Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein is essential for virus lytic replication. ORF57 regulates virus gene expression at multiple levels, enhancing transcription, stability, nuclear export, and translation of viral transcripts. To enhance the nuclear export of viral intronless transcripts, ORF57 (i) binds viral intronless mRNAs, (ii) shuttles between the nucleus, nucleolus, and the cytoplasm, and (iii) interacts with multiple cellular nuclear export proteins to access the TAP-mediated nuclear export pathway. We investigated the implications on the subcellular trafficking, cellular nuclear export factor recruitment, and ultimately nuclear mRNA export of an ORF57 protein unable to bind RNA. We observed that mutation of a carboxy-terminal RGG motif, which prevents RNA binding, affects the subcellular localization and nuclear trafficking of the ORF57 protein, suggesting that it forms subnuclear aggregates. Further analysis of the mutant shows that although it still retains the ability to interact with cellular nuclear export proteins, it is unable to export viral intronless mRNAs from the nucleus. Moreover, computational molecular modeling and biochemical studies suggest that, unlike the wild-type protein, this mutant is unable to self-associate. Therefore, these results suggest the mutation of a carboxy-terminal RGG motif affects ORF57 RNA binding, nuclear trafficking, and multimerization.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 is a gamma-2 herpesvirus associated with multiple AIDS-related malignancies, including Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease (10, 11, 19). Like all herpesviruses, KSHV has two distinct phases within its life cycle: latent persistence and lytic reactivation. During latency the virus expresses a limited subset of genes ensuring the persistence of the genome as a high-copy-number nonintegrated episome but remains undetected by the host immune defense mechanisms (16, 27, 40). After reactivation the virus enters lytic replication, which is typified by a cascade of viral gene expression, culminating in the production of new infectious virus particles (17, 50).
However, in contrast to other oncogenic herpesviruses, where latent gene expression plays a prominent role in tumorigenesis, lytic replication also plays an important part in the tumorigenicity, pathogenesis, and spread of KSHV infection (19). Specifically, lytic replication appears to be a necessary antecedent step in KS development from the primary target of viral infection, the B lymphocyte reservoir, to endothelial cells where tumors are observed. Moreover, lytic gene expression potentially contributes to the development of KS through the expression of lytic viral proteins which mediate paracrine secretion of growth and angiogenic factors that are essential for tumor growth and development (19). In addition, they sustain the population of latently infected cells that would otherwise be reduced due to the poor persistence of the KSHV episome during spindle cell division (24).
Posttranscriptional events that regulate mRNA biogenesis are fundamental to the control of gene expression (37). As a consequence, cells have evolved a “gene expression production line” that encompasses the routing of a nascent transcript through multimeric mRNA-protein complexes that mediate its splicing, polyadenylation, nuclear export, and translation (38). These pathways are particularly important for herpesviruses which replicate in the host-cell nucleus and express numerous lytic intronless mRNAs (6). Due to the reliance of herpesviruses on the host cell machinery for efficient processing of their mRNAs, an immediate issue arises concerning the mechanism by which the viral intronless mRNAs are efficiently exported from the nucleus, given that the majority of cellular bulk mRNA nuclear export is intimately linked, and dependent upon, splicing (32, 49).
To circumvent the problem associated with efficient intronless viral mRNA nuclear export, KSHV encodes a conserved multifunctional protein, ORF57, which enhances this nuclear export and the translation of viral intronless transcripts (3, 4, 35). To this end, KSHV ORF57 binds viral intronless mRNAs, shuttles between the nucleus, nucleolus, and the cytoplasm, and mediates the nuclear export of intronless viral mRNAs via a TAP-mediated pathway (3, 5, 7). These properties are also conserved in ORF57 homologues throughout herpesviruses such as ICP27 from herpes simplex virus type 1 (HSV-1) (12, 13, 28, 43), SM protein from Epstein-Barr virus (EBV) (41, 45), and herpesvirus saimiri (HVS) ORF57 protein (2, 8, 14, 15, 22).
To gain access to the TAP-mediated nuclear export pathway, ORF57 orchestrates the assembly of an export competent viral ribonucleoprotein particle (3). ORF57 specifically recruits the cellular hTREX complex to intronless viral mRNA via a direct interaction with the hTREX protein Aly, which in turn recruits the cellular protein TAP, which in turn leads to the TAP-mediated nuclear export pathway (3). However, results suggest redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless viral mRNAs, since experiments involving small interfering RNA-mediated depletion of Aly report only a limited effect on ORF57-mediated KSHV intronless mRNA export (34). These data correlate with depletion-related studies on the role of Aly in mRNA export in higher eukaryotes where, surprisingly, Aly has been shown to be dispensable in mRNA export (20, 31). Therefore, other cellular mRNA adapter proteins, such as UIF (26), RBM15, and OTT3 (48), which interact with TAP are likely to function in ORF57-mediated nuclear export.
Mutational analyses of the ORF57 sequence have revealed several functional motifs that are required for RNA binding, nuclear localization, nucleolar trafficking, and multimerization (33). ORF57 contains several putative RNA-binding domains, including an amino-terminal arginine-rich region that resembles RNA-binding motifs (ARMs) described in other members of the family, including EBV SM, cytomegalovirus (CMV) UL69, and HVS ORF57. ORF57 also contains two separate arginine-glycine-glycine (RGG) motifs, similar to an RGG box motif, responsible for RNA binding in HSV ICP27 (36). Nuclear targeting is mediated by three functionally independent nuclear localization signals (NLS) (34), and NLS2 or NLS3 in combination with NLS1 are able to function as a nucleolar targeting signal (7). The carboxy-terminal contains a potential leucine zipper motif, a zinc finger-like motif. and a GLFF motif, conserved in gamma herpesvirus ORF57 homologues (21); however, it remains unknown whether these domains are important for ORF57 function.
We investigated here the implications on subcellular trafficking, hTREX complex recruitment, and ultimately nuclear mRNA export of an ORF57 protein unable to bind RNA. We observed that mutation of a carboxy-terminal RGG motif, which prevents RNA binding, affects the subcellular localization and nuclear trafficking of the ORF57 protein, suggesting it forms subnuclear aggregates. Further analysis of the mutant shows that although it still retains the ability to interact with cellular nuclear export proteins, it is unable to export viral intronless mRNAs from the nucleus. Moreover, computational molecular modeling and biochemical studies suggest that, unlike the wild-type protein, this mutant is unable to self-associate. Therefore, these results suggest the mutation of a carboxy-terminal RGG motif affects ORF57 RNA binding, nuclear trafficking, and multimerization.
MATERIALS AND METHODS
Oligonucleotides, plasmids, and antibodies.
Oligonucleotides used in reverse transcription-PCR (RT-PCR) analysis and site-directed mutagenesis are listed in Table 1. To generate pORF57GFP and pORF57-myc, ORF57 cDNA was amplified by PCR and cloned into pEGFP-N1 (BD Biosciences, Ltd.) and pcDNA3.1MycHisB (Invitrogen), respectively. pORF57GFP-RGG1, -RGG2, and -Leu mutants were generated using a QuikChange II site-directed mutagenesis kit (Stratagene). The pORF57GFP-RGG1/2 mutant was subcloned using the 5,574-bp fragment from pORF57GFP-RGG1 and the 494-bp fragment from pORF57GFP-RGG2 after digestion with ApaI/BamHI. The ORF47 gene was cloned into pCDNA3.1+ (Invitrogen). To generate GST-Aly, the Aly open reading frame (ORF) was PCR amplified and cloned into pGEX4T-1 (GE Healthcare). Antibodies to fibrillarin, C23 (Santa Cruz Biotech), green fluorescent protein (GFP; BD Biosciences), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Sigma) were purchased from the respective suppliers. Unless stated all antibodies were used at a dilution of 1:1,000 for Western blot analysis.
Table 1.
Primers used in site-directed mutagenesis of the ORF57 gene and qRT-PCR analysis of the nuclear export assays
| Primer | Sequence |
|---|---|
| 57RGG1F | GGC AGA ACA ACA GCG GCC GGA CAG TGT C |
| 57RGG1R | GAC ACT GTC CGG CCG CTG TTG TTC TGC C |
| 57RGG2F | GCT TAA CTT CGC GGC AGG GCT GCT CTT G |
| 57RGG2R | CAA GAG CAG CCC TGC CGC GAA GTT AAG C |
| 57LeuF | GCA GAG CGA GGC AGC ACT GAT C |
| 57LeuR | GAT CAG TGC TGC CTC GCT CTG C |
| ORF47F | CGG GAT CCA TGG GGA TCT TTG CGC TAT TT |
| ORF47R | CCG CTC GAG TGA CTG ATG GAT AGG GTG CCC |
| GAPDHF | AGG GTC ATC ATC TCT GCC CCC TC |
| GAPDHR | TGT GGT CAT GAG TCC TTC CAC GAT |
Cell culture and transfection.
HEK293T cells were cultured in Dulbecco's modified Eagle medium (Invitrogen, Paisley, United Kingdom) supplemented with 10% fetal calf serum (FCS; Invitrogen), glutamine, and penicillin-streptomycin. Plasmid transfections were carried out with Lipofectamine 2000 (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions.
RNA immunoprecipitation assays.
HEK293T cells were cotransfected with ORF47 and the ORF57 mutants. After 24 h the cells were washed with ice-cold phosphate-buffered saline (PBS) and UV irradiated (900 mJ/cm2) in a Stratalinker. Cells were resuspended in 2mls TAP-lysis buffer (10 mM HEPES, 2 mM MgCl2, 10 mM KCl, 0.5% NP-40, 5% glycerol containing protease inhibitor, and 1 μl of RNaseOUT [Invitrogen]/ml) and lysed on ice for 30 min. Protein A-agarose beads (50 μl of beads/immunoprecipitation) were washed in PBS, followed by incubation with polyclonal anti-GFP antibody (3 μl/IP) at 4°C for 30 min. Then, 50 μl of antibody-bead complex was added to 1 ml of cleared lysate and immunoprecipitated at 4°C with end-over-end mixing for 1 h at 4°C. The beads were washed in ice-cold NT2 wash buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40). The beads were next incubated with 100 μl of NT2 buffer (containing 0.1% sodium dodecyl sulfate [SDS] and 0.5 mg of proteinase K/ml) for 15 min at 55°C. To extract the precipitated RNA, 500 μl of TRIzol reagent (Invitrogen) and 100 μl of chloroform were added, followed by vigorous shaking and centrifugation at 13,000 rpm for 1 min. Next, 3 volumes of isopropanol, 1/10 volume of 3 M sodium acetate (pH 5.5), and 20 μg of glycogen were added to the supernatants, followed by incubation overnight at −20°C to precipitate RNA. The RNA was pelleted by centrifugation at 15,000 rpm for 30 min at 4°C and washed in 80% ethanol. These were again pelleted at 15,000 rpm for 30 min at 4°C before the pellets were air dried and resuspended in 20 μl of diethyl pyrocarbonate in double-distilled H2O plus 0.5 μl of RNaseOUT. RNA was DNase treated using an Ambion DNase-free kit in accordance with the manufacturer's instructions, and 10 μl was used in RT reactions set up using gene-specific primers and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. RT-negative samples were set up alongside. Then, 2 μl of cDNA was used in a nested-PCR with ORF47 specific primers.
Immunofluorescence microscopy and FRAP (fluorescence recovery after photobleaching).
HEK293T cells were grown on polylysine-treated coverslips. Cells were fixed in 4% paraformaldehyde and permeabilized in 1% Triton X-100. Then, the cells were blocked in 1% bovine serum albumin (BSA) made in PBS and incubated at 37°C for 1 h. Primary antibodies, C23 and fibrillarin, were diluted 1:100 in 1% BSA and incubated with the cells for 1 h at 37°C. Texas Red anti-mouse (Vector Laboratories, California) or Alexa Fluor 633 anti-rabbit antibody (Invitrogen) was diluted 1:500 in 1% BSA and incubated with the cells for 1 h at 37°C. Coverslips were mounted in Vectorshield mounting medium (Vector Laboratories), and staining was visualized on an Upright LSM 510 META Axioplan 2 confocal microscope (Zeiss) using LSM imaging software (Zeiss).
For FRAP analysis, HEK293T cells were plated on glass-based 33-mm culture dishes and imaged at 24 h posttransfection using an Upright LSM 510 META Axioplan 2 confocal microscope (Zeiss). Cells were maintained at 37°C and, during imaging, the cell culture medium was exchanged for CO2-independent medium (Gibco). Fluorescence recovery was measured using the ROI mean module of the LSM 510 software.
GST-pulldown and immunoprecipitation assays.
Recombinant glutathione S-transferase (GST) and GST-Aly proteins were expressed and purified from crude bacterial lysates by incubation with glutathione-Sepharose 4B affinity beads according to the manufacturer's specifications (Pharmacia Biotech). The protein-bound beads were then incubated with HEK293T-transfected cell extracts previously lysed with lysis buffer (0.3 M NaCl, 50 mM Tris-HCl [pH 8.0], 1% Triton X-100; 0.1% SDS) containing protease inhibitors (AEBSF; Roche) for 16 h at 4°C. The beads were then pelleted, washed four times in lysis buffer, and resuspended in Laemmli buffer, and the precipitated polypeptides were identified by immunoblot analysis.
To perform coimmunoprecipitation assays, 293T cells were transfected using 2 μg of the appropriate DNAs. After 24 h, the cells were harvested and lysed with lysis buffer (0.3 M NaCl, 1% Triton X-100, 50 mM HEPES buffer [pH 8.0]) containing protease inhibitors (leupeptin and phenylmethylsulfonyl fluoride). For each immunoprecipitation, 20 μl of GFP-Trap-affinity beads (Chromotek) or Myc-Affinity-agarose (Sigma) was incubated with each respective cell lysate for 3 h at 4°C, as directed by the manufacturers. The beads were then pelleted and washed, and the precipitated polypeptides were resolved on a 12% SDS–polyacrylamide gel and analyzed by immunoblotting.
Quantitative RT-PCR.
To assess the mRNA export efficiency, HEK293T cells were cotransfected with ORF47 and ORF57 mutants. After 24 h, RNA was extracted from the total, nuclear, and cytoplasmic fractions by using TRIzol (Invitrogen) as described by the manufacturer. RNA was DNase treated using the Ambion DNase-free kit according to the manufacturer's instructions, and RNA (1 μg) from each fraction was reverse transcribed using SuperScript II (Invitrogen) according to the manufacturer's instructions, using oligo(dT) primers (Promega). Next, 10 ng of cDNA was used as a template in SensiMixPlus SYBR quantitative PCRs (Quantace), as recommended by the manufacturer, using a Rotor-Gene Q 5plex HRM platform (Qiagen), with a standard three-step melt program (95°C for 15 s, 60°C for 30 s, and 72°C for 20 s). With GAPDH as an internal control mRNA, quantitative analysis was performed using the comparative CT method as previously described (44).
RESULTS
Site-directed mutagenesis of carboxy-terminal RGG motif effects ORF57 subnuclear localization.
KSHV ORF57 is a multifunctional protein that (i) binds viral intronless mRNAs, (ii) shuttles between the nucleus, nucleolus, and the cytoplasm, and (iii) mediates the nuclear export of intronless viral mRNAs. To investigate whether RNA binding is linked to the subnuclear trafficking of the ORF57 protein, we analyzed the nuclear localization and trafficking potential of an ORF57 mutant protein, which is unable to bind RNA. Previous work has suggested that deletion of a carboxy-terminal RGG motif affects the ability of ORF57 to bind RNA (39). Therefore, site-directed mutagenesis was performed on the carboxy-terminal RGG box (residues 372 to 374), termed 57RGG2. In addition, two additional site-directed mutants were generated as suitable controls, the first altered a similar amino-terminal RGG motif (residues 138 to 140), termed 57RGG1, whereas the second altered a leucine-rich region (residues 341 to 369), termed 57Leu, previously shown to affect RNA binding (39). Mutagenesis was performed altering the wild-type residues to alanines within pORF57-GFP (3) (Fig. 1 A). Moreover, a double mutant was also produced combining both RGG box mutations. Mutations were performed in pORF57-GFP to allow both indirect immunofluorescence and live subcellular trafficking analysis to be performed. Sequence analysis confirmed the alteration of the wild-type residues to alanines. Moreover, to determine that all of the ORF57 site-directed mutants were expressed at levels similar to the wild-type ORF57, each mutant was transfected into HEK293T cells, and cell lysates assayed by Western blot analysis using a GFP-specific antibody. The results show that all mutants were expressed at levels similar to that for the wild-type ORF57 protein (Fig. 1B).
Fig. 1.
Site-directed mutagenesis of putative RNA binding motifs within the ORF57 protein. (A) Schematic representation of the generated ORF57 site-directed mutants. (B) 293T cells were transfected with either pGFP, pORF57GFP, or the ORF57 mutants. After 24 h, the cell lysates were analyzed by Western blot analysis using GFP and GAPDH-specific antibodies. GAPDH served as a loading control.
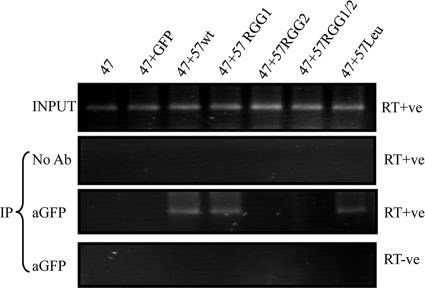
To confirm that the 57RGG2 site-directed mutant was unable to bind RNA, an RNA immunoprecipitation assay was utilized, which we have previously used to show that wild-type ORF57 protein binds to KSHV intronless mRNAs (3). To perform the RNA immunoprecipitation assays, HEK293T cells were cotransfected with a pORF47, in the presence of either GFP, wild-type ORF57, or each ORF57 mutant (57RGG1, 57Leu, 57RGG2, and 57RGG1/2). After 24 h, the cells were UV irradiated to cross-link interacting RNA and protein. A GFP-specific polyclonal antibody was then used to pull down ORF57-RNA complexes, and the isolated RNA was subjected to RT-PCR analysis. The results show that wild-type ORF57, 57RGG1, and 57Leu all bind and precipitate the ORF47 intronless mRNA (Fig. 2). In contrast, both 57RGG2 and 57RGG1/2 are unable to bind ORF47 mRNA, indicating that site-directed mutation of only two amino acid residues within the RGG2 box abrogates the ability of ORF57 to bind intronless mRNAs (Fig. 2). These results are in general agreement with previous deletion analysis of ORF57 RGG motifs, although the results here show that alanine substitutions of the 57Leu region retain the ability to bind RNA, in contrast to a mutation that alters the leucine-rich region with the more disruptive proline residues (39).
Fig. 2.
Site-directed mutagenesis of the carboxy-terminal RGG2 motif inhibits RNA binding. 293T cells were transfected with pORF47 in the absence or presence of either pGFP, pORF57GFP, or the ORF57 mutants. After UV cross-linking, RNA immunoprecipitations were performed using no antibody or a GFP-specific antibody. Total RNA served as a positive control for the PCR (input).
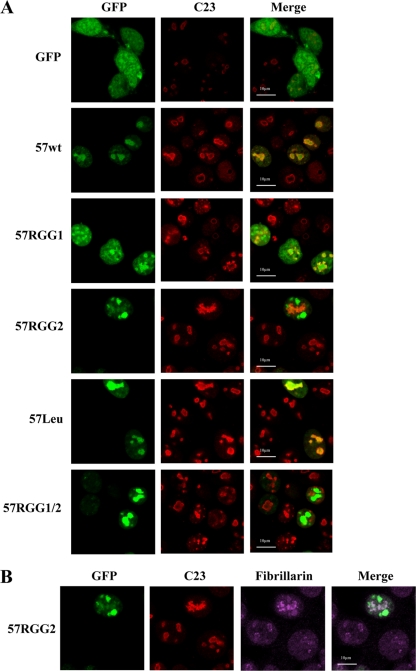
Having produced a site-directed ORF57 mutant that was unable to bind RNA, we next determined whether the inability to bind RNA had any effect on ORF57 subcellular localization and subnuclear trafficking. HEK293T cells were grown on poly-lysine-treated coverslips and transfected with wild-type ORF57 or ORF57 mutants. After 24 h, the cells were fixed and permeabilized, and wild-type ORF57 and ORF57 mutants were visualized directly via GFP fluorescence. In addition, indirect immunofluorescence was used to identify the nucleolus using C23-specific antibodies. C23 or nucleolin is a major nucleolar phosphoprotein that influences synthesis and maturation of ribosomes and is widely used as a nucleolar marker. The results show that wild-type ORF57, 57RGG1, and 57Leu have staining similar to that described previously (7), localizing to the nucleolar and nuclear speckles (Fig. 3). However, it must be noted that 57RGG1 fluorescence seems more diffuse throughout the nucleus, with nuclear speckle and nucleolar localization. Staining of the nucleolus is typified by a halo-like structure due to an inability of the staining antibody to penetrate deep into the dense nucleolus. In contrast, both of the mutants which affect the RNA binding ability of ORF57, 57RGG2 and 57RGG1/2, showed a distinct staining pattern that lacked colocalization with the nucleolar marker C23. This suggests that the alteration of the carboxy-terminal RGG motif which ablates RNA binding may also affect ORF57 subcellular localization, specifically the ability of ORF57 to localize to the nucleolus (Fig. 3A).
Fig. 3.
Site-directed mutagenesis of the carboxy-terminal RGG2 motif affects subcellular localization of the ORF57 protein. 293T cells were transfected with either pGFP, pORF57GFP, or the ORF57 mutants. After 24 h, the cells were fixed and permeabilized, and the GFP fluorescence was analyzed by direct visualization, whereas indirect immunofluorescence was performed using C23-specific antibodies (A) and fibrillarin-specific antibodies (B) to identify the nucleolus.
It is also evident that the classical halo staining pattern of C23 is replaced by a nucleolar-necklace-like staining reminiscent of a disorganized nucleolar structure in the presence of these RGG2 mutants. The nucleolar necklace is formed by rRNA transcription sites extending into the nucleoplasm, where each bead of the necklace corresponds to the nucleolar fibrillar component (FC) and dense fibrillar component (DFC) and contains rDNA, rRNAs, polymerase I, DNA topoisomerase I, nucleolar transcription factor 1 (UBF), and fibrillarin (46). This aberrant localization could be due to 57RGG2 no longer localizing to the nucleolus or to the fact that 57RGG2 still localizes to the nucleolus; however, in their presence C23 is expelled into the nucleoplasm. To further investigate the subcellular localization of 57RGG2, the cells were also stained with a second nucleolar marker, fibrillarin (Fig. 3B). The results demonstrate that fibrillarin shows the same pattern of staining as C23, suggesting that the nucleolus is being stained and that 57RGG2 is no longer localizing to the nucleolus. Therefore, these results suggest that mutation of the ORF57 carboxy-terminal RGG motif, which affects its ability to bind RNA, also affects the ability of ORF57 to localize to the nucleolus and instead the 57RGG2 mutant localizes to distinct nuclear subdomains.
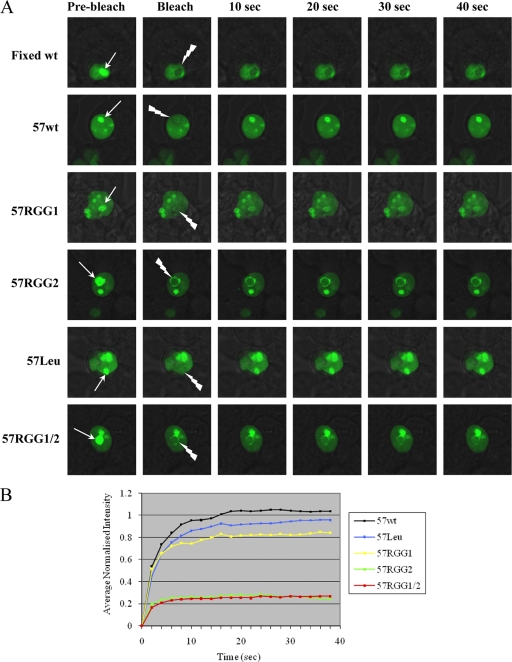
Fluorescence recovery after photobleaching indicates 57RGG2 is unable to traffic in the nucleus.
We have previously shown that ORF57 can shuttle between the nucleus and cytoplasm and also traffic through the nucleolus (7). Therefore, the failure of 57RGG2 to localize to the nucleolus may be due to a defect in its trafficking ability. Hence, FRAP was used to investigate the mobility of 57RGG2 compared to the wild-type protein and other mutants. Using live cell imaging it is possible to gauge and compare ORF57 and mutant protein trafficking rates by photobleaching an area of the localized protein. The fluorescence immediately after the bleach was normalized to 0, and the final fluorescence at equilibrium was designated as 1. 57RGG1 and 57Leu show a rate of mobility similar to that of wild-type ORF57, having almost fully recovered after 20 s (Fig. 4). 57RGG2 and 57RGG1/2, however, display a severe inability to traffic with little recovery of the bleached protein. 57RGG2 was reproducibly less mobile than wild-type ORF57, giving rise to a detectable bleach zone immediately after the bleach (Fig. 4). These results suggest that mutation of the carboxy-terminal RGG motif has a major effect on the trafficking ability of the protein and consequently on its subcellular localization.
Fig. 4.
FRAP analysis performed on 293T cells transfected with either pORF57GFP or the ORF57 mutants. (A) Recovery was assessed over a 40-s period after a photobleaching period. (B) Fluorescence recovery curves. Data were normalized following the bleaching period so that the initial postbleaching was set as 0 and the final intensity was set as 1.
Mutation of the RGG2 motif does not affect ORF57 binding to the hTREX protein, Aly, but has a marked effect on the ability of ORF57 to export viral intronless mRNA from the nucleus.
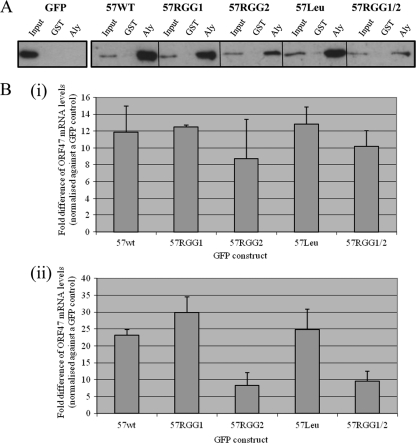
To further investigate the implications of the inability of the 57RGG2 mutant to traffic within the nucleus, each mutant was assessed for its capability to interact with cellular nuclear export factors and also for its ability to export an intronless KSHV mRNA into the cytoplasm. To assess what effect the 57RGG2 mutation had upon known protein-protein interactions required for ORF57-mediated nuclear export, we assessed the potential of each mutant to interact with the cellular protein Aly, which allows the formation of an export-competent ORF57-mediated ribonucleoprotein particle (3). GST-pulldown experiments were performed using recombinant GST and GST-Aly bound to GST-affinity beads and incubated with GFP-, ORF57-, or ORF57 mutant-transfected cell lysates. The results demonstrate that wild-type ORF57 and all ORF57 mutants retain the ability to interact with the hTREX protein, Aly (Fig. 5 A). However, it appears that there may be a reduction in the binding affinity between GST-Aly and the 57RGG2 and 57RGG1/2 constructs compared to the other ORF57 constructs. This suggests that although an alteration in the subcellular localization and trafficking of the 57RGG2 mutant is observed, this does not prevent, but may reduce the affinity of, direct protein-protein interactions with cellular nuclear export adapter proteins.
Fig. 5.
Site-directed mutagenesis of the carboxy-terminal RGG2 motif affects the ability of the ORF57 protein to export intronless mRNAs from the nucleus. (A) Recombinant GST- and GST-Aly-bound glutathione-agarose beads were incubated with pGFP-, pORF57GFP-, or ORF57 mutant-transfected cell lysate. After several washes, the bound proteins were analyzed by Western blotting using a GFP-specific antibody. (B) 293T cells were transfected with pORF47 in the absence or presence of pORF57GFP or mutant pORF57GFP. At 24 h posttransfection, the total (i) and cytoplasmic (ii) RNA was isolated from the cells, and the relative levels were analyzed by quantitative RT-PCR using GAPDH as a reference. The fold increase was determined by using the ΔΔCT method, and statistical significance was determined using an unpaired Student t test. Data from three independent experiments are presented as the fold increase over the GFP control.
To assess the capability of each ORF57 mutant to enhance the nuclear export of a viral intronless mRNA, HEK293T cells were transfected with a plasmid expressing the intronless KSHV ORF47 gene in addition to either GFP, wild-type ORF57, or each ORF57 mutant. After 24 h, RNA was extracted from total and cytoplasmic fractions, and the RNA levels were assessed using quantitative RT-PCR as previously described (7). The total RNA levels were assessed to ensure similar expression levels of the ORF47 mRNA. Figure 5Bi shows that the expression levels from total cell fractions are similar in the presence of wild-type ORF57 and each ORF57 mutant, although there is a slight reduction in the presence of 57RGG2. This is not unexpected since ORF57 RNA binding has been implicated in enhancing the stability of some viral transcripts (42). However, there was a significant change in the levels of ORF47 mRNA in each cytoplasmic fraction (Fig. 5 Bii). The results demonstrate that the wild-type ORF57 protein can enhance the cytoplasmic accumulation of the intronless ORF47 mRNA as previously observed (3). Similarly, site-directed mutagenesis of the RGG1 and leucine-rich region had limited, if any, effect on ORF57-mediated nuclear export. In contrast, however, mutation of the RGG2 box alone or in conjunction with RGG1 had a dramatic effect on the cytoplasmic accumulation of ORF47 mRNA levels (Fig. 5 Bii). This suggests that the alteration in the subcellular localization, nucleolar trafficking, and RNA-binding ability of the 57RGG2 mutant has a dramatic effect on ORF57 function.
57RGG2 is unable to homodimerize.
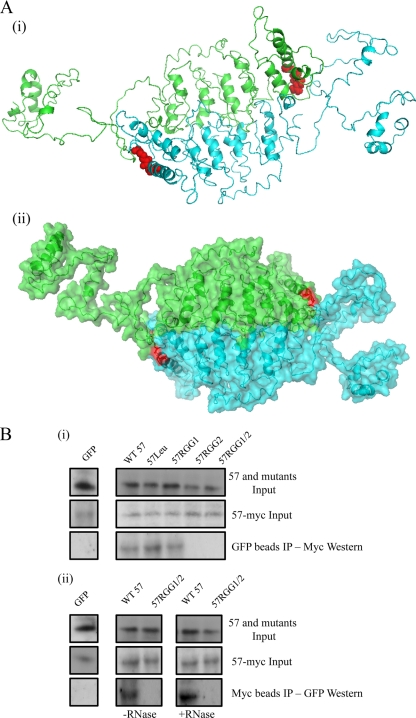
ORF57, similar to its homologues, is thought to have the ability to self-associate. Previous studies have shown that multimerization domains within the ORF57 homologue, ICP27, are contained with the carboxy terminus (21, 51). Therefore, to assess whether alterations within the carboxy-terminal RGG2 motif had any effect on the ability of ORF57 to multimerize, computational molecular modeling studies were performed. The KSHV ORF57 amino acid sequence was analyzed using the I-TASSER database, a three-dimensional structure prediction algorithm. Prediction results demonstrated that ORF57 exhibits a mainly unstructured amino terminus with a packed alpha-helical carboxy terminus. In addition, it contains an unstructured flap containing mainly positively charged residues, which may have a role in stabilizing homodimerization and/or RNA binding. To assess whether the predicted monomer model could undergo self-association, the monomer structural prediction was imported into the Maestro (Schrodinger) prediction algorithm, which assessed possible dimer interfaces. The results demonstrate that ORF57 is most likely to form a head-to-tail concatemer (Fig. 6 A). Moreover, highlighting the position of the carboxy-terminal RGG motif on both monomers (indicated in red) suggests that this region of the carboxy terminus may have an important role in the molecular interactions between the two monomers.
Fig. 6.
Site-directed mutagenesis of the carboxy-terminal RGG2 motif affects the ability of the ORF57 protein to self-associate. (A) The ORF57 amino acid sequence was analyzed using the I-TASSER database to predict the monomer structure. The monomer structural prediction was then imported into Maestro (Schrodinger) and free energy minimized. Possible dimer interfaces were identified manually, and selected structures were subjected to molecular dynamics to aid in the identification of possible conformations. A molecular model of the putative ORF57 dimer (i) and a 50% transparent space-fill model with ribbons (ii) are shown. The RGG motif is indicated in red. (B) For subpanel i, 293T cells were transfected with pORF57-myc in the presence of either pGFP, pORF57GFP, or ORF57 mutants. After 24 h, cell lysates were included with GFP-Trap-Affinity beads. Precipitated proteins were detected by Western blot analysis using myc-specific antibody. For panel ii, 293T cells were transfected with pORF57-myc in the presence of either pGFP, pORF57GFP, or p57RGG2. After 24 h, cell lysates were either mock treated (left panel) or RNase treated (right panel) for 30 min and then precipitated with Myc-Affinity beads. Precipitated proteins were detected by Western blot analysis using GFP-specific antibody.
However, a previous report utilizing a deletion mutant encompassing the RGG2 motif, instead of a site-directed mutant used herein, suggested that deletion of the RGG2 does not affect homodimerization (39). Therefore, to further investigate the possible role of the 57RGG2 motif in ORF57 homodimerization, coimmunoprecipitations were performed as previously described (4, 9, 23, 25). HEK293T cells were cotransfected with pORF57-myc in the presence of either wild-type ORF57 or mutant constructs. After 24 h, cell lysates were incubated with GFP-Trap-Affinity agarose beads, and precipitated proteins were identified by immunoblotting with a Myc-specific antibody (Fig. 6Bi). The results demonstrate that ORF57-myc can interact with wild-type ORF57, 57RGG1, and 57Leu. In contrast, 57RGG2 or 57RGG1/2 were unable to form homodimers, suggesting that the mutation of the C-terminal RGG2 motif affects ORF57 self-association. To confirm these results, reciprocal coimmunoprecipitations were performed using anti-Myc-Affinity beads and immunoblotting with a GFP-specific antibody. Moreover, these experiments were performed in the absence or presence of RNase (Fig. 6 Bii). Again, results show that site-directed mutagenesis of the RGG2 motif affects ORF57 multimerization. Furthermore, wild-type ORF57 can still form multimers in the absence of RNA, suggesting that RNA bridging is not required for multimerization.
DISCUSSION
KSHV ORF57 is a multifunctional regulator of gene expression assuming many roles during the course of the KSHV lytic replication cycle, including transcriptional activation, enhancing viral splicing and viral mRNA accumulation, and viral mRNA translation. Moreover, it has a pivotal role in enhancing the nuclear export of viral intronless mRNAs. Central to this function is the ability of ORF57 to shuttle between the nucleus and the cytoplasm, bind intronless viral transcripts, traffic through the nucleolus, and recruit cellular mRNA export factors forming an export-competent viral ribonucleoprotein particle (6, 35). Although ORF57 has a number of putative RNA-binding domains, it is unclear what region of ORF57 is responsible for recognizing mRNA. Two RGG box motifs are found in ORF57, similar to the motif responsible for binding viral mRNA in HSV-1 ICP27 (29). In addition, ORF57 contains a unique leucine-rich region also implicated in RNA binding (39). Whether these domains are definitive RNA-binding domains or disrupt ORF57 in other ways that have a secondary effect on RNA binding is unknown.
Furthermore, the implications on subcellular trafficking, hTREX complex recruitment and ultimately nuclear mRNA export, of an ORF57 protein unable to bind RNA have not been fully investigated. Therefore, we have assessed here these ORF57 properties using the 57RGG2 mutant, which we have shown fails to interact with viral intronless RNA in RNA immunoprecipitation assays. Interestingly, differences were observed regarding the subcellular localization and nuclear trafficking ability of the ORF57 protein when we compared the wild-type protein and an ORF57 protein unable to bind RNA. Wild-type ORF57, 57RGG1, and 57Leu all localize to the nuclear speckles and nucleolus as previously described (7, 34); however, the ORF57 RNA-binding mutants 57RGG2 and 57RGG1/2 displayed a distinct staining pattern lacking nucleolar localization. In addition to localizing to a distinct nuclear subdomain, 57RGG2 also failed to traffic through the nucleolus and appears to disrupt the structure of the nucleolus. In the presence of 57RGG2, the classical halo structure of the nucleolus is replaced by a necklace-like stain reminiscent of a disorganized nucleolar structure.
The nucleolar necklace is formed by rRNA transcription sites extending into the nucleoplasm, where each bead of the necklace corresponds to the fibrillar nucleolar component (FC and DFC) and contains rDNA, rRNAs, polymerase I, DNA topoisomerase I, UBF, and fibrillarin (46). A similar staining pattern is observed in nucleophosmin/B23-depleted cells (1). Nucleophosmin/B23, like nucleolin/C23, is a nucleolar hub protein that has been suggested to be an important component of nucleolar protein-protein interaction networks that may in turn have a structural role in the formation and maintenance of this dynamic nuclear organelle (18). This is of particular importance since the nucleolus consists of complex protein-protein and protein-nucleic acid interactions and, although it is imaged as a steady-state structure, high-throughput proteomic analysis of purified nucleoli coupled with live-cell imaging of fluorescently labeled nucleolar proteins has revealed a dynamic nuclear compartment, the components of which undergo continuous exchange with the nucleoplasm. Therefore, one possibility for the formation of the nucleolar necklace in the presence of 57RGG2 is that ORF57 binds and sequesters a nucleolar hub protein that gives rise to the disorganized nucleolar structure observed. The interaction between ORF57 and nucleolar hub proteins is now under investigation. The question remains, however, about the specific role of the nucleolus in ORF57-mediated nuclear export. A number of different RNAs are known to be processed during nucleolar trafficking. It is possible that the herpesvirus mRNA is also being modified in a similar fashion. Alternatively, nucleolar trafficking of viral mRNPs could allow the virus to avoid surveillance mechanisms that may otherwise degrade foreign viral intronless mRNAs prior to translation. However, these results suggest that ORF57 mutants, which cannot traffic through this organelle, are deficient in activity.
The results presented here also suggest that KSHV ORF57 has the ability to self-associate. The ability to form multimers is likely to be a conserved property within the herpesvirus ORF57 protein family (30, 51). Moreover, like its homologues, ORF57 self-association is probably not dependent on other functional motifs involved in intracellular transport, key protein-protein interactions with cellular nuclear export factors, or RNA binding. However, the 57RGG2 mutant, which is unable to self-associate, also lacked the ability to bind RNA and traffic within the nucleus, which are both essential for ORF57-mediated nuclear export of viral intronless mRNAs.
Computational molecular modeling suggests that ORF57 most likely forms an intermolecular head-to-tail concatemer. At present, our data suggest that a region of the carboxy terminus, encompassing the RGG2 motif, is important for this concatemer formation. However, whether the site-directed mutagenesis of this motif affects important molecular interactions required for self-association or whether these alterations have generally affected the folding of the carboxy terminus, preventing self-association, is yet to be elucidated. The conserved zinc-finger-like domain has been shown to be important in ICP27 self-association, which suggests that the 57RGG2 mutation has affected the overall structure of the ORF57 carboxy terminus. Notably, we assessed the subcellular localization of additional carboxy-terminal deletions of ORF57 and demonstrated that they also aggregate in subnuclear structures similar to the 57RGG2 mutant, suggesting the alterations within the carboxy terminus have a detrimental effect on the ORF57 protein (unpublished observations). Alternatively, the carboxy-terminal RGG2 motif may undergo a posttranslational modification such as arginine methylation important for protein-protein interactions. Interestingly, the ICP27 RGG box is methylated but, surprisingly, this modification has little effect on the RNA-binding properties of ICP27, instead affecting protein-protein interactions with SRPK1 and Aly (47). Further work is under way to address these questions.
In summary, these results suggest that site-directed mutations of a carboxy-terminal RGG motif has detrimental effects on the ability of the ORF57 protein to bind RNA and to traffic in the nucleus and self-associate, which are essential for the ORF57-mediated nuclear export of viral intronless mRNAs.
ACKNOWLEDGMENTS
We thank Gareth Howell, Leeds Bioimaging and Flow Cytometry Facility, University of Leeds, for useful advice.
This study was supported in part by BBSRC and Wellcome Trust project grants and by an MRC studentship to A.W. AW. is the recipient of a BBSRC Research Development Fellowship.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Amin M. A., Matsunaga S., Uchiyama S., Fukui K. 2008. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem. J. 415:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyne J. R., Colgan K. J., Whitehouse A. 2008. Herpesvirus saimiri ORF57: a posttranscriptional regulatory protein. Front. Biosci. 13:2928–2938 [DOI] [PubMed] [Google Scholar]
- 3. Boyne J. R., Colgan K. J., Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyne J. R., Jackson B., Taylor A., Macnab S., Whitehouse A. 2010. KSHV ORF57 interacts with PYM to enhance the translation of viral intronless mRNAs. EMBO J. 29:1851–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyne J. R., Jackson B. R., Whitehouse A. 2010. ORF57: master regulator of KSHV mRNA biogenesis. Cell Cycle 9:2702–2703 [DOI] [PubMed] [Google Scholar]
- 6. Boyne J. R., Whitehouse A. 2006. Gamma-2 herpesvirus posttranscriptional gene regulation. Clin. Microbiol. Infect. 12:110–117 [DOI] [PubMed] [Google Scholar]
- 7. Boyne J. R., Whitehouse A. 2009. Nucleolar disruption impairs Kaposi's sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 583:3549–3556 [DOI] [PubMed] [Google Scholar]
- 8. Boyne J. R., Whitehouse A. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calderwood M. A., Hall K. T., Matthews D. A., Whitehouse A. 2004. The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J. Gen. Virol. 85:147–153 [DOI] [PubMed] [Google Scholar]
- 10. Chang Y., et al. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 11. Chang Y., Moore P. S. 1996. Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect. Agents Dis. 5:215–222 [PubMed] [Google Scholar]
- 12. Chen I. H., Li L., Silva L., Sandri-Goldin R. M. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen I. H., Sciabica K. S., Sandri-Goldin R. M. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colgan K. J., Boyne J. R., Whitehouse A. 2009. Identification of a response element in a herpesvirus saimiri mRNA recognized by the ORF57 protein. J. Gen. Virol. 90:596–601 [DOI] [PubMed] [Google Scholar]
- 15. Colgan K. J., Boyne J. R., Whitehouse A. 2009. Uncoupling of hTREX demonstrates that UAP56 and hTHO-complex recruitment onto herpesvirus saimiri intronless transcripts is required for replication. J. Gen. Virol. 90:1455–1460 [DOI] [PubMed] [Google Scholar]
- 16. Dittmer D., et al. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dourmishev L. A., Dourmishev A. L., Palmeri D., Schwartz R. A., Lukac D. M. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emmott E., Hiscox J. A. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep. 10:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Invest. 120:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gatfield D., Izaurralde E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159:579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goodwin D. J., et al. 2000. The carboxy terminus of the herpesvirus saimiri ORF 57 gene contains domains that are required for transactivation and transrepression. J. Gen. Virol. 81:2253–2658 [DOI] [PubMed] [Google Scholar]
- 22. Goodwin D. J., Whitehouse A. 2001. A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J. Biol. Chem. 276:19905–19912 [DOI] [PubMed] [Google Scholar]
- 23. Gould F., Harrison S. M., Hewitt E. W., Whitehouse A. 2009. Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J. Virol. 83:6727–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grundhoff A., Ganem D. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 113:124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall K. T., et al. 2002. The herpesvirus saimiri open reading frame 73 gene product interacts with the cellular protein p32. J. Virol. 76:11612–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hautbergue G. M., et al. 2009. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19:1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedes D. H., Lagunoff M., Renne R., Ganem D. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100:2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koffa M. D., et al. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lengyel J., Guy C., Leong V., Borge S., Rice S. A. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 76:11866–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lischka P., Thomas M., Toth Z., Mueller R., Stamminger T. 2007. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J. Gen. Virol. 88:405–410 [DOI] [PubMed] [Google Scholar]
- 31. Longman D., Johnstone I. L., Caceres J. F. 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo M. J., Reed R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. U. S. A. 96:14937–14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majerciak V., et al. 2008. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 82:2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Majerciak V., Yamanegi K., Nie S. H., Zheng Z. M. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 35. Majerciak V., Zheng Z. M. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mears E., Rice S. A. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA binding activity and determines in vivo methylation. J. Virol. 70:7445–7453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514–1518 [DOI] [PubMed] [Google Scholar]
- 38. Moore M. J., Proudfoot N. J. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 [DOI] [PubMed] [Google Scholar]
- 39. Nekorchuk M., Han Z., Hsieh T. T., Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rainbow L., et al. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruvolo V., Gupta A. K., Swaminathan S. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 75:6033–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sahin B. B., Patel D., Conrad N. K. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 6:e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sandri-Goldin R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241–5256 [DOI] [PubMed] [Google Scholar]
- 44. Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 45. Sergeant A., Gruffat H., Manet E. 2008. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 13:3798–3813 [DOI] [PubMed] [Google Scholar]
- 46. Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. 2008. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129:13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Souki S. K., Sandri-Goldin R. M. 2009. Arginine methylation of the ICP27 RGG box regulates the functional interactions of ICP27 with SRPK1 and Aly/REF during herpes simplex virus 1 infection. J. Virol. 83:8970–8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uranishi H., et al. 2009. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J. Biol. Chem. 284:26106–26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Valencia P., Dias A. P., Reed R. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 105:3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. West J. T., Wood C. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150–5163 [DOI] [PubMed] [Google Scholar]
- 51. Zhi Y., Sciabica K. S., Sandri-Goldin R. M. 1999. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 257:341–351 [DOI] [PubMed] [Google Scholar]