Abstract
Herpes simplex virus (HSV) immediate-early protein ICP0 is a transcriptional activator with E3 ubiquitin ligase activity that induces the degradation of ND10 proteins, including the promyelocytic leukemia protein (PML) and Sp100. Moreover, ICP0 has a role in the derepression of viral genomes and in the modulation of the host interferon response to virus infection. Here, we report that ICP0 interacts with SIAH-1, a cellular E3 ubiquitin ligase that is involved in multiple cellular pathways and is itself capable of mediating PML degradation. This novel virus-host interaction profoundly stabilized SIAH-1 and recruited this cellular E3 ligase into ICP0-containing nuclear bodies. Moreover, SIAH-1 mediated the polyubiquitination of HSV ICP0 in vitro and in vivo. After infection of SIAH-1 knockdown cells with HSV, higher levels of ICP0 were produced, ICP0 was less ubiquitinated, and the half-life of this multifunctional viral regulatory protein was increased. These results indicate an inhibitory role of SIAH-1 during lytic infection by targeting ICP0 for proteasomal degradation.
INTRODUCTION
ICP0 is a member of the class of immediate-early gene products of herpes simplex virus 1 and 2 (HSV-1 and -2), with homologs in other herpesvirus subfamilies. ICP0 is required for the efficient initiation of viral lytic infection and reactivation from latently infected neurons (reviewed in references 22, 34, 35, and 65). ICP0 appears to be a multifunctional regulator of gene expression that directly and indirectly interacts with numerous viral and cellular proteins. For example, these include the viral transcriptional activator ICP4, the cellular transcription factor BMAL1, the translation initiation factor 1α, cell cycle regulators such as cyclins, and the tumor suppressor p53 (6, 45–47, 81). ICP0 is also a component of viral particles, where it can be detected in a salt-resistant fraction of the tegument, suggesting it to be an inner tegument protein closely associated with the capsid (21, 53, 66).
Early in infection, newly synthesized ICP0 localizes to discrete subnuclear structures, which are characterized by the presence of the promyelocytic leukemia protein (PML) and are variously referred to as nuclear domain 10 (ND10), Kremer bodies, PML nuclear bodies, or PML oncogenic domains (PODs). ND10 has been implicated in a variety of cellular processes, including the regulation of growth control, senescence, apoptosis, transformation, and antiviral responses (for reviews, see references 4, 15, and 24). During the early stage of infection, HSV genomes locate preferentially in the periphery of ND10, and these nuclear areas are considered to be the sites where viral transcription initiates (23, 55). Importantly, later in infection the ND10 components PML and Sp100, which is another major and constitutive component of ND10 (7, 12, 27), are rearranged and disrupted by the E3 ubiquitin ligase activity of ICP0, which is mediated by a zinc-binding RING finger domain near its amino terminus (8, 28, 56, 57). Nevertheless, ubiquitination of PML by ICP0 was not reconstituted in vitro (7), suggesting additional proteins involved in this activity. ICP0 itself autoubiquitinates via its RING domain, a process counteracted by the ubiquitin-specific protease USP7, which is in turn subject to ICP0-mediated degradation (5, 10).
Furthermore, ICP0 counteracts silencing of viral DNA by dissociating the histone deacetylases (HDAC) 1 and 2 from the repressor complex CoREST/REST, permitting progression of viral gene expression from the immediate-early to the early and late stages (33, 68). Nevertheless, these functions appear to be dispensable for ND10 breakdown and the reactivation of quiescent genomes (25, 29).
The cellular E3 ubiquitin ligase SIAH has been described to target PML (31). In fact, SIAH-1-mediated PML degradation results in the loss of the transcriptional coactivating properties of PML and the significant reduction of the number of ND10 (31). SIAH family members represent mammalian homologs of the Drosophila SINA (seven in absentia) protein (11), of which the two human homologs SIAH-1 and SIAH-2 have been described (41). Through their intrinsic E3 ubiquitin ligase activity, they play an important role in the proteasome-mediated degradation of various proteins involved in transcriptional regulation, cell growth, tumorigenesis, and hypoxia signaling (32, 38, 41, 51, 54, 59, 62, 82). SIAH-1 targets a constantly growing number of proteins, including itself, for ubiquitination and proteasome-dependent degradation (18, 31, 40, 41, 51, 54, 77, 80). Moreover, for many target proteins of SIAH, a consensus binding motif was identified (36, 37).
SIAH-1 is expressed in various tissues, including neurons (39, 61, 77). Interestingly, its highly related Drosophila homolog SINA targets the transcriptional repressor Tramtrack, which is a potent repressor of neuronal cell fate (11, 48, 73). It therefore appears that SIAH-1 and its homologs fulfill important regulatory functions, particularly in neuronal tissues, the site where HSV latent infection is established and reactivation is initiated (69).
In the present study, we identified a consensus SIAH-1 interaction motif in the HSV immediate-early gene product ICP0. By coimmunoprecipitation, we found ICP0 to be a novel SIAH-1 interaction partner. Transient overexpression of ICP0 or infection with HSV profoundly stabilized SIAH-1 by a posttranslational mechanism. The specific interaction of SIAH-1 and ICP0 resulted in polyubiquitination of the latter, whereas there was no apparent influence of SIAH-1 binding on the ability of ICP0 to mediate PML degradation. By constitutively silencing SIAH-1, we observed improved stability of ICP0 during infection. Therefore, we postulate that the interaction of ICP0 and SIAH-1 results in ubiquitination and proteasome-mediated degradation of the viral ICP0 regulatory protein. The implication of this so-far-unknown virus-host interaction with respect to the viral life cycle is discussed.
MATERIALS AND METHODS
Cells and viruses.
HEK293T (ATCC CRL-11268) and U2OS (ATCC HTB-96) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, and 3.75 mg/ml sodium bicarbonate. Vero (ATCC CCL-81) and BHK-21 (ATCC CCL-10) cells were maintained in MEM supplemented with 10% FCS, 2 mM l-glutamine, 10 mM HEPES, and 2.25 mg/ml sodium bicarbonate.
HSV-1, strain 17+, and the ICP0-negative dl1403 mutant (HSV-1ΔICP0) (26) were provided by Roger D. Everett (MRC, Glasgow, United Kingdom); HSV-2, strain US, was provided by Andreas Sauerbrei and Peter Wutzler (Universität Jena, Germany). Wild-type HSV was propagated in BHK-21 cells, and HSV-1ΔICP0 in U2OS cells. All viruses were plaque-titrated on U2OS cells in the presence of 25 μg/ml pooled human IgG (19). For a synchronized infection, cells were inoculated for 2 h on ice in a small volume of virus to allow attachment of virions followed by washing and incubation at 37°C for different amounts of time.
Expression constructs.
The HSV-1 and HSV-2 ICP0 coding sequences (17) were synthesized in vitro (Geneart, Regensburg, Germany) and ligated as carboxy-terminal Flag or green fluorescent protein (GFP) fusions between the BamHI and XhoI site of the eukaryotic expression vector pcDNA3 (Invitrogen, Darmstadt, Germany). The proteins will be referred to as ICP0(1) and ICP0(2), respectively. Various internal ICP0(2) deletion mutants were constructed by removal of the respective nucleotides by PCR or via restriction sites, which were introduced during the in vitro synthesis, and religation using the appropriate oligonucleotide linkers. The ICP0(2)ΔRING mutant was derived by eliminating nucleotides corresponding to codons 126 to 166 in the ICP0(2) coding sequence. For bacterial expression of the amino-terminal hexahistidine-tagged fusion protein, the ICP0(2) coding sequence was inserted between the NheI and XhoI sites of pET-28b (Novagen, Darmstadt, Germany). The plasmids encoding hemagglutinin (HA)-ubiquitin, N-terminally HA-tagged SIAH-1, and inactive HA–SIAH-1–C44S, the bacterial expression construct for GST–SIAH-1, were described elsewhere (74, 75, 80).
DNA transfection.
293T cells were transfected using polyethylenimine (PEI; Polysciences, Eppelheim, Germany) at a microgram ratio of PEI and DNA of 6:1. Vero cells were transfected using Lipofectamine 2000 (Invitrogen) or TransIT (Mirus Bio, Madison, WI) according to the manufacturer's protocol. When required, the amount of total input DNA in each transfection was kept constant by the addition of the respective parental vector.
Reagents and primary antibodies.
Protein G Sepharose was obtained from GE Healthcare (Freiburg, Germany). GFPtrapA beads consist of a single-chain GFP antibody immobilized on agarose beads (Chromotek; Martinsried, Germany). Proteasome inhibitor MG132 was purchased from Sigma (Munich, Germany), and TriFast reagent was purchased from Peqlab (Erlangen, Germany). The following primary antibodies were used: anti-Flag M2, anti-tubulin DM1A, mouse IgG (all from Sigma); anti-GFP antibody (Innogenetics, Gent, Belgium); anti-PML H-238, anti-HA Y-11, anti-SIAH-1 N-15, anti-HSV-1 ICP0 11086, anti-HSV-1/2 ICP4 H943, anti-ubiquitin P4D1 (Santa Cruz, CA); anti-HSV-1/2 VP5 H1.4 (Acris, Herford, Germany); and anti-GST-DX700, anti-HA-DX700, and anti-His-DX-700 antibody (IRDye-labeled antibodies; Rockland Immunochemicals, Gilbertsville, CA).
Immunoprecipitation studies.
293T cells were harvested 1 or 2 days posttransfection and washed twice in phosphate-buffered saline (PBS). Subsequently, the cells were resuspended in E1A lysis buffer (300 mM NaCl, 50 mM HEPES [pH 7.5], 5 mM EDTA, 0.5% NP-40) or radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-Cl [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), both containing protease inhibitors (5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, and 125 μg/ml Pefabloc), and incubated on ice for 30 min. The lysates were centrifuged for 15 min at 20,000 × g, and the supernatant was transferred to a new tube. For preclearing, protein G Sepharose beads, which were three times equilibrated in E1A wash buffer (150 mM NaCl, 50 mM HEPES [pH 7.5], 5 mM EDTA, 0.5% NP-40), were added to the lysates and the tubes were placed on an overhead mixer at 4°C. After 60 min, the beads were sedimented by centrifugation at 100 × g and the supernatant was divided and transferred to new tubes. Fresh protein G Sepharose beads, together with 5 μg of specific antibody, were added to one half of the supernatant. As a control, 5 μg rabbit or mouse IgGs and fresh beads were added to the other half. Alternatively, washed GFPtrapA beads were added to the cell lysates after centrifugation without further preclearing. After overnight incubation on an overhead mixer at 4°C, the beads were sedimented by centrifugation at 100 × g and washed three times in E1A wash buffer. SDS sample buffer (2×) was added to the precipitates and boiled for 10 min at 95°C, and analysis was performed by SDS-PAGE and immunoblotting.
Pulse-chase experiments.
At 24 h posttransfection, 293T cells were washed once with cell culture medium without cysteine and methionine and incubated further in the same medium for 1 h before 20 μCi TRAN35S-LABEL (MP Biomedicals, Eschwege, Germany) was added for 30 min. Subsequently, cells were washed twice and further incubated in DMEM. At various time points, cells were lysed and equal amounts of the respective total protein extracts were used for immunoprecipitation as described before. The labeled proteins were analyzed by SDS-PAGE and autoradiography. The degree of SIAH-1 expression was quantified by densitometric analysis of the respective signals using AIDA image software (Raytest, Straubenhardt, Germany).
Western blot analyses.
For whole-cell lysates, cells were washed twice with PBS and lysed. After sonication, 1× SDS sample buffer was added to the cell lysates and equal amounts of total protein were separated by SDS-PAGE. Alternatively, cells were washed with PBS and directly scraped into hot sample buffer containing protease inhibitors. Proteins were transferred to nitrocellulose membrane (Schleicher & Schuell) by immunoblotting, followed by enhanced chemiluminescence (ECL) detection (GE Healthcare) after applying the appropriate antibodies. Alternatively, IRDye700- or IRDye800-labeled secondary antibody (Rockland) was applied and fluorescence detected with an Odyssey Imager (LI-COR Biosciences, Bad Homburg, Germany).
Overexpression and purification of recombinant ICP0(2) and SIAH-1.
Overnight-grown Escherichia coli BL21(DE3) CodonPlus-RIPL cells, harboring the pET28b-ICP0 plasmid, were taken to inoculate 500 ml of LB medium containing kanamycin (100 μg/ml) and grown for 2 h at 37°C. The culture was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated for another 6 h at 22°C. The purification was carried out according to Canning and coworkers (10) with slight modifications. Briefly, the cells from the induced culture were harvested by centrifugation and resuspended in buffer A (100 mM Tris-Cl [pH 8.0], 500 mM NaCl, 10% glycerol, 2 mM dithiothreitol [DTT], 0.1% Nonidet P-40, and 0.1 Triton X-100) in the presence of complete EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany) and lysed by sonication. Insoluble debris was removed by centrifugation, and subsequently cleared lysate was filtered through a 0.22-μm filter. The filtrate was directly loaded to a 2-ml Ni-nitrilotriacetic acid (NTA) bead column, preequilibrated with buffer A. The column was washed with 100 ml of buffer B (100 mM Tris-Cl [pH 8.0], 250 mM NaCl, 5% glycerol, 20 mM imidazole [pH 8.0]). The protein was eluted from the column in 5 ml of buffer C (100 mM Tris-HCl [pH 8.0], 250 mM NaCl, 5% glycerol, 2 mM DTT, and 300 mM imidazole [pH 8.0]). The 5-ml fraction was concentrated to 1 ml using an Amicon Ultra-15 (Millipore) membrane with a molecular weight cutoff of 50. Imidazole was removed by dialyzing the protein sample twice in 1 liter of buffer D (50 mM Tris-Cl [pH 8.0], 300 mM NaCl, 10% glycerol, 2 mM DTT, and 2.5 mM MgCl2) and stored at −20°C until further use. The purity of the protein was determined by 10% SDS-PAGE.
For purification of SIAH-1, overnight cultures of E. coli BL21 Gold (DE3) pLysS harboring the plasmid encoding GST–SIAH-1 were used to inoculate 500 ml of LB medium containing ampicillin (100 μg/ml) and grown for 2 h at 37°C (optical density at 600 nm [OD600] = 0.65). The cultures were induced with 0.5 mM IPTG and incubated for another 4 h at 30°C. The cells from the induced cultures were harvested by centrifugation and resuspended in 20 ml of lysis buffer (1× PBS [pH 7.4], containing complete protease inhibitor cocktail tablets from Roche). The cells were lysed by sonication, and the lysate was then centrifuged for 30 min at 14,000 rpm at 4°C. To the supernatants, 500 μl of glutathione S-transferase (GST) beads preequilibrated with PBS (pH 7.4) was added, and the mixture was incubated for 1 h at 4°C in an end-to-end rotor. After 1 h, the mixture was loaded to gravity flow columns and the unbound sample was collected. The column containing beads was then washed with 2 column volumes of PBS. The column was further washed with 3 column volumes of 50 mM Tris-Cl (pH 8.0) and 300 mM NaCl. Finally, the protein was eluted in 2 ml with 50 mM Tris-Cl (pH 8.0), 300 mM NaCl, and 20 mM reduced glutathione. The purified fractions were dialyzed against 50 mM Tris-Cl (pH 7.4), 5 mM MgCl2, and 2 mM DTT.
In vitro ubiquitination assay.
Recombinant human ubiquitin-activating enzyme UBE1, ubiquitin-conjugating enzyme UbcH5b, and ubiquitin were purchased from Boston Biochem (Cambridge, MA). The in vitro ubiquitination reaction was set up in a total volume of 30 μl with 100 nM UBE1, 392 nM UbcH5b, 39.2 μM ubiquitin, 8.3 ng/μl SIAH-1, 17 to 170 μg/μl ICP0 in reaction buffer (50 mM NaCl, 2 mM DTT, 5 mM MgCl2, 50 mM Tris-Cl [pH 7.4], 5 mM ATP) (10, 18, 80). After 1 h of incubation at 37°C, 6 μl 6× SDS-PAGE buffer was added and the samples were analyzed by SDS-PAGE and Western blotting.
Immunofluorescence staining and microscopy.
Vero cells were seeded onto 10-mm glass coverslips in 24-well plates at a density of 7.5 × 104 cells per well. After transfection and/or infection with HSV, they were washed with PBS and subsequently fixed in 3% paraformaldehyde (PFA) for 20 min. Following washing with 50 mM NH4Cl-PBS, cells were permeabilized with 0.1% Triton X-100–PBS for 5 min and blocked with 0.5% bovine serum albumin (BSA)-PBS for 30 min. Proteins were immunolabeled in 0.5% BSA-PBS using the respective primary antibodies, followed by fluorescence-coupled secondary antibodies (Molecular Probes, Invitrogen). Nuclear DNA was visualized by Draq5 (Enzo Life Sciences, Lörrach, Germany). Samples were analyzed on an Axiovert 200 M microscope equipped with an LSM 510 META confocal laser scanning unit (Zeiss, Jena, Germany) using a Plan-Apochromatic ×63 oil immersion objective lens with a 1.4 numeric aperture. Image acquisition and processing were performed by using the Zeiss LSM imaging software and Adobe Photoshop CS3 (Adobe Systems, San Jose, CA).
Viral complementation assay. (i) Viral gene expression.
293T cells were plated at 5 × 105 cells per well in a 32-mm-diameter plate. The next day, the cells were transfected using PEI with 0.5 μg of HSV-1ΔICP0 DNA isolated from capsids (52) and 0.5 μg of pcDNA3 encoding either GFP, ICP0(2)-GFP, or ICP0(2)Δ410-420–GFP. After 2 days, cells were harvested and analyzed by Western blotting.
(ii) Virus reconstitution.
The capability of ICP0(2) to complement HSV-1ΔICP0 was analyzed as described before (9, 16). Vero cells were plated at 2 × 105 cells per well in a 6-well plate. The next day, the cells were transfected using Fugene 6 (Roche) with a total of 3 μg DNA [0.5 μg HSV-1ΔICP0 DNA, salmon sperm DNA, and 0.075 μg of pcDNA3 encoding either GFP, ICP0(2)-GFP, or ICP0(2)Δ410-420–GFP]. Transfected cells were harvested 48 h after transfection, lysed by freeze-thawing, and plaque titrated on U2OS cells. Alternatively, the cells were cultured after transfection for 48 h in medium with 25 μg/ml pooled human IgG, and plaques were immunostained after methanol fixation using anti-VP5 antibody together with IRDye detection (Li-COR Biosciences).
RNA isolation, cDNA synthesis, and quantitative PCR.
Total RNA was isolated using TriFast (Peqlab) according to the manufacturer's instructions. RNA samples were reversely transcribed using the first-strand cDNA synthesis kit for reverse transcription (RT)-PCR (Moloney murine leukemia virus [MMLV]; Promega) according to the manufacturer's instructions. Primers and probes for cDNA analysis by quantitative PCR (TaqMan; ABI) were purchased from Sigma. For glyceraldehyde-3-phosphate dehydrogenase (GAPDH) detection, we used GTC ATC AAT GGA AAT CCC ATC A and TGG TTC ACA CCC ATG ACG AA as primers and 5′-(6-tetramethyl rhodamire [FAM])-TCT TCC AGG AGC GAG ATC CCT C-(tetramethylrhodamine [TAMRA])-3′ as the probe. For SIAH-1 detection, we used TAA ATG GTC ATA GGC GAC GA and GCA ATG CTG GTG TCA AAG AC as primers and 5′-(FAM)-CGA GGA GTC GCT TCC CAA GTC A-(TAMRA)-3′ as the probe.
Lentiviral-based RNAi.
Replication-incompetent and self-inactivating lentiviral vectors (adapted from references 20 and 70) that express GFP together with either SIAH-1-specific short hairpin RNA (shRNA) (targeting nucleotides 203 to 221 in the SIAH-1 open reading frame [ORF]; GenBank accession no. NM_003031) or off-target (luciferase-specific) shRNA under the control of the histone 1 gene promoter were used for RNA interference (RNAi). For production of lentiviral pseudotypes, 2.5 × 106 293T cells were cotransfected with either 3 μg of the SIAH-1 or luciferase-specific lentiviral vector together with 1.5 μg pMDLg/pRRE, 0.75 μg pRSV-Rev, and 0.75 μg pCMV–VSV-G (vesicular stomatitis virus glycoprotein) (3, 20) using PEI. At 48 h posttransfection, the culture supernatants containing lentiviral particles were harvested and passed through 0.45-μm-pore-size filters, and viral p24Gag antigen levels were determined by enzyme-linked immunosorbent assay (ELISA; Innogenetics N.V.). Subsequently, Vero cells were transduced by spinoculation as previously described (63) using virus-containing supernatants equivalent to 500 ng of p24 antigen per 5 × 105 cells. GFP-positive cells were enriched by their fluorescence on a BD FACSAria cell sorter (BD, Heidelberg, Germany).
RESULTS
ICP0 associates with SIAH-1.
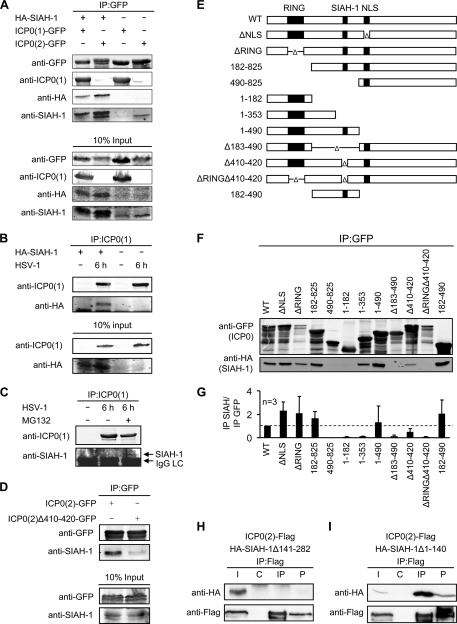
The ICP0 homologs in the genus Simplexvirus in the Alphaherpesvirinae subfamily of herpesviruses contain the consensus SIAH-1 binding motif PXAXVXPXXR, which is present in many functionally diverse SIAH-1 targets. The most important core residues, VXP (36, 37), are conserved in all homologs as analyzed by ClustalW alignment (see Fig. S1A in the supplemental material). Therefore, we analyzed whether the ICP0 homologs of herpes simplex virus 1 and 2, ICP0(1) and ICP0(2), respectively, formed a complex with SIAH-1 by coimmunoprecipitation assays. 293T cells were transfected with expression vectors encoding GFP-tagged ICP0 together with HA-tagged SIAH-1 or empty vector (Fig. 1A). If not otherwise stated, the transfections were conducted in the presence of the proteasome inhibitor MG132 to increase the amount of SIAH-1, as this protein is very unstable, due to self-ubiquitination (40) (see below). HA–SIAH-1 was clearly detected in immunoprecipitates of both ICP0 homologs. Endogenous SIAH-1 was also detected in the immunoprecipitates, suggesting a physiological relevance of this interaction. With ICP0(1), however, less (HA)SIAH-1 coimmunoprecipitated than with ICP0(2). Due to the low abundance and detection difficulties of endogenous SIAH-1, most experiments analyzing the interaction of SIAH-1 and ICP0 were performed with HA-tagged SIAH-1.
Fig. 1.
ICP0 associates with SIAH-1. (A) SIAH-1 interacts with ICP0. 293T cells (2.5 × 106) were transfected with 3 μg expression vector encoding either ICP0(1)-GFP or ICP0(2)-GFP and 3 μg of HA–SIAH-1. Alternatively, the cells were transfected alone with 6 μg expression vector encoding either ICP0(1)-GFP or ICP0(2)-GFP. The cells were harvested after 2 days. Eight hours before cell lysis, 25 μM MG132 was added to the cultures. Immunoprecipitation was performed using anti-GFP immunobeads. Proteins in the immunoprecipitates and the input controls were detected by Western blotting with anti-GFP, anti-ICP0(1), anti-HA, or anti-SIAH-1 antibodies. (B) HA–SIAH-1 interacts with ICP0 of HSV-1 during infection. 293T cells were transfected overnight with HA–SIAH-1 or left untreated and infected with HSV-1 at an MOI of 10 for 6 h. Immunoprecipitation was performed with anti-ICP0(1) antibody, and precipitates were subjected to SDS-PAGE. Proteins in the immunoprecipitates or the input controls were analyzed by Western blotting using anti-ICP0(1) and anti-HA antibodies. (C) ICP0 also precipitates endogenous SIAH-1 after HSV-1 infection. 293T cells were infected with HSV-1 at an MOI of 10 for 6 h in the presence or absence of 25 μM MG132. After ICP0(1) immunoprecipitation, proteins in the immunoprecipitates or the input controls were analyzed by Western blotting using anti-ICP0(1) and anti-SIAH-1 antibodies. (D) Amino acid residues 410 to 420 of ICP0(2) are necessary for efficient binding to endogenous SIAH-1. 293T cells (2.5 × 106) were transfected with 6 μg expression vector encoding either ICP0(2)-GFP or ICP0(2)Δ410-420 and analyzed by immunoprecipitation and Western blotting as described before. Eight hours before cell lysis, 25 μM MG132 was added to the cultures. (E) Schematic representation of HSV-2 ICP0 mutants tested for their ability to bind SIAH-1. The amino-terminal RING finger domain, the predicted SIAH-1 binding motif, and the nuclear localization signal (NLS) of ICP0 are indicated by solid boxes. WT, wild type; Δ, internal deletion. (F) 293T cells (2.5 × 106) were cotransfected with 3 μg ICP0-GFP and 3 μg of HA–SIAH-1 expression vector. Before harvesting, the cells were exposed to 25 μM MG132 for 8 h. The cell lysates were subjected to immunoprecipitation (IP) using anti-GFP beads. Immunoprecipitates were examined by Western blotting using HA and GFP antibodies. (G) Quantification of SIAH-1 binding capability. The signal ratios of coimmunoprecipitated ICP0(2)-GFP and HA–SIAH-1 from three independent experiments were determined using the Li-COR software. (H, I) The C-terminal substrate binding domain of SIAH-1 is required and sufficient for interaction with ICP0. 293T cells (2.5 × 106) were cotransfected with 3 μg ICP0(2)-Flag and 3 μg of HA–SIAH-1Δ141-282 (H) or HA–SIAH-1Δ1-140 (I) expression vector. The cell lysates were subjected to immunoprecipitation using anti-Flag antibody or control mouse IgGs. Western blots were examined by Western blot analysis using Flag and HA antibodies. I, input; C, control IgG; IP, Flag; P, insoluble pellet.
We next infected HA–SIAH-1-transfected 293T cells with HSV-1 at a multiplicity of infection (MOI) of 10 for 6 h to probe the interaction of ICP0(1) and SIAH-1 in the context of viral infection. This experiment was conducted in the absence of MG132. Also from infected cell lysates, immunoprecipitation using an antibody against ICP0(1) revealed an interaction of HSV-1 ICP0 and SIAH-1 (Fig. 1B). Endogenous SIAH-1 was detected in ICP0(1) immunoprecipitates of infected cells, after the intracellular amount of SIAH-1 was enhanced by application of the proteasome inhibitor MG132 (25 μM) (Fig. 1C).
The interaction of ICP0 and SIAH-1 is specific.
The predicted SIAH-1 binding motif is located at amino acid residues 410 to 420 in ICP0(2). We constructed a GFP-tagged mutant lacking these residues, ICP0(2)Δ410-420, and analyzed whether it was still able to interact with SIAH-1. Deletion of residues 410 to 420 resulted in a reduced amount of endogenous SIAH-1 in GFP immunoprecipitates compared to that in full-length ICP0(2) (Fig. 1D). This suggests that the predicted interaction motif is essential for the efficient interaction of both proteins.
As shown in Fig. 1E, we tested further GFP-tagged ICP0(2) mutants with deletions of the RING domain, the nuclear localization signal (NLS), the predicted SIAH-1 interaction motif, or larger truncations in a coimmunoprecipitation assay with HA–SIAH-1. HA–SIAH-1 was clearly detected in GFP immunoprecipitates of those ICP0(2) mutants containing the predicted SIAH-1 interaction motif (Fig. 1F). The extent of interaction was determined by calculating the ratio of coimmunoprecipitated HA–SIAH-1 and GFP-ICP0(2) from three independent experiments (Fig. 1G). For the ICP0(2)1-353 and ICP0(2)Δ410-420 mutants, a residual interaction was observed in the Western blots (Fig. 1F) or in comparison to the amount of immunoprecipitated ICP0(2)-GFP (Fig. 1G). Although both lacked the SIAH-1 interaction motif at residues 410 to 420, this interaction may be mediated by a PAVAAVVPRVA motif at residues 311 to 320, which shares some similarity with the “canonical” binding motif and might thus provide weak binding to SIAH-1 (see Fig. S1B and S2 in the supplemental material).
The substrate binding domain of SIAH-1 is located in the C-terminal portion of the protein (37). When we cotransfected Flag-tagged ICP0(2) and an SIAH-1 mutant with a C-terminal deletion (HA–SIAH-1Δ141-282), no interaction of the two proteins was observed after Flag-specific immunoprecipitation (Fig. 1H). On the other hand, the C-terminal fragment (HA–SIAH-1Δ1-140) was sufficient for binding to ICP0(2)-Flag (Fig. 1I). The C-terminal substrate binding domain of human SIAH-1 shares 90% sequence homology to the human SIAH-2 homolog (see Fig. S3 in the supplemental material). Residues important for efficient interaction, such as T156, L158, L166, and M180 (37), are conserved between the two homologs. Thus, we predict that SIAH-2 should also interact with ICP0.
Taken together, the viral ICP0 and the cellular SIAH-1 interact specifically via the substrate binding domain in SIAH-1 and a conserved SIAH-1 interaction motif in ICP0.
ICP0 affects the stability of SIAH-1.
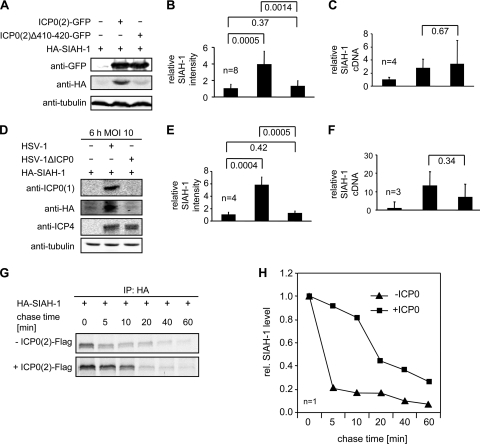
During the immunoprecipitation experiments shown above, we noted a potential influence of ICP0 on SIAH-1 abundance. To examine this, we cotransfected 293T cells with HA–SIAH-1 and either full-length ICP0(2)-GFP or the mutant lacking the SIAH-1 binding motif ICP0(2)Δ410-420. All these experiments were conducted without proteasome inhibition. Whole-cell lysates were analyzed by Western blotting (Fig. 2A). In the absence of ICP0(2), SIAH-1 was barely detectable, whereas an SIAH-1-specific signal was clearly visible when full-length ICP0(2) but not when ICP0(2)Δ410-420 was coexpressed. Quantification of SIAH-1 signals from 8 independent experiments indicated that SIAH-1 abundance in the presence of full-length ICP0(2) was increased 4-fold, whereas with ICP0(2)Δ410-420, the amount was not significantly increased (Fig. 2B). ICP0 has been previously described to act as a transactivator of gene expression at the level of initiation of mRNA synthesis (22, 44). This suggested that the observed increase of SIAH-1 in response to overexpression of ICP0(2) may have resulted from ICP0-mediated activation of the cytomegalovirus immediate-early (CMV-IE) promoter in our HA–SIAH-1 expression vector. To test this possibility, we determined the amount of SIAH-1 mRNA in the presence of full-length and mutant ICP0(2) by quantitative real-time RT-PCR. Indeed, in our experimental setup, SIAH-1 transcript levels were enhanced in the presence of ICP0(2). However, this was observed to the same extent in the presence of ICP0(2)Δ410-420 (Fig. 2C). Therefore, the ICP0-dependent detection of SIAH-1 observed before appears to involve a posttranscriptional mechanism and depends on the efficient interaction of the two proteins.
Fig. 2.
ICP0 affects the stability of SIAH-1. (A) 293T cells (5 × 105) were cotransfected with 0.5 μg HA–SIAH-1 and 0.5 μg of either empty vector, full-length ICP0(2)-GFP, or the ICP0(2)Δ410-420–GFP mutant. At 1 day after transfection, cells were harvested and equal amounts of the respective cellular extracts were subjected to Western blot analyses using anti-GFP, anti-HA, and anti-tubulin antibodies. (B) The corresponding HA–SIAH-1 signal from 8 independent experiments was quantified using the Li-COR software and normalized to the HA–SIAH-1 signal obtained without coexpression of ICP0(2). The statistical significance (P values as indicated) was determined by the Student's t test. (C) Quantification of SIAH-1 cDNA (n = 4) by quantitative real-time RT-PCR. The values were adjusted to GAPDH cDNA and normalized. (D) Vero cells (5 × 105) were transfected with 2 μg of HA–SIAH-1 expression vector. At 16 h posttransfection, the transfected cells were infected with HSV-1 or HSV-1ΔICP0 at an MOI of 10 for 6 h. Western blotting of cell lysates was performed with antibodies recognizing HA, ICP0(1), ICP4, or tubulin. SIAH-1 was detected predominantly in HSV-1-infected cells. (E) Quantification of the corresponding HA–SIAH-1 signal intensities (n = 4). (F) Quantification of GAPDH-adjusted SIAH-1 cDNA by quantitative real-time RT-PCR (n = 4). (G) ICP0(2) increases the half-life of SIAH-1. 293T cells (2.5 × 106) were cotransfected with 3 μg HA–SIAH-1 and either 3 μg of the empty vector or ICP0(2)-Flag vector. At day 1 posttransfection, the cell cultures were pulsed with [35S]methionine for 30 min. Subsequently, the cells were washed and further cultured in [35S]methionine-free medium (chase). Cellular extracts were prepared at the indicated time points and analyzed by fluorography after immunoprecipitation of HA-SIAH-1. (H) Quantification of the obtained data.
The increased amount of SIAH-1 observed during transient expression of ICP0(2) prompted us now to investigate the level of SIAH-1 expression during viral infection. Vero cells transfected with HA–SIAH-1 were infected for 6 h at an MOI of 10 with HSV-1 or with the ICP0 deletion mutant HSV-1ΔICP0. Subsequently, viral infection was analyzed by Western blotting using antibodies directed against the immediate-early proteins ICP0(1) and ICP4 (Fig. 2D). As expected, extracts derived from cells infected with HSV-1ΔICP0 lacked an ICP0-specific signal. HA–SIAH-1 itself was strongly detected migrating at 34 kDa in HSV-1-infected but not in HSV-1ΔICP0-infected cells (Fig. 2D and E). The staining of ICP4 revealed a comparable onset of infection with the two viruses. Thus, the levels of SIAH-1 were also increased in an ICP0-dependent manner during viral infection. The quantification of SIAH-1 transcripts further supported the finding that HSV-1 infection of HA–SIAH-1-transfected cells resulted in increased SIAH-1 mRNA levels. Importantly, this effect was also observed using ICP0-deficient HSV-1 (HSV-1ΔICP0), suggesting that this effect is caused by an ICP0-unrelated viral activity.
To examine the possibility that ICP0 caused elevated SIAH-1 levels by slowing down the turnover of this cellular protein, we performed a pulse-chase experiment using transfected 293T cells (Fig. 2G). Quantification of the obtained data revealed that in the absence of ICP0(2), SIAH-1 turns over rapidly, resulting in a half-life (t1/2) of ∼3 min (Fig. 2H). However, coexpression of ICP0(2) obviously decreased the SIAH-1 turnover rate, which is indicated by an elevated protein half-life of ∼18 min.
In conclusion, the ICP0-dependent abundance of SIAH-1 protein appears to involve a posttranscriptional mechanism, most likely by inhibition of SIAH-1 autoubiquitination.
SIAH-1 colocalizes with ICP0 in nuclear compartments.
SIAH-1 has been previously reported to localize to both the nuclear and cytoplasmic compartments (11, 41). In contrast, early in infection, ICP0 particularly accumulates in ND10 and subsequently disrupts these specific nuclear structures (28, 56, 57). Afterwards (at ∼5 to 9 h postinfection), ICP0 translocates to the cytoplasm (2).
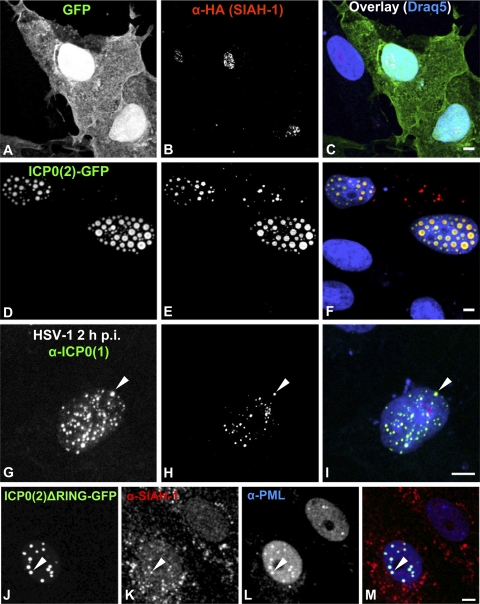
Since the data raised in the present study demonstrated an interaction of SIAH-1 and ICP0 (Fig. 1), we next analyzed the localization of SIAH-1 by immunofluorescence microscopy. We therefore cotransfected Vero cells with HA–SIAH-1 and either GFP as the control (Fig. 3A to C) or ICP0(2)-GFP (Fig. 3D to F). Again, we were unable to detect the highly unstable SIAH-1 protein in the absence of ICP0 (Fig. 3B; the minor nucleolar staining appears to be a cross-reaction of the HA antibody). Cotransfection of ICP0(2) strongly stabilized SIAH-1, and HA–SIAH-1 perfectly colocalized with ICP0(2) in nuclear speckles (Fig. 3D to F). Moreover, HA–SIAH-1 was present also in cytoplasmic spots, which did not colocalize with ICP0(2). To investigate the stabilization and interaction of SIAH-1 and ICP0(1) in HSV-1-infected cells, we transfected Vero cells with HA–SIAH-1. At 24 h posttransfection, cells were infected with HSV-1 at an MOI of 10 for 2 h before fixation and immunofluorescence labeling using HA- and ICP0(1)-specific antibodies (Fig. 3G to I). As shown, HA–SIAH-1 and viral ICP0(1) also colocalized in the nucleus in infected cells, although some ICP0 spots did not colocalize with HA–SIAH-1.
Fig. 3.
SIAH-1 colocalizes with ICP0 in nuclear compartments. Vero cells (7.5 × 104) were cotransfected on coverslips in 24-well plates with 0.5 μg HA–SIAH-1 and either 0.5 μg GFP (A to C) or 0.5 μg of ICP0(2)-GFP (D to F) expression vector. At 24 h posttransfection, cells were fixed, permeabilized, immunolabeled against HA (B and E; red labels in panels C and F), and examined by confocal laser scanning microscopy. SIAH-1 colocalizes with ICP0(2) in the nucleus. The nucleolar HA signal (B) is a cross-reaction of the HA antibody. Alternatively, cells were transfected with 1 μg of HA–SIAH-1 expression vector and infected the next day with HSV-1 at an MOI of 10 PFU/cell for 2 h (G to I). Immunolabeling was carried out against ICP0(1) (G; green label in panel I) and HA (H; red label in panel I). The arrowhead indicates colocalization of ICP0(1) and HA–SIAH-1. Endogenous SIAH-1 colocalizes with ICP0 at PML nuclear bodies. Vero cells (7.5 × 104) were transfected on coverslips in 24-well plates with 1 μg ICP0(2)ΔRING-GFP expression vector (J; green label in panel M). At 24 h posttransfection, cells were fixed, permeabilized, immunolabeled against SIAH-1 (K; red label in panel M) and against PML (L; blue label in panel M), and examined by confocal laser scanning microscopy. The arrowhead indicates colocalization of ICP0(2)ΔRING and SIAH-1 at a PML nuclear body. Scale bars, 5 μm.
The exact localization of the ICP0- and SIAH-1-containing nuclear bodies was determined by staining cells transfected with ICP0(2)ΔRING-GFP against endogenous SIAH-1 and the PML protein (Fig. 3J to M). For this, we utilized the ICP0(2)ΔRING mutant to prevent dispersal and degradation of PML nuclear bodies by the E3 ligase activity of ICP0. ICP0(2)ΔRING, SIAH-1, and PML colocalized, suggesting that the observed nuclear interaction of ICP0 and SIAH-1 occurs at PML nuclear bodies.
Thus, this intracellular virus-host interaction does not only stabilize the otherwise highly labile SIAH-1 but also recruits this cellular E3 ubiquitin ligase into ICP0-containing nuclear compartments.
SIAH-1 mediates polyubiquitination of ICP0 in vivo and in vitro.
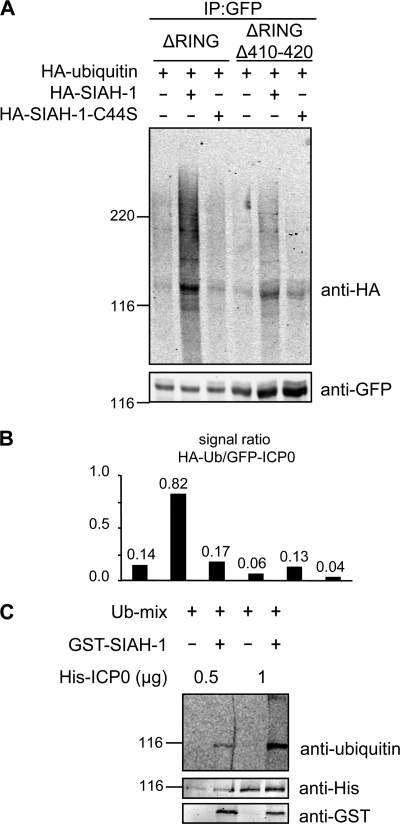
SIAH-1 has been demonstrated to specifically mediate degradation of its substrate proteins through the proteasomal pathway (41). In light of the data raised above, we next investigated whether SIAH-1 is also able to mediate ubiquitination of ICP0. Importantly, it has been previously shown that ICP0 of HSV-1 is able to ubiquitinate itself via its RING domain (5, 10). Therefore, to prevent autoubiquitination of ICP0, we employed the RING domain-deficient mutants ICP0(2)ΔRING and ICP0(2)ΔRINGΔ410-420 in the following experiment. Cultures of 293T cells were transfected with plasmids expressing the respective GFP fusion proteins together with a vector encoding HA-tagged ubiquitin. In addition, control (empty) vector or constructs expressing either HA–SIAH-1 or the inactive mutant HA–SIAH-1–C44S (80) were cotransfected in various combinations. After overnight culturing, the transfected cells were lysed and ICP0(2) was enriched by binding to GFP immunobeads. Bound proteins were subsequently analyzed by Western blotting using GFP- and HA-specific antibodies. Inspection of the GFP-specific signals revealed that larger amounts of ICP0(2) protein were recovered from ICP0(2)ΔRINGΔ410-420–GFP-expressing cultures than in cells expressing the ICP0(2)ΔRING-GFP protein (cf. Fig. 4A and 1E). When HA-specific antibodies were applied, a weak signal of comparable molecular mass (134 kDa) and high-molecular-weight signals were observed in the experiment expressing either the ICP0(2)ΔRING-GFP fusion protein alone or together with the inactive SIAH-1 mutant (C44S). In contrast, this signal was clearly enhanced when intact SIAH-1 was overexpressed. Furthermore, analysis of ICP0(2)ΔRINGΔ410-420–GFP resulted generally in weaker HA-specific signals, particularly when the above-mentioned differences in the overall recovery of GFP fusion protein were considered. This became even more obvious when these signals were quantified and adjusted to ICP0(2)-GFP in each reaction (Fig. 4B). These data therefore indicated that the viral ICP0 may serve as a target for SIAH-1-mediated ubiquitination.
Fig. 4.
SIAH-1 mediates ubiquitination of ICP0 in vivo and in vitro. (A) 293T cells (2.5 × 106) were cotransfected with expression vectors encoding GFP-tagged versions of ICP0(2)ΔRING or ICP0(2)ΔRINGΔ410-420. The respective cell lysates were immunoprecipitated (IP) using GFPtrapA beads and subjected to Western blotting using GFP- and HA-specific antibodies. (B) Quantification of the data shown in panel A depicting the signal ratio of HA-ubiquitin/GFP-ICP0(2). (C) For in vitro ubiquitination analysis, recombinant His-tagged ICP0(2) and GST-tagged SIAH-1 were incubated with purified components of the ubiquitin conjugation machinery, including recombinant ubiquitin (Ub-mix). Reactions were analyzed by SDS-PAGE and immunoblotting using antibodies specific for His, GST, and ubiquitin.
To directly verify this notion, we next analyzed ICP0 ubiquitination in vitro. His-tagged ICP0 of HSV-2 and GST–SIAH-1 were expressed and purified from E. coli and incubated with recombinant ubiquitin, E1 ubiquitin-activating enzyme, and the E2 ubiquitin transferase Ubc5B in the presence of ATP (Fig. 4C). Reactions were analyzed by Western blotting using His-, GST-, and ubiquitin-specific antibodies. As shown, a pronounced signal representing ubiquitinated ICP0(2) was observed only in the reaction mixtures containing SIAH-1.
Taken together, the data raised in transfected cells and by enzyme assays demonstrated that the cellular E3 ligase SIAH-1 is able to ubiquitinate the ICP0 regulator protein of herpes simplex virus.
SIAH-1 binding is dispensable for ICP0-mediated PML degradation.
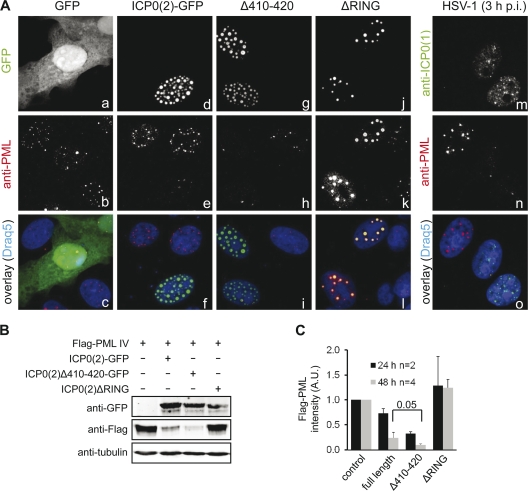
Our finding that ICP0 interacts and stabilizes the cellular E3 ubiquitin ligase SIAH-1 and a previous report demonstrating that SIAH-1 is able to degrade PML by direct association with this protein (31) prompted us to investigate whether SIAH-1 plays a role in the ICP0-induced PML redistribution and degradation (8, 12, 27, 60, 64). Hence, we transfected Vero cells with expression vectors encoding full-length and mutant GFP-tagged ICP0(2) and immunolabeled the respective cultures for endogenous PML (Fig. 5A). In confocal sections of control cells transfected with GFP alone, we observed around 5 to 20 PML bodies per nucleus (Fig. 5A, panels a to c). After expression of ICP0(2)-GFP, the number of PML bodies was drastically reduced, and only isolated PML accumulations colocalizing with ICP0 were observed, whereas the nucleus was filled with ICP0-containing dots (Fig. 5A, panels d to f). The same result was obtained after expression of the SIAH-1 binding-deficient mutant ICP0(2)Δ410-420 (Fig. 5A, panels g to i). As a negative control, we transfected RING domain-deficient ICP0 [ICP0(2)ΔRING], which was unable to reduce the number of PML bodies, although it colocalized with PML in large nuclear dots (Fig. 5A, panels j to l). Infection with HSV-1 at an MOI of 10 for 3 h (Fig. 5A, panels m to o) resulted in the same dispersal of PML as that observed after transfection with the ICP0(2) expression vector (Fig. 5A, panels d to f).
Fig. 5.
SIAH-1 binding is dispensable for ICP0-mediated PML degradation. (A) Vero cells (7.5 × 104) were transfected with 1 μg of a vector expressing GFP (a to c), ICP0(2)-GFP (d to f), or various ICP0(2)-GFP mutants (g to l; green label in panels f, i, and l). After 24 h, the cells were fixed and immunolabeled for endogenous PML protein (b, e, h, k; red label in panels c, f, i, l) and analyzed by confocal laser scanning microscopy. Cells expressing ICP0(2) or ICP0(2)Δ410-420 displayed reduced numbers of PML nuclear bodies, whereas in cells expressing ICP0(2)ΔRING, PML size was enlarged but no degradation occurred. Likewise, cells were infected with HSV-1 at an MOI of 10 PFU/cell for 3 h (m to o) and immunolabeled for ICP0(1) (m; green label in panel o) and PML (n; red label in panel o). Cellular DNA was stained with Draq5 (blue label in panels c, f, i, l, o). (B) Vero cells (2.5 × 106) were cotransfected with an expression vector encoding the Flag-tagged PML isoform IV and either ICP0(2)-GFP vector or ICP0(2)Δ410-420–GFP vector. At 48 h posttransfection, whole-cell lysates were analyzed by Western blotting using antibodies directed against Flag, GFP, or tubulin (loading control). (C) Quantitative analysis of three independent experiments. The Flag-PML signal in the absence of ICP0 was normalized to 1. The statistical significance (P value as indicated) was determined by the Student t test.
To visualize the degradation of PML in an independent experiment, we cotransfected Vero cells with an expression vector encoding the Flag-tagged PML isoform IV (Flag-PML IV) together with a vector encoding either ICP0(2)-GFP or ICP0(2)Δ410-420–GFP. Protein expression was analyzed at 48 h after transfection by Western blotting using Flag-, GFP-, and tubulin-specific antibodies. As shown (Fig. 5B), PML degradation clearly occurred in an ICP0-dependent manner but was more efficient in the presence of the ICP0(2)Δ410-420 mutant, which lacks the SIAH-1 core binding motif. As expected, the ICP0(2)ΔRING mutant failed to reduce Flag-PML levels. Quantitative analysis of several transfection experiments at different time points revealed that the enhanced efficiency in PML degradation of ICP0(2) lacking the SIAH-1 binding motif was significant (Fig. 5C).
Taken together, the disruption of PML nuclear bodies occurred in an ICP0-dependent but SIAH-1-independent manner. As reported previously (8, 28, 56, 57), the N-terminal RING domain of ICP0 was strictly required. Importantly, a lack of SIAH-1 interaction was beneficial for the PML degradation capability of ICP0.
The SIAH-1 binding motif in ICP0 is located in a region involved in transcriptional activation.
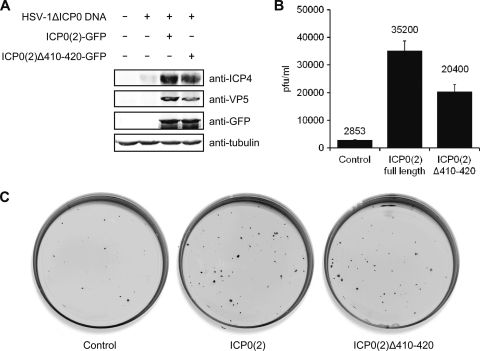
We tested the ability of ICP0(2) to induce expression of HSV-1 genes and whether deletion of the SIAH-1 binding motif influenced this activity. Therefore, naked viral DNA of the HSV-1ΔICP0 mutant was cotransfected together with plasmids encoding GFP, ICP0(2)-GFP, or ICP0(2)Δ410-420–GFP. After 2 days, cell lysates were analyzed by Western blotting for ICP4, VP5, GFP, and tubulin (Fig. 6A). In the absence of ICP0(2), very little expression of ICP4 and no VP5 expression were observed, whereas both were clearly detected with ICP0(2)-GFP. ICP0(2)Δ410-420 induced expression to a somewhat lesser extent than full-length ICP0(2).
Fig. 6.
Complementation of HSV-1ΔICP0 by ICP0(2). (A) 293T cells (5 × 105) in 32-mm-diameter-well dishes were transfected with 0.5 μg of naked HSV-1ΔICP0 DNA and 0.5 μg of pcDNA3 encoding GFP or the indicated GFP-tagged ICP0(2) variants. After 2 days, the cells were harvested and analyzed by Western blotting. (B) Vero cells in 32-mm-diameter dishes were transfected with 0.25 μg HSV-1ΔICP0 DNA and 0.0375 μg of pcDNA3 encoding GFP or the indicated GFP-tagged ICP0(2) variants. Two days after transfection, the cells were harvested, freeze-thawed, and plaque titrated in triplicates on U2OS cells. (C) Here, the Vero cells were incubated for 2 days after transfection with 25 μg/ml human IgGs, before they were fixed with methanol and immunolabeled against VP5. Infected cell foci and plaques were detected using IRDye fluorescence (Li-COR).
Both ICP0(2) variants were also tested for their ability to induce productive infection from naked viral DNA. Therefore, Vero cells were transfected with HSV-1ΔICP0 DNA together with plasmids encoding GFP, ICP0(2)-GFP, or ICP0(2)Δ410-420–GFP. After 2 days, the cells were harvested and plaque titers were determined on U2OS cells (Fig. 6B). Transfected together with empty control plasmid, HSV-1ΔICP0 DNA resulted in a titer of 2.9 × 103 PFU/ml after 2 days. On the other hand, ICP0(2) enhanced the production of infectious particles to 3.5 × 104 PFU/ml, whereas for ICP0(2)Δ410-420 the titer was 2.0 × 104 PFU/ml (Fig. 6B). In a parallel experiment, the Vero cells were fixed and immunostained against VP5 to visualize the plaques and foci of infected cells (Fig. 6C). The number of infected cell foci on transfected cells was increased in the presence of ICP0(2). Again, ICP0(2)Δ410-420 complemented less well than the full-length protein (Fig. 6C).
In summary, we have shown that, like ICP0(1), ICP0(2) is also able to complement an HSV-1ΔICP0 mutant. The region in ICP0(2) containing the SIAH-1 binding motif is not essential for induction of viral gene expression, although its deletion resulted in reduced activation and infectious titers.
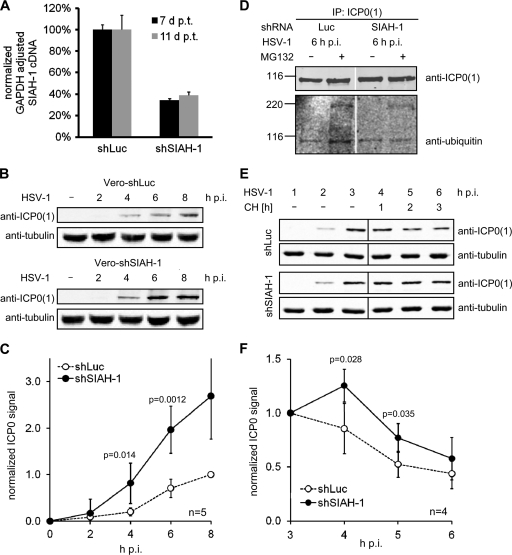
SIAH-1-mediated ubiquitination affects the half-life of ICP0 during infection.
To analyze HSV infection in the presence of reduced SIAH-1 levels, we knocked down SIAH-1 in Vero cells using VSV-G-pseudotyped lentiviral vectors expressing GFP and shRNA either directed against SIAH-1 mRNA or luciferase mRNA as the control. After transduction, the cells were sorted for GFP expression. Since SIAH-1 is highly labile and, therefore, at steady state, difficult to detect (Fig. 1A and 2A and D), we employed quantitative real-time PCR to demonstrate reduced levels of SIAH-1 transcripts in these lentiviral vector-transduced cells (Fig. 6A). SIAH-1 transcript levels were stably reduced to ∼40% at days 7 to 11 after transduction, the time period in which the following experiments were conducted (Fig. 7A). We aimed to analyze ICP0 levels during infection in the presence or absence of SIAH-1. Therefore, we infected our stable SIAH-1 knockdown or control Vero cells with HSV-1 at an MOI of 1 for up to 8 h. At different time points after infection, cells were harvested and analyzed for ICP0(1) expression by Western blotting (Fig. 7B). ICP0(1) was detected as early as 2 h postinfection (p.i.), and the levels increased constantly during the following hours. Compared to the control cells, the increase in ICP0(1) was more pronounced in cells in which SIAH-1 was knocked down (Fig. 7B). This became particularly evident when the Western blots from five independent experiments were quantified (Fig. 7C), demonstrating significantly enhanced ICP0(1) levels in SIAH-1 knockdown cells.
Fig. 7.
SIAH-1-mediated ubiquitination affects the half-life of ICP0 during infection. (A) Vero cells were transduced with lentiviral pseudotypes encoding hairpin shRNA directed against SIAH-1 (shSIAH-1) or luciferase (shLuc). The extent of SIAH-1 knockdown was estimated by quantitative real-time PCR of cDNA preparations at 7 or 11 days after transduction (d p.t.). SIAH-1 was knocked down to ∼ 40%. (B) Control cells (5 × 105) or stable SIAH-1 knockdown cells were inoculated with HSV-1 at an MOI of 1 PFU/cell for up to 8 h. Cells were harvested at the indicated time points p.i., and lysates were analyzed by Western blotting with antibodies raised against ICP0 and tubulin. (C) Quantification of ICP0 signal intensity from 5 independent experiments as shown in panel A. The ICP0 intensity was adjusted to tubulin signal strength and normalized to the 8-h-p.i. signal of ICP0 in control cells. P values were determined using the Student t test. (D) Control cells (2.5 × 106) or stable SIAH-1 knockdown cells were inoculated with HSV-1 at an MOI of 5 PFU/cell for up to 6 h in the presence or absence of 25 μM MG132. Cells were lysed, and ICP0(1) was immunoprecipitated. The immunoprecipitates were analyzed by Western blotting against ICP0(1) and ubiquitin. (E) Control or SIAH-1 knockdown Vero cells were inoculated with HSV-1 at an MOI of 5 PFU/cell. At 3 h p.i., 100 μg/ml cycloheximide was added to the medium, and the cells were incubated for an additional 3 h. At the indicated time points, cells were harvested and lysates were analyzed by Western blotting with antibodies specific for ICP0 and tubulin. (F) Quantification of ICP0 signal intensity from 4 independent experiments, as shown in panel A. The ICP0 intensity was adjusted to tubulin signal strength and normalized to the 3-h-p.i. signal. P values were determined using the Student t test.
We thus assumed that SIAH-1 knockdown could result in a reduced ubiquitination of ICP0 during HSV-1 infection. When we immunoprecipitated ICP0(1) from control or SIAH-1 knockdown cells at 6 h p.i. and analyzed the immunoprecipitates for endogenous ubiquitin, we noted that in cells with reduced SIAH-1 levels, the ICP0-associated ubiquitin-specific signal (compare Fig. 4A and C) was also diminished (Fig. 7D). In these experiments, we employed the proteasome inhibitor MG132 to enhance the amount of ubiquitinated ICP0 species.
SIAH-1-mediated ubiquitination and subsequent proteasomal degradation should affect the half-life of ICP0 in HSV-1-infected cells. We therefore infected the SIAH-1 knockdown or control Vero cells with HSV-1 at an MOI of 5 for 3 h. At this time point, the respective cultures were supplemented with 100 μg/ml cycloheximide to arrest de novo protein synthesis for up to 3 h (chase time). ICP0 levels were then analyzed at the individual time points by Western blotting, as before (Fig. 7E). Quantification of these Western blots (Fig. 7F) revealed that after the addition of cycloheximide, ICP0 levels developed differently in SIAH-1 knockdown and control cells. In SIAH-1 knockdown cells, ICP0 levels increased for 1 h before dropping, whereas the relative ICP0 levels in control cells declined immediately with a half-life of ∼2 h.
In sum, the presented data suggest that SIAH-1 restricts the amount of ICP0 in infected cells by lowering its half-life, which appears to be caused by SIAH-1-mediated ubiquitination of ICP0 and its subsequent proteasomal degradation.
DISCUSSION
The presented data identify a novel and up-to-now-unknown interaction between the HSV regulator protein ICP0 and cellular SIAH-1. Both proteins belong to the group of E3 RING ubiquitin ligases which induce proteasome-dependent degradation of several cellular proteins (42, 43, 76). Our study demonstrated reciprocal activities between ICP0 and SIAH-1. ICP0 stabilized SIAH-1 by specific interaction and colocalized with this cellular protein in nuclear domains. In the absence of ICP0, or in the presence of ICP0 mutants lacking the consensus binding motif (36, 37), SIAH-1 was almost undetectable due to its high turnover, which is caused by dimerization, self-ubiquitination, and subsequent proteasomal degradation (40). In contrast, SIAH-1 facilitated via its RING finger domain the polyubiquitination of ICP0 via specific interaction. This ubiquitination adds up to the previously reported intrinsic autoubiquitination of ICP0, which is counteracted by the cellular ubiquitin-specific protease USP7 (5, 10). Already a modest knockdown of SIAH-1 by RNAi resulted in increased amounts of ICP0 produced during early infection, concomitant with an increased protein half-life.
Initial analyses of ICP0 function in the absence of an SIAH-1 binding motif revealed that dispersal of PML nuclear bodies by ICP0 was independent of ICP0–SIAH-1 interaction. This finding was further supported by the observation that an ICP0 mutant lacking the SIAH-1 binding motif was even more efficient in mediating PML degradation after transfection.
It is well established that incoming viral genomes and early replication compartments are associated with ND10 (58). However, the exact function of ND10 in the context of HSV replication is still largely unknown. It has been proposed that disruption of ND10 by ICP0 releases host cell factors that are required for productive infection or, alternatively, that this process prevents the establishment of an antiviral host cell status that is otherwise maintained by the interferon-induced expression of the major ND10 component PML (reviewed in references 15, 22, and 67). Our analysis of the role of SIAH-1 during HSV infection was originally triggered by a report showing that this protein is able to bind and target PML to degradation by the 26S proteasome, which thereby reduces noticeably the cellular abundance of ND10 (31). Furthermore, it has been also reported that no ubiquitination of PML by ICP0 could be observed in in vitro assays using purified components (7), suggesting that additional factors are required. Therefore, we originally speculated that SIAH-1 may be involved in the ICP0-mediated degradation of PML. However, our analyses revealed that SIAH-1 does not directly participate in ICP0-mediated PML degradation. In that respect, the effect of SIAH-1 appears to be rather indirect and modest by affecting ICP0 stability.
In a recent study, RNAi technology was employed to deplete PML in human fibroblasts (30). That investigation demonstrated that the lack of PML significantly increases HSV-1 gene expression and plaque formation in the absence of ICP0. Therefore, PML appears to be a component of an innate virus restriction mechanism that targets viral genomes and, in the case of HSV, is antagonized by ICP0 (24). Taking our results into account, SIAH-1 then again appears to counteract and to slow down HSV infection by inducing degradation of ICP0, thus posing another barrier for HSV lytic infection.
The SIAH-1 interaction motif (36, 37) is conserved in the Simplexvirus ICP0 homologs and does not seem to be subject to evolutionary pressure to evade this putative cellular defense mechanism. Notably, the region of ICP0 in which the consensus SIAH-1 binding motif is located does not overlap with ICP0 domains reported to be important for E3 ubiquitin ligase activity, nuclear localization, derepression of latent genomes, or binding to USP7 or CoREST (29, 33). The ICP0 homolog of varicella-zoster virus (human herpesvirus 3), another alphaherpesvirus from the Varicellovirus genus, lacks a “canonical” SIAH-1 interaction motif, as do the betaherpesviruses human cytomegalovirus and human herpesvirus 6 in their respective IE1 proteins, which have been described to exhibit functions similar to those of ICP0 of herpes simplex virus (60, 72). SIAH-1, however, has been recently shown to interact with and ubiquitinate the ORF45 protein of Kaposi's sarcoma-associated herpesvirus (KSHV) (1), an immediate-early protein that appears to antagonize type I interferon antiviral responses (83). We observed that ICP0(2) with a deleted SIAH-1 binding motif is slightly impaired in supporting gene expression of an HSV-1ΔICP0 mutant. This is consistent with previous data of ICP0(1) mutants with deletions in that region. In HSV-1, the SIAH-1 binding motif is located at amino acids 400 to 410 (see Fig. S2 in the supplemental material). Previous studies have shown that an ICP0 (1)Δ400-460 mutant had slight defects in inducing expression from different classes of HSV-1 promoters (14). Moreover, replacement of ICP0 in HSV-1 with this mutant resulted in 2- to 10-fold-reduced titers dependent on cell type and MOI (13). Although this region in ICP0 is not essential for transcriptional activity, its deletion appears to have adverse effects on viral fitness. This may explain why this region, containing the binding site for a potentially antiviral protein like SIAH-1, is so well conserved between different simplexviruses.
Another role of the interaction of ICP0 and SIAH-1 could be proposed for the transition from latent infection to lytic reactivation, a mechanism strictly requiring ICP0 (22, 34, 35, 65). Of note, it was reported that lytic infection with HSV induces a DNA damage response in the host cell (49, 50, 71, 78). ICP0 can inhibit the function of the checkpoint kinase ATR by dissociating it from its recruitment factor ATRIP and thus blocks a cellular damage response during lytic infection (79). On the other hand, HSV takes advantage of the ATM DNA damage signaling machinery in establishing viral replication compartments in a newly infected cell (49). Both ATM and ATR induce the phosphorylation of SIAH-1 at amino acid residue S19 upon DNA damage, followed by its dissociation from the cell death regulator HIPK2 (80), which in turn is stabilized due to absence of SIAH-mediated ubiquitination and proteasomal degradation. One may now speculate that DNA damage may trigger escape of HSV from latency by a similar mechanism, in which the ubiquitination-mediated suppression of ICP0 by SIAH-1 is counteracted.
In summary, we postulate that during lytic infection the interaction of cellular SIAH-1 with the HSV immediate-early protein ICP0 followed by the polyubiquitination and likely degradation of the latter may counteract efficient viral gene expression and virus-induced ND10 disruption. Clearly, the identification of SIAH-1 as being a so-far-unknown cellular target of ICP0 provides new possibilities to analyze now in more detail how HSV interferes with the metabolism of its host cell and hijacks the cellular DNA damage response for the transition from latent to productive infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mel Smith for critical comments on the manuscript. We are also grateful to Rudolph Reimer (Heinrich Pette Institute) for assistance with confocal laser scanning microscopy and to Arne Düsedau and Cordula Grüttner (Heinrich Pette Institute) for fluorescence-activated cell sorting (FACS). We thank Roger D. Everett (MRC, Glasgow) and Andreas Sauerbrei and Peter Wutzler (Universitätsklinikum Jena) for providing virus isolates and Beate Sodeik (Hannover Medical School) for providing cell lines.
The Heinrich Pette Institute is supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health. This work was supported in part by the Stiftung zur Bekämpfung neuroviraler Erkrankungen (NVE).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 1 June 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Abada R., et al. 2008. SIAH-1 interacts with the Kaposi's sarcoma-associated herpesvirus-encoded ORF45 protein and promotes its ubiquitylation and proteasomal degradation. J. Virol. 82:2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Advani S. J., Hagglund R., Weichselbaum R. R., Roizman B. 2001. Posttranslational processing of infected cell proteins 0 and 4 of herpes simplex virus 1 is sequential and reflects the subcellular compartment in which the proteins localize. J. Virol. 75:7904–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beyer W. R., Westphal M., Ostertag W., von Laer D. 2002. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J. Virol. 76:1488–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borden K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boutell C., Canning M., Orr A., Everett R. D. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 79:12342–12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boutell C., Everett R. D. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278:36596–36602 [DOI] [PubMed] [Google Scholar]
- 7. Boutell C., Orr A., Everett R. D. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boutell C., Sadis S., Everett R. D. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai W. Z., Schaffer P. A. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canning M., Boutell C., Parkinson J., Everett R. D. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 279:38160–38168 [DOI] [PubMed] [Google Scholar]
- 11. Carthew R. W., Rubin G. M. 1990. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63:561–577 [DOI] [PubMed] [Google Scholar]
- 12. Chelbi-Alix M. K., de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 13. Chen J., Silverstein S. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J. X., Zhu X. X., Silverstein S. 1991. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology 180:207–220 [DOI] [PubMed] [Google Scholar]
- 15. Ching R. W., Dellaire G., Eskiw C. H., Bazett-Jones D. P. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118:847–854 [DOI] [PubMed] [Google Scholar]
- 16. Davido D. J., von Zagorski W. F., Lane W. S., Schaffer P. A. 2005. Phosphorylation site mutations affect herpes simplex virus type 1 ICP0 function. J. Virol. 79:1232–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davison A. J., Wilkie N. M. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55:315–331 [DOI] [PubMed] [Google Scholar]
- 18. Dimitrova Y. N., et al. 2010. Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J. Biol. Chem. 285:13507–13516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Döhner K., et al. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dull T., et al. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott G., Hafezi W., Whiteley A., Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761–770 [DOI] [PubMed] [Google Scholar]
- 23. Everett R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–7273 [DOI] [PubMed] [Google Scholar]
- 24. Everett R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365–374 [DOI] [PubMed] [Google Scholar]
- 25. Everett R. D. 2010. Depletion of CoREST does not improve the replication of ICP0 null mutant herpes simplex virus type 1. J. Virol. 84:3695–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Everett R. D., Boutell C., Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Everett R. D., et al. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Everett R. D., Maul G. G. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Everett R. D., Parsy M. L., Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Everett R. D., et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fanelli M., et al. 2004. The coiled-coil domain is the structural determinant for mammalian homologs of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J. Biol. Chem. 279:5374–5379 [DOI] [PubMed] [Google Scholar]
- 32. Germani A., et al. 2000. SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene 19:5997–6006 [DOI] [PubMed] [Google Scholar]
- 33. Gu H., Roizman B. 2009. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J. Virol. 83:4376–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hagglund R., Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halford W. P., et al. 2006. ICP0 antagonizes Stat 1-dependent repression of herpes simplex virus: implications for the regulation of viral latency. Virol. J. 3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. House C. M., et al. 2003. A binding motif for Siah ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 100:3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. House C. M., et al. 2006. Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure 14:695–701 [DOI] [PubMed] [Google Scholar]
- 38. House C. M., Möller A., Bowtell D. D. 2009. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 69:8835–8838 [DOI] [PubMed] [Google Scholar]
- 39. Hu G., et al. 1997. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46:103–111 [DOI] [PubMed] [Google Scholar]
- 40. Hu G., Fearon E. R. 1999. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu G., et al. 1997. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11:2701–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson P. K., et al. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429–439 [DOI] [PubMed] [Google Scholar]
- 43. Joazeiro C. A., Weissman A. M. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552 [DOI] [PubMed] [Google Scholar]
- 44. Jordan R., Schaffer P. A. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kawaguchi Y., Bruni R., Roizman B. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J. Virol. 71:1019–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kawaguchi Y., et al. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. U. S. A. 98:1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kawaguchi Y., Van Sant C., Roizman B. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li S., Li Y., Carthew R. W., Lai Z. C. 1997. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 90:469–478 [DOI] [PubMed] [Google Scholar]
- 49. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., Weitzman M. D. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lilley C. E., Schwartz R. A., Weitzman M. D. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119–126 [DOI] [PubMed] [Google Scholar]
- 51. Liu J., et al. 2001. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927–936 [DOI] [PubMed] [Google Scholar]
- 52. MacLean A. R. 1998. Preparation of HSV-DNA and production of infectious virus. Methods Mol. Med. 10:19–25 [DOI] [PubMed] [Google Scholar]
- 53. Maringer K., Elliott G. 2010. Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle. J. Virol. 84:4682–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsuzawa S. I., Reed J. C. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915–926 [DOI] [PubMed] [Google Scholar]
- 55. Maul G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660–667 [DOI] [PubMed] [Google Scholar]
- 56. Maul G. G., Everett R. D. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223–1233 [DOI] [PubMed] [Google Scholar]
- 57. Maul G. G., Guldner H. H., Spivack J. G. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679–2690 [DOI] [PubMed] [Google Scholar]
- 58. Maul G. G., Ishov A. M., Everett R. D. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67–75 [DOI] [PubMed] [Google Scholar]
- 59. Möller A., et al. 2009. Inhibition of Siah ubiquitin ligase function. Oncogene 28:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Müller S., Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nagano Y., et al. 2003. Siah-1 facilitates ubiquitination and degradation of synphilin-1. J. Biol. Chem. 278:51504–51514 [DOI] [PubMed] [Google Scholar]
- 62. Nakayama K., Qi J., Ronai Z. 2009. The ubiquitin ligase Siah2 and the hypoxia response. Mol. Cancer Res. 7:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. O'Doherty U., Swiggard W. J., Malim M. H. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parkinson J., Everett R. D. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Preston C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1–19 [DOI] [PubMed] [Google Scholar]
- 66. Radtke K., et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Regad T., Chelbi-Alix M. K. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286 [DOI] [PubMed] [Google Scholar]
- 68. Roizman B., Gu H., Mandel G. 2005. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle 4:1019–1021 [DOI] [PubMed] [Google Scholar]
- 69. Roizman B., Knipe D. M. 2001. Herpes simplex viruses and their replication, p. 2399–2459 In Knipe D. M., Howley P., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 70. Rubinson D. A., et al. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401–406 [DOI] [PubMed] [Google Scholar]
- 71. Shirata N., et al. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336–30341 [DOI] [PubMed] [Google Scholar]
- 72. Stanton R., Fox J. D., Caswell R., Sherratt E., Wilkinson G. W. 2002. Analysis of the human herpesvirus-6 immediate-early 1 protein. J. Gen. Virol. 83:2811–2820 [DOI] [PubMed] [Google Scholar]
- 73. Tang A. H., Neufeld T. P., Kwan E., Rubin G. M. 1997. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell 90:459–467 [DOI] [PubMed] [Google Scholar]
- 74. Tiedt R., Bartholdy B. A., Matthias G., Newell J. W., Matthias P. 2001. The RING finger protein Siah-1 regulates the level of the transcriptional coactivator OBF-1. EMBO J. 20:4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Treier M., Staszewski L. M., Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787–798 [DOI] [PubMed] [Google Scholar]
- 76. Tyers M., Willems A. R. 1999. One ring to rule a superfamily of E3 ubiquitin ligases. Science 284:601–604 [DOI] [PubMed] [Google Scholar]
- 77. Wheeler T. C., Chin L. S., Li Y., Roudabush F. L., Li L. 2002. Regulation of synaptophysin degradation by mammalian homologs of seven in absentia. J. Biol. Chem. 277:10273–10282 [DOI] [PubMed] [Google Scholar]
- 78. Wilkinson D. E., Weller S. K. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wilkinson D. E., Weller S. K. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Winter M., et al. 2008. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 10:812–824 [DOI] [PubMed] [Google Scholar]
- 81. Yao F., Schaffer P. A. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158–8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang J., Guenther M. G., Carthew R. W., Lazar M. A. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhu F. X., Sathish N., Yuan Y. 2010. Antagonism of host antiviral responses by Kaposi's sarcoma-associated herpesvirus tegument protein ORF45. PLoS One 5:e10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.