binding region of gp120 and was enhanced by, but did not require, gp120 association with CD4. This peptide, previously described as pΔE51, is referred to here as CCR5mim, reflecting its homology with the tyrosine-sulfated CCR5 amino terminus and, in particular, its arrangement of aspartic acids and sulfotyrosines (Fig. 1A). In parallel, we modified a 27-amino-acid CD4 mimetic peptide, CD4M33, developed by Carlo Vita and colleagues (23, 32), to include only naturally occurring residues. This modified peptide is referred to here as CD4mim (Fig. 1A). The use of natural amino acids permits its study as an Fc fusion protein and enables its subsequent improvement by phage display technology, but it lowers its affinity for gp120 (38). The original peptide induces in gp120 a conformation nearly identical to that induced by CD4 (23). CCR5mim and CD4mim are represented in Fig. 1 as both the monomers and as Fc fusion proteins (CCR5mim-Ig and CD4mim-Ig) used in most of the subsequent experiments.
Fig. 1.
Receptor-mimetic constructs. (A) CCR5mim (green bar, previously described as pΔE51 [12]) and CD4mim (red circle, a natural amino acid form of the previously described CD4-mimetic peptide CD4M33 [23, 32]) are shown with their sequences to the right. At least four of the tyrosines of CCR5mim are modified by sulfate (SO4), as indicated. Red indicates residues of CCR5mim and CD4mim identical to CCR5 and CD4, respectively, and critical for gp120 association. The disulfide bonds of CD4mim are indicated by connecting lines. (B) DM1, a peptide composed of CD4mim and CCR5mim linked by a (GSGGG)2 linker, is represented. Fusions of these peptides with the Fc region of human IgG1 (CCR5mim-Ig, CD4mim-Ig, and DM1-Ig), as well as with domains 1 and 2 of CD4 (CD4-Ig), are also shown.
CCR5mim-Ig binds HIV-1 envelope glycoprotein trimers, but not monomeric gp120, more efficiently than CD4mim-Ig.
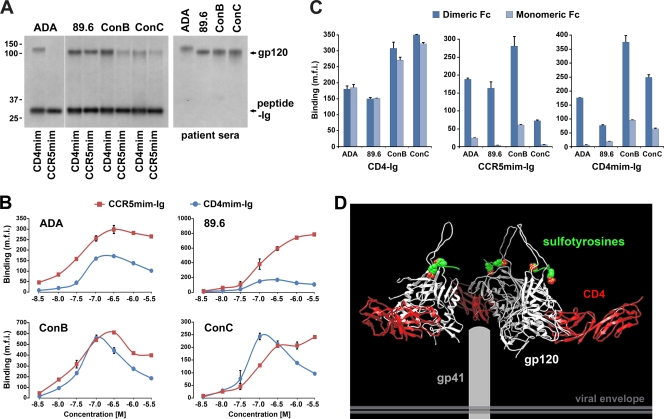
We first compared the abilities of CD4mim-Ig and CCR5mim-Ig to immunoprecipitate the gp120 molecules of the clade B R5 isolate ADA, the clade B R5X4 isolate 89.6, and gp120 molecules composed of consensus amino acid sequences of clade B or clade C isolates (ConB and ConC, respectively). CD4mim-Ig precipitated each gp120 molecule more efficiently than CCR5mim-Ig (Fig. 2A). Figure 2A also demonstrates the variation in binding efficiency among the isolates: CD4mim-Ig bound better to clade B gp120 molecules than to ConC gp120, and CCR5mim-Ig bound most efficiently to gp120 of the clade B R5X4 isolate 89.6. We also investigated by flow cytometry the ability of CD4mim-Ig and CCR5mim-Ig to bind the envelope glycoprotein trimer expressed on the cell surface (Fig. 2B). In contrast to the precipitation results, in three cases (ADA, 89.6, and ConB) CCR5mim-Ig recognized the trimeric form of the envelope glycoprotein more efficiently than CD4mim-Ig, suggesting that, relative to CD4mim, the access of CCR5mim to gp120 is less occluded in the trimer. Alternatively, dimeric CCR5mim-Ig binds with higher avidity to the envelope glycoprotein trimer than to a gp120 monomer, whereas this effect may be less pronounced with CD4mim-Ig. We also observed that higher concentrations of CD4mim-Ig resulted in reduced binding. This effect sometimes was observed with CCR5mim-Ig. This observation further raised the possibility that, at lower concentrations, these Fc fusion proteins bound two gp120 molecules of the trimer, whereas at higher concentrations each peptide-Ig bound only a single gp120 monomer. This lower-avidity association at higher concentrations likely would be more susceptible to wash steps preceding flow-cytometric analysis.
Fig. 2.
CCR5mim-Ig but not CD4-Ig binds two monomers of the HIV-1 envelope glycoprotein trimer. (A) gp120 molecules of the indicated HIV-1 isolates were transfected into 293T cells and metabolically labeled, and cell supernatants were precipitated with metabolically labeled CD4mim-Ig, CCR5mim-Ig, or a cocktail of sera from HIV-1-positive individuals, as indicated. gp120 and mimetic peptides are indicated at the right, and molecular mass (in kDa) is indicated at the left. (B) 293T cells were transfected with the envelope glycoproteins of the indicated HIV-1 isolates, stained 2 days later with the indicated concentrations of CD4mim-Ig (red) or CCR5mim-Ig (blue), and analyzed by flow cytometry. (C) CD4-Ig, CCR5mim-Ig, CD4mim-Ig, or variants thereof which express as monomeric proteins were incubated at approximately half-maximal concentrations (5 nM for CD4-Ig, 50 nM for CD4mim-Ig and CCR5mim-Ig) with 293T cells expressing the indicated envelope glycoproteins and analyzed by flow cytometry. Differences between monomeric and dimeric forms of CCR5mim-Ig and CD4mim-Ig in all cases are significant (P < 0.005) by Student's t test. (D) A model of the HIV-1 envelope glycoprotein trimer (white) with domains 1 and 2 (D1D2) of CD4 bound (red) and sulfotyrosines (green-red) shown associated with their gp120 pockets. Note the proximity of the sulfotyrosine pocket of different monomers and, in contrast, the distance between C termini of the bound CD4 D1D2 domains. We suggest that this difference explains the data shown in panel C. Experiments shown in panels A and B are representative of three experiments with similar results. The experiment shown in panel C is representative of two experiments with similar results. Error bars indicate standard errors from triplicates within experiments.
CCR5mim and CD4mim bind two gp120 monomers of the HIV-1 envelope glycoprotein.
To explore these possibilities directly, we modified the Fc domain to prevent its dimerization and compared the binding of monomeric forms of these Fc-fusion proteins to their original dimeric forms. We observed that the monomeric and dimeric forms of CD4-Ig bound cell-expressed HIV-1 envelope glycoprotein trimers with comparable efficiency (Fig. 2C). In contrast, dimeric forms of both CCR5mim-Ig and CD4mim-Ig bound cell-expressed envelope glycoprotein markedly more efficiently than their monomeric forms. A possible basis for the difference between CD4-Ig and CCR5mim-Ig in this assay is shown in Fig. 2D. Specifically, the C terminus of domain 2 of CD4 is extended far from the central axis of the envelope glycoprotein trimer, preventing two CD4 molecules of CD4-Ig from binding the same trimer. In contrast, the sulfotyrosine-binding pockets of the envelope glycoprotein lie close to the axis of the trimer, easily permitting two CCR5mim molecules of CCR5mim-Ig to associate. Similarly, the distance between the C termini of CD4mim may be close enough to allow CD4mim-Ig to associate with two gp120 monomers of the trimer. The experiments shown in Fig. 2 thus indicate that CD4mim-Ig associates more efficiently than CCR5mim-Ig with soluble gp120 but less efficiently with cell-expressed HIV-1 envelope glycoprotein trimer. They also show that both CD4mim-Ig and CCR5mim-Ig bind two monomers of the envelope glycoprotein trimer, whereas CD4-Ig binds only one. Finally, they suggest that the CD4mim-binding site is less accessible on the trimer than the same site on the monomer and on the CCR5mim-binding site.
CCR5mim stabilizes association of CD4mim with the HIV-1 envelope glycoprotein.
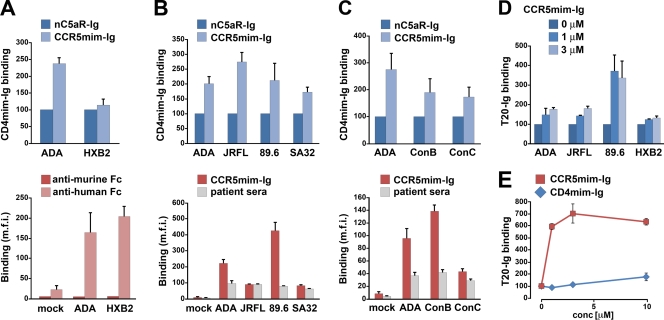
Virion-associated envelope glycoprotein typically binds CD4 before it binds CCR5, and this association facilitates subsequent binding to CCR5. Similarly, CD4 induces a conformation in monomeric gp120 that more efficiently associates with CCR5, with tyrosine-sulfated CD4i antibodies such as E51 and 412d, and with CCR5mim-Ig (6, 12, 40, 45). The comparable binding affinities of CD4mim-Ig and CCR5mim-Ig allowed us to explore the possibility that this induction is bidirectional. To do so, we exchanged the Fc region of CD4mim-Ig with its murine ortholog, thus permitting the recognition of CCR5mim-Ig and CD4mim-Ig by distinct secondary antibodies. We then sought to measure the effect of CCR5mim-Ig on CD4mim-Ig binding to the envelope glycoprotein trimer. We observed that CCR5mim-Ig, but not an Fc fusion of a control sulfated peptide (nC5aR-Ig), markedly enhanced the association of CD4mim-Ig with the ADA envelope glycoprotein but poorly enhanced CD4mim-Ig binding to the envelope glycoprotein of the X4 isolate HXB2 (Fig. 3A, top). This difference is not due to differences in the ability of CCR5mim-Ig to bind each envelope glycoprotein in the absence of CD4mim-Ig, as both envelope glycoproteins bound CCR5mim-Ig efficiently (Fig. 3A, bottom). The enhancement of CD4mim-Ig association in the presence of CCR5mim-Ig also is not due to the cross-reactivity of the secondary antibodies: an anti-murine Fc did not recognize CCR5mim-Ig bound to the HIV-1 envelope glycoprotein in the absence of CD4mim-Ig (Fig. 3A, bottom). CCR5mim-Ig enhanced the association of CD4mim-Ig with the envelope glycoproteins of a range of isolates, including those of the R5 isolate JRFL, R5X4 isolate 89.6, and X4 isolate SA32 (Fig. 3B). The enhancement of CD4mim-Ig association also was observed for consensus ConB and ConC envelope glycoproteins (Fig. 3C). In some cases (JRFL, SA32, and ConC), this enhancement occurred despite the poor binding of CCR5mim-Ig in the absence of CD4mim-Ig (Fig. 3B and C, bottom), which is consistent with the enhanced binding of CCR5mim-Ig in the presence of CD4mim-Ig. Figures 3A to C thus show that CCR5mim-Ig induces or stabilizes a conformation preferred by CD4mim-Ig, indicating that induction can be bidirectional.
Fig. 3.
CCR5mim promotes conformational changes in the HIV-1 envelope glycoprotein. (A to C, top) 293T cells transfected to express the indicated envelope glycoproteins on their surface were incubated on ice with 10 nM CD4mim-mIg (a fusion of CD4mim with a murine Fc region) and 300 nM either CCR5mim-Ig or a control sulfopeptide Fc fusion (nC5aR-Ig). Cells were washed and analyzed by flow cytometry using a secondary antibody recognizing the murine Fc domain of CD4mim-mIg. Results shown are normalized to CD4mim-mIg binding in the presence of nC5aR-Ig. All shown comparisons between CCR5mim-Ig and nC5aR-Ig are significant (P < 0.01), except those with HXB2. (A, bottom) Binding of CCR5mim-Ig in the absence of CD4mim-mIg. Aliquots of the cells were incubated on ice with CCR5mim-Ig and analyzed by flow cytometry using a secondary antibody recognizing the murine or human Fc domain, as indicated. (B and C, bottom) Envelope glycoprotein expression and CCR5mim-Ig binding were assessed using an aliquot of the cells shown in the respective top panels. Specifically, cells were incubated with CCR5mim-Ig or sera from patients and analyzed by flow cytometry using a secondary antibody recognizing human Fc. (D) 293T cells transfected to express the indicated envelope glycoproteins were incubated on ice with CCR5mim-Ig at the indicated concentrations and 10 nM T20-mIg (a fusion of the T20 peptide with the murine Fc region). Cells were washed, incubated with a secondary antibody recognizing T20-mIg, and analyzed by flow cytometry. Differences between 0 μM CCR5mim-Ig and 1 or 3 μM are significant (P > 0.01). (E) Experiment similar to that shown in panel D, except that 293T cells expressing envelope glycoprotein of the 89.6 isolate were incubated with T20-mIg and the indicated concentrations of CD4mim-Ig or CCR5mim-Ig. Experiments are representative of at least three (A to D) or two (E) experiments with similar results. Error bars indicate standard errors from triplicates within experiments. Representative experiments used to generate panels A to E are shown in Fig. S1 in the supplemental material.
CCR5mim promotes exposure of gp41 HR1.
CD4 association induces the exposure of HR1 of gp41, as indicated by the enhanced binding of peptides based on HR2, such as T20 (enfuvirtide) or T2635, to cell-expressed envelope glycoprotein in the presence of soluble CD4 (21, 37). We hypothesized that CCR5mim-Ig similarly promotes the exposure of HR1. To study this possibility, we generated a fusion of T20 with a murine Fc region and measured its association with cell-expressed envelope glycoprotein in the presence of various concentrations of CCR5mim-Ig (Fig. 3D). CCR5mim-Ig enhanced the association of T20-Ig with envelope glycoproteins of several HIV-1 isolates, most markedly with that of the 89.6 isolate. Indeed, for this isolate, CCR5mim-Ig enhanced T20-Ig association more efficiently than did CD4mim-Ig (Fig. 3E), likely reflecting the higher affinity of CCR5mim-Ig for this envelope glycoprotein (Fig. 2B). Collectively, the data of Fig. 3 indicate that CCR5mim-Ig can induce or stabilize conformational transitions in the HIV-1 envelope glycoprotein similar to those reported for CD4 and CD4mim-like peptides.
DM1, a combination of CCR5mim and CD4mim, efficiently binds gp120.
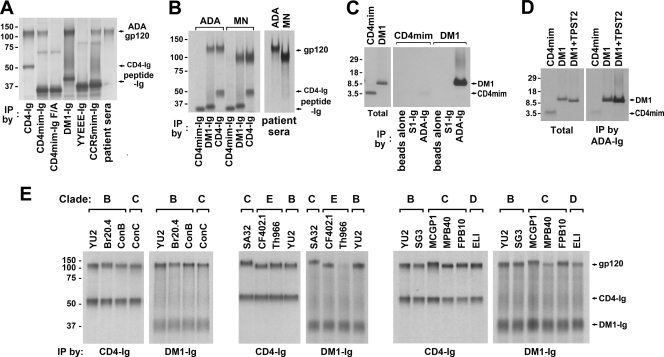
The ability of CD4mim-Ig and CCR5mim-Ig to cooperate bidirectionally suggested that a combination of both peptides would bind the envelope glycoprotein and neutralize HIV-1 entry more efficiently than either peptide alone. To explore this possibility, we linked CD4mim to CCR5mim with a 9-amino-acid glycine linker, and we expressed the resulting double-mimetic peptide (DM1) as an Fc fusion (DM1-Ig) (Fig. 1). We compared the abilities of DM1-Ig, CD4-Ig, CD4mim-Ig, and CCR5mim-Ig to immunoprecipitate the gp120 molecule of the ADA isolate (Fig. 4A). DM1-Ig precipitated gp120 with efficiency comparable to that of CD4-Ig and markedly more efficiently than CD4mim-Ig or CCR5mim-Ig alone. Control variants of each peptide (CD4mim-Ig F/A and CCR5mim-Ig YYY/EEE [12]) did not associate with gp120. Similar results were obtained with gp120 of the X4 isolate MN (Fig. 4B). We also explored the ability of CD4mim and DM1 peptides alone, without their respective Fc regions, to bind gp120. To do so, we engineered a thrombin cleavage site between the peptide and the Fc domain, prepared the monomeric peptide by treating the peptide-Ig forms with thrombin, and purified the cleaved peptide by removing the Fc domain and thrombin by filtration. These peptides were incubated with an Fc fusion with the ADA gp120 (ADA-Ig) or with a control Fc fusion protein with the SARS coronavirus S1 domain (S1-Ig). DM1 bound ADA-Ig specifically and with markedly greater efficiency than CD4mim (Fig. 4C). This association depends in part on the sulfotyrosines of DM1's CCR5mim components, as indicated by the lower association of DM1 to ADA-Ig in the absence of exogenous tyrosyl protein sulfotransferase 2 (TPST2), an enzyme responsible for modifying the tyrosines of CCR5 (Fig. 4D). Finally, we compared the ability of CD4-Ig and DM1-Ig to bind gp120 molecules from a range of clade B, C, D, and E isolates. DM1 bound 9 of 12 gp120 molecules with efficiencies comparable to that of CD4-Ig. DM1-Ig also precipitated gp120 molecules of at least two of the remaining three isolates (MPB40 and ELI). The data shown in Fig. 4 indicate that DM1 binds a range of envelope glycoproteins more efficiently than either of its component peptides and comparably to CD4-Ig.
Fig. 4.
High-affinity association between a double-mimetic peptide (DM1) and the gp120 molecules of diverse isolates. (A) Metabolically labeled CD4-Ig, CD4mim-Ig, DM1-Ig, CCR5mim-Ig, and patient sera were used to immunoprecipitate (IP) labeled HIV-1 gp120 (ADA isolate). Specifically, supernatants of labeled 293T cells expressing each peptide-Ig variant were mixed with supernatants from gp120-expressing 293T cells, precipitated with protein A-Sepharose, and analyzed by SDS-PAGE. Two additional control peptides were used: a CD4mim-Ig F/A variant in which phenylalanine 23, critical to association with gp120, was altered to alanine, and a previously described inactive YYEEE-Ig variant of CCR5mim-Ig. (B) Experiments were similar to those described for panel A, except that gp120 of the ADA or MN isolate was precipitated with patient sera, CD4mim-Ig, DM1-Ig, or CD4-Ig as indicated. (C) Monomeric CD4mim and DM1 peptides were produced from metabolically labeled CD4mim-Ig and DM1-Ig by thrombin digestion and filtration; CD4mim-Ig and DM1-Ig were bound to protein A-Sepharose, washed, and digested with thrombin in 5 mM Tris-150 mM NaCl2, pH 8, and released peptides were eluted through Ultra YM10 (Millipore) filter units. For quantification, aliquots of these peptides were directly analyzed by SDS-PAGE (total, left panel). Separate aliquots were incubated with purified, unlabeled ADA-Ig or S1-Ig (a control protein of the SARS coronavirus S1 domain), precipitated with protein A-Sepharose, and analyzed by SDS-PAGE (right panel). (D) The experiment shown is similar to that shown in panel C, except that DM1 was produced in the presence or absence of tyrosyl protein sulfotransferase 2 (TPST2). (E) Metabolically labeled gp120 molecules of the indicated HIV isolates were precipitated with CD4-Ig or DM1-Ig and analyzed by SDS-PAGE. Clades of the indicated isolates are indicated at the top.
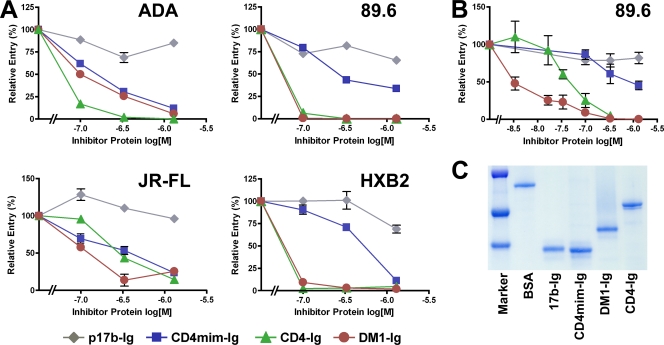
DM1-Ig efficiently inhibits HIV-1 entry.
To determine whether DM1 or DM1-Ig could serve as a lead peptide for a new peptide inhibitor of HIV-1 entry, we compared it to CD4-Ig for its ability to prevent the infection of four well-characterized clade B HIV-1 isolates. DM1-Ig neutralized the entry of the R5 isolate JRFL, the R5X4 isolate 89.6, and the X4 isolate HXB2 comparably to CD4-Ig, whereas it neutralized the R5 ADA isolate less efficiently (Fig. 5A). Figure 5B underscores one advantage of DM1-Ig over CD4-Ig. Specifically, DM1-Ig does not enhance infection at any tested concentration, whereas, as previously reported, neutralization by CD4-Ig at low concentrations reflects some enhancement of infection, presumably by promoting virion association with CCR5. Figure 5C shows a Coomassie stain of purified fusion proteins used in Fig. 5A and B, indicating the relative absence of contaminants. Figure 5 thus indicates that DM1-Ig neutralizes some HIV-1 isolates comparably to or more efficiently than CD4-Ig and suggests that, with improvements in its CD4mim and CCR5mim components, peptides like DM1 will make potent inhibitors of HIV-1 entry.
Fig. 5.
DM1 efficiently neutralizes R5, R5X4, and X4 isolates. (A) HIV-1 of the indicated isolates was incubated with GHOST-CCR5 (ADA, JR-FL, and 89.6) or GHOST-CXCR4 (HXB2) cells in the presence of 0, 120, and 400 nM or 1.6 μM indicated peptide-Ig variants. Cells were washed 2 h postincubation, and viral entry was measured by flow cytometry 2 days later. The experiment shown is representative of three experiments with similar results. (B) The experiment shown is similar to that shown in panel A, except that lower concentrations (4 and 20 nM) of inhibitor were included to detect enhancement effects, if any, at low concentrations. The experiment is representative of two experiments with similar results. (C) Coomassie-stained gel used to normalize expression of proteins used in panels A and B. Error bars in panels A and B indicate the range of two replicates within experiments.
DISCUSSION
The only accessible and conserved regions of HIV-1 gp120 are its CD4 and coreceptor-binding sites (46). These conserved regions are smaller than the epitopes of most neutralizing antibodies, and therefore it is relatively easy for HIV-1 to escape neutralization from any single antibody (22, 25–27, 46). Several CD4 and CCR5-mimetic peptides have been described whose association with gp120 is limited to their respective receptor-binding regions (12, 19, 23, 32). Here, we explore the properties of two of these peptides, CCR5mim and CD4mim.
We show that, compared to CCR5mim-Ig, CD4mim-Ig more efficiently binds monomeric gp120 in solution, whereas CCR5mim-Ig more efficiently associates with cell surface-expressed HIV-1 envelope glycoprotein trimers. There are at least two possible reasons for this difference. First, as cryoelectron microscopy studies indicate (30), the sulfotyrosine-binding pockets of gp120 are readily accessible at the top (membrane-distal) region of the envelope glycoprotein, whereas the CD4-binding site is partially occluded in the trimer, perhaps slowing the on-rate of CD4mim-Ig. Second, because the sulfotyrosine-binding pockets are near the trimer axis, CCR5mim-Ig can readily bind two of the three gp120 molecules of the trimeric spike. We confirmed this second possibility by comparing CCR5mim-Ig with a monomeric form of the same molecule: the association of the envelope glycoprotein trimer with monomeric CCR5mim-Ig was markedly less efficient than with its original dimeric form. In contrast, monomeric and dimeric forms of CD4-Ig bound the envelope glycoprotein trimer with comparable efficiencies, demonstrating that dimeric CCR5mim-Ig, but not CD4-Ig, can access two gp120 molecules of the trimer. However, our data show that, like CCR5mim-Ig and unlike CD4-Ig, CD4mim-Ig can bridge two gp120 monomers of the envelope glycoprotein trimer. Nonetheless, cryoelectron microscopy structures of the CD4-liganded and unliganded envelope glycoprotein trimers (30) suggest that the binding of the second CD4mim peptide comes at a higher energetic price than in the case of CCR5mim-Ig, explaining in part the more efficient binding of dimeric CCR5mim-Ig to the trimer. This price may arise from steric interference between the envelope glycoprotein and the Fc domain, or it may arise because the two CD4mim domains are held too closely to permit the more open CD4-liganded conformation of the envelope glycoprotein (30).
We also describe the ability of CCR5mim-Ig to promote and stabilize the association of CD4mim-Ig. It is unclear whether CCR5mim-Ig enhances the on-rate of CD4mim-Ig by inducing the CD4-bound conformation of the envelope glycoprotein, or whether it simply stabilizes that conformation, decreasing the off-rate of CD4mim-Ig. One observation, which suggests both mechanisms are relevant, is that CCR5mim-Ig can promote the exposure of HR1 of gp41 in envelope glycoproteins that it binds avidly, notably that of the 89.6 isolate (Fig. 3D and E). This observation suggests that CCR5mim-Ig can induce quaternary changes in the envelope glycoprotein similar to those observed with soluble CD4 (21, 37). It is possible that these changes both promote and stabilize the association of CD4mim-Ig with the envelope glycoprotein. The ability of CCR5mim-Ig to expose HR1 also raises the possibility that future variants of the peptides described here can be used effectively in concert with T20/enfuvirtide.
In the experiments shown, the dual-tropic 89.6 envelope glycoprotein bound CCR5mim-Ig more efficiently than most other isolates (Fig. 2B). CCR5mim-Ig also exposed the HR1 region of this envelope glycoprotein more effectively than the other envelope glycoproteins tested (Fig. 3D and E). Moreover, it was the only isolate observed to be neutralized by DM1-Ig more efficiently than by CD4-Ig (Fig. 5A and B). The 89.6 isolate also is among the most sensitive to perturbations of individual CCR5 sulfotyrosines (17). These observations suggest that the sulfotyrosine-binding pockets of the 89.6 envelope glycoprotein are exposed and of especially high affinity. This property may facilitate dual tropism by permitting high-affinity interaction with CCR5 while simultaneously enabling adaptation to the extracellular loop domains of CXCR4 (31). If so, inhibitors that include tyrosine-sulfated CCR5-mimetic peptides might slow the emergence of R5X4 and X4 isolates.
The relatively high avidity of CCR5mim-Ig for the envelope glycoprotein and the bidirectional cooperativity between CCR5mim-Ig and CD4mim-Ig suggested that a fusion of these two peptides would be especially efficient in inhibiting HIV-1 entry. Our first combination of these peptides, DM1, appears promising. DM1-Ig precipitated gp120 molecules from a range of isolates with efficiencies comparable to those of CD4-Ig and markedly better than those of either receptor-mimetic peptide alone. DM1 peptide, lacking an Fc domain, also bound gp120 much more efficiently than the CD4mim peptide or, presumably, the CCR5mim peptide. DM1-Ig also neutralized several HIV-1 isolates comparably to CD4-Ig, and notably it did not enhance infection at low concentrations, a persistent limitation of the use of soluble CD4 and CD4-Ig as a therapy (39). Unlike other constructs that similarly target the receptor- and coreceptor-binding sites of gp120 (1, 10, 43), DM1 is compact and made of natural amino acids, facilitating its further improvement by phage-based selection.
Although promising, DM1-Ig still has significant limitations as a therapeutic. First, CD4-Ig and DM1-Ig neutralize HIV-1 much less efficiently than the best neutralizing antibodies, so considerable improvements in affinity would be necessary for DM1 or DM1-Ig to be effective in vivo. Indeed, each component of DM1-Ig must be made effective independently to limit escape through the evasion of a single component. Because both components of DM1 are composed of natural amino acids, including sulfotyrosines, that are amenable to phage display (4, 29), such improvements are possible. Second, despite the fact that DM1 binds the two most conserved regions of gp120 and very little else, it does not efficiently neutralize many non-clade B isolates, presumably because each component of DM1 was selected against a clade B isolate (6, 32) and because they are imperfect mimetics of the receptors they mimic. Phage-based selection for binding gp120 molecules from other clades could enhance the breadth of DM1. Finally, the CD4mim and CCR5mim components of DM1 cannot bind the same gp120, as indicated by the fact that a DM1-Ig variant without a linker between its CD4mim and CCR5mim components bound gp120 and neutralized HIV-1 as efficiently as DM1-Ig itself (not shown). This observation indicates that the 9-amino-acid linker is insufficient to reach both receptor-binding sites. The inspection of the structure of gp120 suggests that if the order of mimetics can be reversed, the linked mimetics could bind a single gp120 monomer of the trimer. This change requires the elimination of the first disulfide bond of CD4mim (32) and subsequent optimization through phage display. The resulting construct likely would substantially enhance the association of CD4mim with the envelope glycoprotein trimer, because all four mimetic domains of DM1-Ig could bind the trimer. It therefore is both possible and necessary to make the effects of DM1 broader and more potent.
How can an improved DM1 or DM1-Ig be used therapeutically? In most contexts, peptides and proteins are less practical as therapies than orally available small molecules with similar activities. However, a recent study has shown that a one-time injection with an adeno-associated virus (AAV)-based vector can deliver antibody-like molecules at high titers for several years and protect rhesus macaques from an SIV challenge (24). It is likely that an improved version of DM1-Ig, or perhaps DM1-Ig itself, would be similarly protective against HIV-1. Nonetheless, an AAV-based prophylaxis faces considerable technical and especially safety challenges. These hurdles may be somewhat less daunting for an AAV-delivered therapeutic, but such a therapy faces at least two challenges: the possibility of immune clearance by anti-inhibitor antibodies and the problem of viral escape. However, peptides (for example, enfuvirtide/T20) elicit a less robust antibody response than larger proteins (33), and this response is attenuated in late-stage HIV-1 infections. Thus, peptides such as DM1 might avoid clearance by the immune system. Also, in contrast to HIV-1 neutralizing antibodies, the peptides we describe here bind solely the conserved CD4- and coreceptor-binding sites (12, 23), limiting the available pathways of viral escape. It therefore is possible that improved versions of CCR5mim, CD4mim, or DM1 can be optimized to anticipate these limited pathways and to stably suppress viral replication.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI080324 (H.C. and M.F.) and by New England Primate Research Center Base grant RR000168 (M.F.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Baleux F., et al. 2009. A synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry. Nat. Chem. Biol. 5:743–748 [DOI] [PubMed] [Google Scholar]
- 2. Carr C. M., Kim P. S. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823–832 [DOI] [PubMed] [Google Scholar]
- 3. Choe H., Farzan M. 2009. Chapter 7. Tyrosine sulfation of HIV-1 coreceptors and other chemokine receptors. Methods Enzymol. 461:147–170 [DOI] [PubMed] [Google Scholar]
- 4. Choe H., Farzan M. 2006. Tyrosine sulfate trapped by amber. Nat. Biotechnol. 24:1361–1362 [DOI] [PubMed] [Google Scholar]
- 5. Choe H., et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 6. Choe H., et al. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161–170 [DOI] [PubMed] [Google Scholar]
- 7. Connor R. I., Sheridan K. E., Ceradini D., Choe S., Landau N. R. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cormier E. G., Tran D. N., Yukhayeva L., Olson W. C., Dragic T. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Decker J. M., et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dey B., Del Castillo C. S., Berger E. A. 2003. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J. Virol. 77:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doms R. W., Peiper S. C. 1997. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology 235:179–190 [DOI] [PubMed] [Google Scholar]
- 12. Dorfman T., Moore M. J., Guth A. C., Choe H., Farzan M. 2006. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 281:28529–28535 [DOI] [PubMed] [Google Scholar]
- 13. Dragic T., et al. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673 [DOI] [PubMed] [Google Scholar]
- 14. Eckert D. M., Kim P. S. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810 [DOI] [PubMed] [Google Scholar]
- 15. Farzan M., et al. 2002. The role of post-translational modifications of the CXCR4 amino-terminus in SDF-1a association and HIV-1 entry. J. Biol. Chem. 28:28. [DOI] [PubMed] [Google Scholar]
- 16. Farzan M., et al. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farzan M., et al. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676 [DOI] [PubMed] [Google Scholar]
- 18. Farzan M., et al. 2001. Sulfated tyrosines contribute to the formation of the C5a docking site of the human C5a anaphylatoxin receptor. J. Exp. Med. 193:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farzan M., et al. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516–33521 [DOI] [PubMed] [Google Scholar]
- 20. Feng Y., Broder C. C., Kennedy P. E., Berger E. A. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 21. Furuta R. A., Wild C. T., Weng Y., Weiss C. D. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 22. Huang C. C., et al. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C. C., et al. 2005. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure 13:755–768 [DOI] [PubMed] [Google Scholar]
- 24. Johnson P. R., et al. 2009. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15:901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlsson Hedestam G. B., et al. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143–155 [DOI] [PubMed] [Google Scholar]
- 26. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Labrijn A. F., et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao H. X., et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu C. C., Choe H., Farzan M., Smider V. V., Schultz P. G. 2009. Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry 48:8891–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu Z., et al. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. U. S. A. 94:6426–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin L., et al. 2003. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 21:71–76 [DOI] [PubMed] [Google Scholar]
- 33. Matthews T., et al. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215–225 [DOI] [PubMed] [Google Scholar]
- 34. Pejchal R., et al. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rizzuto C. D., et al. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 36. Rucker J., et al. 1996. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437–446 [DOI] [PubMed] [Google Scholar]
- 37. Si Z., et al. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:5036–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stricher F., et al. 2005. A high-throughput fluorescence polarization assay specific to the CD4 binding site of HIV-1 glycoproteins based on a fluorescein-labelled CD4 mimic. Biochem. J. 390:29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sullivan N., et al. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trkola A., et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 41. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 43. West A. P., Jr., et al. 2010. Evaluation of CD4-CD4i antibody architectures yields potent, broadly cross-reactive anti-human immunodeficiency virus reagents. J. Virol. 84:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong S. K., Li W., Moore M. J., Choe H., Farzan M. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu L., et al. 1996. CD4-induced interactions of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 184:179–183 [DOI] [PubMed] [Google Scholar]
- 46. Wyatt R., et al. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.