Abstract
Hepatitis C virus (HCV) is characterized by a narrow host range and high interindividual variability in the clinical course of infection. Both of these traits are thought to be largely due to genetic variation between species and between individual hosts. The tight junction component occludin (OCLN) is essential for HCV entry into host cells, and the differences between human and murine OCLN are thought to account in part for the inability of HCV to infect mice and hence preclude their use as a convenient small-animal model. This study assesses the impact of genetic variation in OCLN on cell culture-grown HCV (HCVcc) using a newly generated and characterized OCLNlow subclone of the Huh-7.5 cell line (Huh-7.5 subclone in which endogenous OCLN expression has been downregulated by a short hairpin RNA). We report the frequency of coding nonsynonymous single nucleotide polymorphisms, i.e., polymorphisms resulting in amino acid exchanges, present in the human population and determine their ability to function as HCV (co)receptors. Moreover, we show that murine OCLN can sustain HCVcc entry, albeit with about 5-fold reduced efficiency compared to that of human OCLN. This reduction in efficiency is due solely to two amino acid residues previously identified by others using an HCV pseudoparticle approach. Finally, we use the Huh-7.5/OCLNlow cell line to show that HCV spread between neighboring cells is strictly dependent on OCLN.
INTRODUCTION
The hepatitis C virus (HCV) is a small enveloped viral pathogen of the Flaviviridae family with a single plus-strand RNA genome (26). About 130 million individuals worldwide are currently chronically HCV infected and thus at risk for the development of cirrhosis, end-stage liver disease, and hepatocellular carcinoma (34).
HCV primarily infects and replicates in hepatocytes. All stages of its replication cycle are dependent on various host-encoded factors. Completion of the earliest stage of the replication cycle, HCV cell entry, requires at least four host factors on the hepatocyte surface. These include the tetraspanin CD81 (30), scavenger receptor class B type I (SR-BI) (32), and more recently described, the tight junction components claudin 1 (CLDN1) (13) and occludin (OCLN) (27, 31). It is thought that the viral envelope glycoproteins E1 and E2 interact with these four (co)receptors sequentially and that these interactions are orchestrated by cell signaling processes (6, 14, 28). SR-BI may act early and the tight junction components may act later in this process (11, 13, 21, 24, 42). The exact mechanisms and sequence of the interactions remain to be determined, but the result is known to be the uptake of the virion or a virion-coreceptor complex into an endosomal compartment. Acidification then triggers fusion of the viral envelope with the endosomal membrane (20, 24, 40). This releases the nucleocapsid into the cytosol, completing the cell entry process.
Only humans and chimpanzees are natural hosts of HCV. This is thought to be because HCV is unable to utilize other species' homologues of several essential host factors. Such restrictions seem to exist at all stages of the viral replication cycle. It is a major goal of HCV research to define and overcome these restrictions in order to create a convenient small-animal, ideally murine, model of HCV infection (25). At the cell entry stage, it has been shown that human and murine SR-BI and CLDN1 can serve as HCV (co)receptors with about equal efficiency (8, 13). Conversely, CD81 and OCLN have been reported to confer species restriction since the murine homologues of these entry factors are significantly less efficient than their human counterparts (16, 29, 31). In both cases, the second of two extracellular loops has been identified as the critical region determining species restriction. In the case of OCLN, this characteristic has been pinpointed to two amino acid residues (29). However, a limitation is that while in the case of CD81, this notion has been substantiated in several studies, including ones using authentic cell culture-grown HCV (HCVcc) as a model system (4, 16), data on the species specificity of OCLN stems purely from experiments with lentiviruses pseudotyped with HCV E1E2 (HCV pseudoparticles [HCVpp]), because a cell line that is OCLN negative yet permissive to HCV replication is not available. While HCVpp have overall proven to be reasonably good models of HCV cell entry, their structure and especially the packing of E1E2 complexes on their surfaces are very different from authentic HCVcc (5).
Within humans, HCV's natural host, the course of infection is highly variable (33, 43), and again, genetic variation in host factors seems to be causative. Acute infection can be clinically silent or present as mild to moderate and sometimes even severe hepatitis. The outcome of acute infection is spontaneous resolution in about a quarter of cases, whereas the majority progress to chronic infection. When treated with the current regimes, chronically infected individuals resolve infection in about 50% of cases, whereas therapy fails for the other half. Finally, among those who remain chronically infected, some remain asymptomatic for life, while others require liver transplantation or die from HCV-associated end-stage liver disease or liver cancer. Two genetic variations in the interleukin-28B (IL-28B) gene have recently been shown to account at least in part for whether or not spontaneous or treatment-induced resolution of infection is achieved (17, 36, 37). However, additional genetic factors must clearly be involved. The occludin gene encoding the OCLN coreceptor displays a fair amount of variation and a remarkable amount of alternative splice variants (23). At least three coding nonsynonymous single-nucleotide polymorphisms (SNPs) in the occludin coding region have been reported (http://www.ncbi.nlm.nih.gov/snp); however, there is little information on their frequency, and their impact on HCV cell entry is unknown.
The aim of the present study was to determine the impact of intra- and interspecies genetic variation on the OCLN coreceptor. To this end, we have used a short hairpin RNA (shRNA) targeting all known occludin transcripts to create an OCLNlow subclone of the Huh-7.5 cell line (Huh-7.5 subclone in which endogenous OCLN expression has been downregulated by a short hairpin RNA) that supports the full HCV replication cycle upon restoration of OCLN expression.
MATERIALS AND METHODS
Cell culture and cell lines.
Huh-7.5 cells and subclones, 786-O cells, and 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal bovine serum, l-glutamine, nonessential amino acids, penicillin, and streptomycin. Cells harboring shRNA constructs were kept in the presence of 5 μg/ml blasticidin.
DNA constructs.
Transgenes were expressed from the previously described lentiviral pTRIP vector, a self-inactivating lentiviral genome that expresses no HIV proteins but drives transgene expression from an internal cytomegalovirus (CMV) promoter (35). pTRIP constructs encoding N-terminally green fluorescent protein (GFP)-tagged wild type CLDN1 and human OCLN (31), N-terminally Venus-tagged human and mouse OCLN, and the tagRFP-nuclear localization sequence (NLS)-interferon-β promoter stimulator protein (IPS) reporter (22) (see below) were kind gifts from C. Rice (The Rockefeller University, New York, NY). OCLN SNPs were recreated in the GFP-OCLN context by standard PCR-based mutagenesis, in which in a first round, two overlapping PCR fragments are created with internal oligonucleotides encoding the desired changes and, in a second round, the first-round fragments are combined with outside oligonucleotides that span unique restriction sites (MscI, XhoI). These enzymes were then used to clone the mutant fragment back into the likewise digested pTRIP vector (detailed cloning strategies are available upon request). The human-mouse A223G-plus-A224G and G223A-plus-G224A changes were introduced into the Venus-OCLN context by the same approach. The DNA sequences of all newly modified constructs were confirmed by direct sequencing (Eurofins MWG Operon, Ebersberg, Germany).
For the generation of pseudoparticles carrying a firefly luciferase reporter, we created CSFlucW2 as a derivative of CSGW (12) by introducing a unique SpeI site downstream of the GFP open reading frame (ORF) by PCR-based mutagenesis as described above. Then the firefly luciferase ORF was amplified using oligonucleotides containing BamHI and SpeI sites and ligated into the likewise digested CSGW derivative with the SpeI site.
The HCV reporter genome Luc-Jc1, an intragenotypic HCV chimera consisting of Japanese fulminant hepatitis 1 (JFH1) (genotype 2a; GenBank accession no. AB047639) and J6/CF (genotype 2a; GenBank accession no. AF177036) genome segments, has been described (24). The intergenotypic H77/JFH (genotype 1a/2a) genome used in some experiments was a kind gift from Jens Bukh (Copenhagen University Hospital, Copenhagen, Denmark) (18).
Short hairpin RNA's targeting occludin was constructed in the context of the shRNA expression vector pLenti-3′-U6-EC-EP7, a blasticidin selectable lentiviral genome vector expressing the shRNA of interest from an internal U6 promoter (41). The target specific sequence used for the creation of an OCLNlow cell line was 5′-GGGCATTGCTCATCCTGAAGA-3′, which is contained in the 5′-untranslated region of all known occludin transcripts in humans. An analogous construct containing an irrelevant sequence not predicted to target any human transcript was used as a negative control.
Genotyping and retrieval of genetic data.
Use of anonymous human DNA samples from healthy volunteers to determine allele frequencies in the respective populations were approved by the institutional review boards at Hannover Medical School (Hannover, Germany) and at German University in Cairo (Cairo, Egypt). Genotyping was performed using custom-made LightSNiP assays for rs28562785, rs28418826, and rs17852716 that were purchased from TIB Molbiol (Berlin, Germany) and run on a LightCycler 480 (Roche Diagnostics, Mannheim, Germany). Briefly, following PCR amplification of the genomic region of interest, LightSNiP assays (TIB Molbiol) employ fluorescently labeled probes to create a genotype-specific melting curve pattern when running a temperature ramp.
Allele frequencies in HapMap panels were retrieved from the project's website (http://hapmap.ncbi.nlm.nih.gov/) using HapMap Genome Browser release 28 (phases 1, 2, and 3—merged genotypes and frequencies) (1).
Pseudotyping of lentiviral particles and transduction of target cells.
Pseudoparticles were generated as previously described (13). Three plasmids carrying (i) a provirus containing either a firefly luciferase reporter (CSFlucW2) or one of various transgenes (pTRIP constructs), (ii) HIV Gag-Pol, and (iii) the appropriate viral envelope glycoprotein(s) were cotransfected into 293T cells. To generate HCVpp, the HCV glycoproteins E1 and E2 of strain H77 were used (15). For transduction experiments, the G protein of vesicular stomatitis virus (VSV-G) was employed. 293T cells were seeded at 8 × 105 cells per well into a poly-l-lysine (Sigma-Aldrich, Munich, Germany)-coated 6-well plate. Transfection of a total of 1.5 μg of DNA was performed the following day using FuGene (Roche Diagnostics, Mannheim, Germany), according to the manufacturer's protocol. Supernatants were collected at 48 and 72 h posttransfection and passed through a 0.45-μm-pore-size filter. For pseudovirus transduction, filtered supernatants supplemented with 4 μg/ml Polybrene were added to the target cells for 6 h. Luciferase activity was measured after 72 h. Transgene-expressing cells were used for subsequent experiments at 48 h or later. Transduction levels were determined by flow cytometry.
HCVcc infection.
Huh7.5 cells were transfected with an HCV genome (Jc1 or H77/JFH1 chimera) by electroporation as previously described (9) and seeded into 6-well plates. Where desired, HCV RNA replication was quantified by measuring the luciferase activity. To harvest HCVcc for subsequent infection experiments, supernatants were collected at 48 h after electroporation, filtered through 0.45-μm-pore-size filters, and used to infect target cells. Chimeric H77/JFH1 HCVcc was additionally concentrated 10-fold using Amicon ultrafiltration devices (Millipore, Billerica, MA) prior to use in infection experiments. Target cells were seeded at 8 × 104 per well in 12-well plates (luciferase reporter assay) or at 1 ×104 per well in 96-well plates (50% tissue culture infective dose [TCID50]/ml assay). The following day, cells were inoculated with 500 or 100 μl virus containing supernatant. HCV infection was quantified by measuring luciferase activity or staining for HCV NS5A 48 h later. Luciferase measurements were performed as described previously (9); all luciferase assays were performed in duplicate (for replication) or quadruplicate (for infection). For NS5A staining, supernatant from the 9E10 hybridoma line at a dilution of 1:2,000 was used, and the TCID50/ml was calculated according to the method of Spearman and Kärber as described previously (9).
Detection of HCV cell-to-cell transmission.
To measure HCV infection by direct cell-to-cell transmission between adjacent cells, we performed a modified version of the agarose overlay assay recently described by Baldick and colleagues (3) in which transmission from HCV-positive donor cells to HCV-negative target cells in coculture was assessed. Donor Huh-7.5 cells were infected with supernatant containing an excess of HCVcc (H77/JFH1 chimera [18]). A subfraction of the donor cells was fixed and stained for NS5A to confirm that >99% of cells were HCV positive. Target cells were transduced with the pTRIP-tagRFP-NLS-IPS1 reporter construct recently described by Jones and colleagues (22). Briefly, this construct contains a fusion protein of the red fluorescent protein tagRFP, a simian virus 40 (SV40) nuclear localization sequence (NLS), and the C-terminal part (amino acids 462 to 540) of the interferon-β promoter stimulator protein 1 (IPS1). The IPS1 C terminus contains a recognition site for the HCV NS3/4A protease upstream of the mitochondrial targeting sequence and a transmembrane segment that is physiologically lodged in the mitochondrial membrane. Thus, in the absence of HCV infection, the tagRFP reporter is tethered to the mitochondrial membrane, resulting in a punctate cytosolic distribution of fluorescence. In HCV-infected cells, NS3/4A cleaves the tagRFP-NLS fusion from its mitochondrial anchor and, driven by the NLS, the reporter relocates to the nucleus. Donor and target cells were mixed at a 1:1 ratio and seeded at 4 × 104 cells per well into 12-well plates. The cell mixture was overlaid with medium containing 1% SeaPlaque low-melting-temperature agarose to prevent virus spread through the cell-free route. Redistribution of the tagRFP reporter was monitored through fluorescent microscopy, and the percentage of HCV-positive target cells, as indicated by nuclear localization of tagRFP, was determined after 96 h.
Flow cytometry.
Cells were trypsinized and resuspended in phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum (FCS) and 0.5% paraformaldehyde. Expression of GFP and Venus tags was quantified using a BD FACSCanto II cytometer (Becton Dickinson, Heidelberg, Germany). Data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Immunoblot analysis.
Cells were washed once with PBS and lysed in sample buffer containing 2% sodium dodecyl sulfate (SDS). Proteins were resolved by standard electrophoresis, blotted onto a polyvinylidene difluoride membrane, and detected with OCLN (clone OC-3F10; Zymed/Invitrogen, Darmstadt, Germany)-, CLDN1 (clone 2H10D10; Zymed/Invitrogen)-, or beta-actin (Sigma-Aldrich, Munich, Germany)-specific antibody, horseradish peroxidase-coupled anti-mouse IgG secondary antibody (Sigma), and the ECL Plus Western blotting detection system (GE Healthcare Europe, Freiburg, Germany).
Statistical analyses.
Numerical data were analyzed using Excel (Microsoft, Redmond, WA). All experiments were repeated on separate occasions, and each repetition was done in multiple replicates. Unless stated otherwise, the mean ± standard deviation of the replicates in one representative experiment is shown, with the number of replicates indicated. Significant differences were identified using the unpaired two-sided Student t test. P values below 0.05 were considered significant and are indicated by an asterisk. Bonferroni correction for multiple comparisons was used to adjust significance levels and avoid false positives where appropriate.
RESULTS
Frequency of coding nonsynonymous SNPs in the occludin gene and impact on HCVpp.
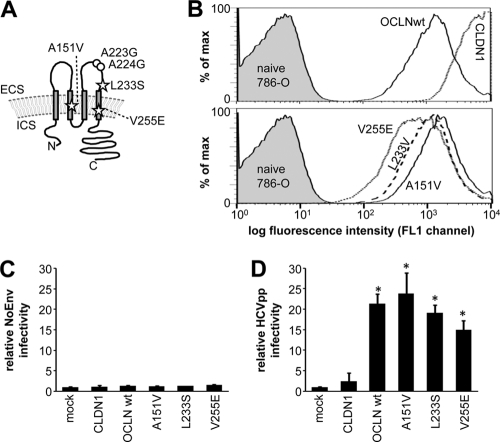
Three coding nonsynonymous SNPs in the occludin gene causing amino acid changes in the second transmembrane segment (A151V), the second extracellular loop (L233S), and the fourth transmembrane segment (V255E) have been reported (Fig. 1A). While changes in transmembrane segments have the potential to disrupt the topology of membrane proteins, L233S lies in the C-terminal half of the second extracellular loop of OCLN, previously reported to be critical for species selectivity and in close proximity to two residues (alanines 223 and 224) that are responsible for the more efficient coreceptor function of human OCLN rather than murine OCLN in HCVpp assays (29). The frequency of the L233S SNP, but not that of the A151V or V255E SNP, has been determined to be about 1% in African and Chinese HapMap panels (Table 1) (1). In addition, we measured the allele frequencies of all three SNPs in a Caucasian (German ancestry, n = 100) and an Oriental (Egyptian ancestry; n = 100) population sample. Thus, SNPs resulting in amino acid changes in the OCLN coreceptor occur with 1 to 2.5% allele frequency in all human populations assessed, with the L233S exchange being most common.
Fig. 1.
HCVpp can utilize human OCLN variants for efficient cell entry. (A) Schematic representation of OCLN, with stars indicating the positions of amino acid changes caused by known nonsynonymous SNPs in the occludin coding region and circles indicating the positions of two alanine residues that are replaced by glycine in the mouse homologue and responsible for the inferior (co)receptor function of murine OCLN in HCVpp assays. (B) Flow cytometric analysis of GFP-expression in 786-O cells transduced to express GFP-tagged wild-type OCLN, human OCLN variants, and CLDN1 at the time when susceptibility to HCVpp entry was assessed. (C, D) Above-mentioned 786-O cells expressing OCLN variants were challenged with envelope-deficient pseudoparticles (NoEnv), HCVpp strain H77, and VSV-Gpp. Cells were lysed, and luciferase activity was determined 3 days after infection. NoEnv (C) and HCV (D) infectivity relative to VSV-G infectivity (mean ± standard deviation) in a representative experiment, with 3 replicates per condition, is shown, with the value determined in mock-transduced cells set to 1. Statistically significant (P < 0.05) differences from mock-transduced cells are indicated by asterisks. ECS, extracellular space; ICS, intracellular space; wt, wild type.
Table 1.
Allele frequencies of coding nonsynonymous SNPs in the occludin gene in different populations
| SNP | Change | Allele frequency (%)a |
||||
|---|---|---|---|---|---|---|
| ASW (HapMap) | Caucasian (this study) | CHB (HapMap) | GIH (HapMap) | Oriental (this study) | ||
| rs28562785 | A151V | ND | 0 | ND | ND | 1 |
| rs17852716 | L233S | 1 | 2.5 | 1 | 0 | 1 |
| rs28418826 | V255E | ND | 0 | ND | ND | 0 |
Data were obtained from the HapMap project (hapmap.ncbi.nlm.nih.gov/) (1) or were assessed as part of this study by genotyping 100 individuals of German (Caucasian) and 100 individuals of Egyptian (Oriental) ancestry. ASW, African ancestry in southwest United States (n = 57); CHB, Han Chinese in Beijing, China (n = 136); GIH, Gujarati Indians in Houston, TX (n = 101); ND, not done.
We introduced the three SNPs into lentiviral OCLN expression constructs that carry an N-terminal green fluorescent protein (GFP) tag and used these to transduce the HCV replication-incompetent 786-O cell line that lacks detectable levels of OCLN. GFP-tagged CLDN1 was transduced in parallel as a negative control. Transduction efficiency was assessed by flow cytometry, and >99% transduction rates were detected in all cases, with CLDN1 consistently showing higher fluorescence intensity that those of the OCLN constructs (Fig. 1B). Subsequently, transduced 786-O cells were challenged with HCVpp (strain H77, genotype 1a), as well as glycoprotein-less pseudoparticles (NoEnv) and pseudoparticles bearing the G protein of vesicular stomatitis virus (VSV-Gpp) as negative and positive controls, respectively. Results are shown as NoEnv (Fig. 1C) and HCVpp (Fig. 1D) entry into 786-O cells relative to the signal obtained with VSV-Gpp. While expression of OCLN variants did not alter the uptake of NoEnv or VSV-Gpp, HCVpp entry was significantly enhanced compared to that in mock transduced cells by all OCLN variants tested, while expression of the CLDN1 coreceptor had no effect. HCVpp entry mediated by OCLN V255E appeared to be slightly less efficient than that of the wild type and the A151V and L233S variants, but this did not reach statistical significance and was not observed in subsequent experiments with HCVcc (see below).
Generation and characterization of a fully replication-competent OCLNlow Huh-7.5 subclone.
To be able us to study the impact of variations in the OCLN coreceptor on HCVcc, we generated a lentivirus-borne shRNA targeting a sequence contained in the second exon of the occludin gene that is part of the 5′-untranslated region of all occludin mRNA transcripts described so far (23). Transduction of Huh-7.5 cells resulted in a near complete knockdown of OCLN protein expression (Fig. 2A) and a marked reduction in HCV susceptibility, comparable to the effect seen when CD81 or CLDN1 are targeted by RNA interference (RNAi) (data not shown). In order to have a stable cellular background for subsequent testing of OCLN variants, we expanded subclones from the Huh-7.5/OCLNlow population harboring the shRNA. A subclone (designated clone#1) that showed very low OCLN levels, fast growth kinetics, and low HCVpp susceptibility while retaining good susceptibility to VSV-Gpp was chosen for subsequent experiments (Fig. 2A and data not shown). Expression of CLDN1, another tight junction-associated coreceptor of HCV, was unchanged in clone#1 cells compared to that in Huh-7.5 cells expressing an irrelevant shRNA (Fig. 2B). In comparison to parental Huh-7.5 cells, clone#1 showed >500-fold reduced susceptibility to infection with HCVcc Jc1 carrying a firefly luciferase reporter (Fig. 2C) and a significantly reduced ability to support HCVpp entry in absolute terms as well as relative to VSV-Gpp (Fig. 2D). Intracellular HCV RNA replication was largely unaltered (<2-fold change) (Fig. 2E). Release of infectious HCVcc particles following electroporation of HCV RNA was about 10-fold reduced compared to that in parental Huh-7.5 but remained readily detectable (Fig. 2F). Thus, clone#1 is an OCLNlow Huh-7.5 subclone that is resistant to HCV entry while remaining fully permissive to replication and largely permissive to virus production.
Fig. 2.
Characterization of clone#1, an OCLNlow yet HCV replication- and assembly-competent Huh-7.5 subclone. (A) Immunoblot for OCLN and beta-actin in parental Huh-7.5 cells, Huh-7.5 cells expressing an irrelevant shRNA (sh-irrelevant) or occludin-specific shRNA (sh-OCLN), and clone#1, a subclone from the Huh-7.5/occludin-specific shRNA population chosen for subsequent experimentation. (B) Immunoblot for CLDN1 and beta-actin in Huh-7.5 cells expressing an irrelevant shRNA and clone#1. (C) Parental Huh-7.5 and clone#1 cells were inoculated with supernatant containing Jc1-Luc reporter virus. Luciferase activity was assayed 48 h postinfection. Mean values and the standard deviations from a representative experiment performed in triplicate are shown. (D) Parental Huh-7.5 and clone#1 cells were inoculated with HCVpp and VSV-Gpp. HCVpp infectivity relative to VSV-Gpp infectivity, with the value detected in Huh-7.5 cells set to 100% (means and standard deviations of 3 replicates), is shown. (E) Parental Huh-7.5 (open squares) and clone#1 (closed squares) cells were electroporated with Jc1-Luc RNA transcripts, and luciferase activity over the following 72 h was monitored as indicated. Mean values from a representative experiment done in duplicate are shown. (F) To assess efficiency of viral particle production, supernatant from Huh-7.5 and clone#1 cells electroporated with Jc1-Luc RNA transcripts was collected after 72 h, filtered, and used to inoculate naïve Huh-7.5 cells. Luciferase activity was assessed 48 h after inoculation. “Mock” indicates the background luciferase reading detected in uninfected cells. Mean values and the standard deviations from a representative experiment done in quadruplicate are shown. Significant differences from parental Huh-7.5 are indicated by asterisks. RLU, relative light units.
Effect of OCLN overexpression in Huh-7.5 cells on HCV permissiveness.
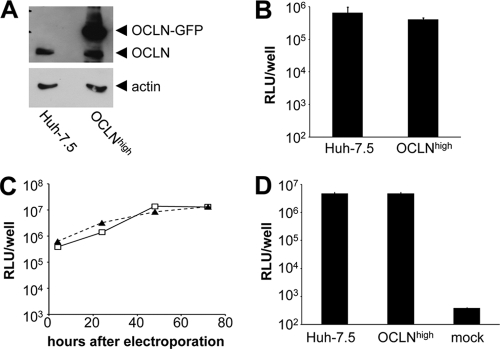
Before beginning overexpression experiments intended to characterize the phenotype of OCLN variants in the clone#1 cell line, we tested whether overexpression of OCLN in parental Huh-7.5 cells had an effect on HCV permissiveness. To this end, we transduced Huh-7.5 cells with pTRIP encoding GFP-tagged wild-type human OCLN (Fig. 3A). As expected, parental Huh-7.5 cells and Huh-7.5 cells expressing GFP-OCLN showed similar levels of susceptibility to HCVcc infection (Fig. 3B), HCV RNA replication following electroporation (Fig. 3C), and release of progeny HCVcc (Fig. 3D). Thus, raising the expression of OCLN protein above the endogenous levels does not seem to enhance HCV permissiveness.
Fig. 3.
OCLN overexpression does not alter HCV permissivity of Huh-7.5 cells. (A) Immunoblot for OCLN and beta-actin in parental Huh-7.5 cells and Huh-7.5 cells transduced to overexpress OCLN-GFP (OCLNhigh). (B) Parental Huh-7.5 and OCLNhigh cells were inoculated with supernatant containing Jc1-Luc reporter virus. Luciferase activity was assayed at 48 h postinfection. Mean values and the standard deviations from a representative experiment performed in triplicate are shown. (C) Parental Huh-7.5 (open squares) and OCLNhigh (closed triangles) cells were electroporated with Jc1-Luc RNA transcripts, and luciferase activity over the following 72 h was monitored as indicated. Mean values from a representative experiment done in duplicate are shown. (D) Supernatant from Huh-7.5 and OCLNhigh cells electroporated with Jc1-Luc RNA transcripts was collected after 72 h, filtered, and used to inoculate naïve Huh-7.5 cells. The mock value represents background luciferase readings in uninfected cells. Luciferase activity was assessed 48 h after inoculation. Mean values and the standard deviations from a representative experiment done in duplicate are shown.
Impact of human OCLN variants on HCVcc.
To determine the impact of SNPs in the occludin gene on HCVcc entry, we expressed GFP-tagged OCLN variants in the context of clone#1 (Fig. 4A). In keeping with the above-mentioned pseudovirus data, all OCLN SNPs restored the susceptibility of clone#1 to genotype 2 (Fig. 4B) and genotype 1 (Fig. 4C) HCVcc, with an efficiency comparable to that of wild-type OCLN, while expression of CLDN1 had no effect. Thus, the known OCLN SNPs are unlikely to have an effect on HCV infection, even in homozygous carriers. To rule out an effect of fluorescent tags on OCLN localization, we performed immunofluorescence staining of OCLN in parental Huh-7.5 cells, clone#1 cells, and clone#1 cells expressing either untagged or GFP- or Venus-tagged OCLN. The observed subcellular distributions were similar, showing both intracellular and membrane staining, most notably in areas where neighboring cells interfaced. Untransduced clone#1 cells remained largely unstained (data not shown).
Fig. 4.
HCVcc can utilize human OCLN variants for efficient cell entry. (A) Flow cytometric analysis of GFP expression in clone#1 cells transduced to express GFP-tagged wild-type OCLN, human OCLN variants, and CLDN1 at the time when susceptibility to HCVcc was assessed. (B) Above-mentioned clone#1 cells expressing OCLN variants were challenged with genotype 2a Jc1-Luc HCVcc, and luciferase activity was measured 48 h after inoculation. Mean values and the standard deviations from three independent experiments each done in duplicate are shown. Significant differences from mock-transduced cells are indicated by asterisks. (C) Transgene-expressing cells were challenged with genotype 1a/2a (structural region of genotype 1a) H77/JFH1 chimeric HCVcc in a standard limiting dilution assay (12 dilution steps, 8 wells per dilution). Cells were fixed and stained for NS5A expression 48 h after inoculation, and the TCID50/ml was determined. Results from a representative experiment are shown.
Impact of mouse-specific residues on OCLN coreceptor function.
OCLN, together with CD81, has been suggested to be a determinant of host restriction of HCV entry (16, 31). In a detailed mutagenesis study using HCVpp and 786-O cells as a cellular background, the better coreceptor function of human OCLN has been pinpointed to two residues, alanines 223 and 224, in the second extracellular loop (29). Murine OCLN has two glycines in positions 223 and 224. Exchange of these two residues between human and mouse OCLN reproduces the phenotype of the respective other species in an HCVpp assay. We tested wild-type human OCLN and murine OCLN as well as the reported chimeras [human OCLN(A223G A224G) and murine OCLN(G223A G224A)] in the context of clone#1 (Fig. 5A) for their ability to support HCVcc entry. We observed full restoration of HCVcc entry when human OCLN or murine OCLN(G223A G224A) was expressed, while murine OCLN and human OCLN(A223G A224G) showed partial rescue (Fig. 5B). The difference between human OCLN and murine OCLN, as judged by the luciferase reporter signal, was about 6-fold. Next we tested the susceptibility of the same cells to a variant of Jc1 that had been adapted to use murine CD81 with the same efficiency as human CD81 (4). This variant carries a total of three amino acid exchanges in E1 (L216F) and E2 (V388G and M405T) and has been shown to be capable of entering cells expressing only murine HCV (co)receptors. We found that, similar to Jc1, the mouse CD81-adapted Jc1 variant was able to utilize murine OCLN and human OCLN(A223G A224G) with an efficiency that was 5 to 8-fold below that of human OCLN or murine OCLN(G223A G224A) (Fig. 5C). Thus, murine OCLN does support HCVcc entry, albeit with an efficiency that is reduced compared to that of human OCLN. As in the case of HCVpp, this difference is fully attributable to the two alanine residues in positions 223 and 224.
Fig. 5.
HCVcc can utilize murine OCLN for cell entry with reduced efficiency. (A) Flow cytometric analysis of Venus expression in clone#1 cells transduced to express Venus-tagged human or murine OCLN variants as well as chimeric OCLN with residues 223 and 224 exchanged between the human and murine homologues at the time when susceptibility to HCVcc was assessed. (B, C) Above-mentioned clone#1 cells expressing the respective OCLN variants were challenged with genotype 2a Jc1-Luc HCVcc (B) and a Jc1-Luc variant adapted to utilize murine CD81, with efficiency equal to that of human CD81 (C) (4). Luciferase activity was measured 48 h after inoculation. Mean values and the standard deviations from at least three independent experiments each done in duplicate are shown. Significant differences from mock- and human OCLN (hOCLN)-transduced cells are indicated by asterisks and pound signs, respectively. mOCLN, murine OCLN.
Importance of OCLN for cell-to-cell spread of HCV.
Finally, HCV has been reported to be transmitted in cell culture both as cell-free virions and in a direct cell-to-cell manner that does not seem to involve a cell-free virion stage. There is some controversy as to whether cell-to-cell transmission is mechanistically different from cell entry by extracellular virions, in that it may not be dependent on the presence of CD81 on the target cell. Very recently, cell-to-cell transmission has been reported to be reduced when OCLN is downregulated in an assay in which neutralizing antibodies against E2 present in the culture medium were used to block cell-free transmission (7). We were interested to test the susceptibility of clone#1 to cell-to-cell transmission in an independent assay and to assess to what extent OCLN variants support cell-to-cell transmission. To this end, we infected a population of Huh-7.5 cells with an HCVcc (H77/JFH chimera) (18) at a high multiplicity of infection and confirmed that >99% were HCV NS5A positive by immunofluorescence staining (data not shown). These “donor cells” were cocultured with Huh-7.5 or clone#1 “target cells” that had additionally been transduced with a reporter construct in which HCV infection is detected at the single-cell level in living cells by relocation of the red fluorescent protein tagRFP from a mitochondrial (uninfected) to a nuclear (infected) localization (22). In addition, target cells were transduced to express mock or different OCLN variants. The cell mixture was overlaid with medium containing 1% agarose to prevent virus spread through the cell-free route as recently reported (3) (see Materials and Methods for more assay details). Among parental Huh-7.5 target cells, we detected a high percentage of infected cells on day 4 postinfection, while among clone#1 target cells, not a single infected cell was seen in any experiment, even in areas where donors and acceptors interfaced directly (Fig. 6A). When using clone#1 cells transduced to express human OCLN wild type, A151V, L233S, or V255E as target cells, cell-to-cell infection of target cells was readily detectable and not significantly different from that of parental Huh-7.5. Murine OCLN was also clearly able to support cell-to-cell spread at lower levels than that supported by parental Huh-7.5 or clone#1 expressing human OCLN, but this difference did not reach statistical significance. Thus, in this assay, direct spread between neighboring cells is strongly dependent on the presence of OCLN.
Fig. 6.
Cell-to-cell spread of HCV is undetectable in the absence of OCLN. (A) As detailed in the text, a fully H77/JFH1-infected population of nonfluorescent Huh-7.5 donor cells was put in coculture with uninfected target cells (parental Huh-7.5 [left] or clone#1 [right]) harboring a tagRFP-NLS-IPS reporter (22). Briefly, in uninfected cells tagRFP fluorescence is localized in the cytoplasm in a punctuate mitochondrial pattern; upon HCV infection, the tagRFP-NLS is cleaved from its IPS anchor in the mitochondrial membrane, and fluorescence relocates to the nucleus driven by the NLS fused to the tagRFP. Nuclei were costained with DAPI (4′,6-diamidino-2-phenylindol). (B) Percentage of infected cells, as indicated by nuclear localization of tagRFP per well after 96 h of coculture using as target cells Huh-7.5, clone#1, or clone#1 expressing different OCLN variants. Means and standard deviations from a representative experiment done in duplicate are shown, and significant differences from parental Huh-7.5 are indicated by asterisks.
DISCUSSION
This study is the first to describe how cell entry by HCVcc is affected by genetic variation in the essential OCLN coreceptor. To enable us and others in the field to address this topic, we have created an OCLNlow Huh-7.5 subclone harboring a highly efficient shRNA suppressing all known occludin transcripts. Upon reintroduction of OCLN, this cell line becomes again permissive to all stages of the HCV replication cycle. We report the frequency of SNPs present in the human population that change the amino acid sequence of critical regions of OCLN and are the first to test their impact on HCVpp and HCVcc. Moreover, we show that murine OCLN can serve as a coreceptor for HCV, although with reduced efficiency compared to that of its human homologue, and that the loss in efficiency is attributable to the same two alanine residues previously reported to be critical for HCVpp entry (29). Lastly, our data suggest that OCLN not only enhances but also is essential for spread of HCV between adjacent cells (cell-to-cell route) and that different OCLN variants, including murine OCLN, support this route of viral spread.
HCV cell entry is a complex multistep process. Many or maybe all of the essential host factors involved are known, but our understanding of what each factor contributes, how they interact with the virus and with one another, and how these interactions are orchestrated is limited. Variation within and between host and nonhost species is thought to account for both HCV's narrow host range and the variations in the clinical course of hepatitis C. The study of both of these is more than an academic exercise: variation between species, such as the entry factors CD81 and OCLN, is thought to be key to overcome the species barrier and create better small-animal models than those currently available. Understanding the impact of variation within the human species is key to predicting treatment response and may in the future be used to individualize treatment strategies.
Over recent years, robust in vitro systems for the study of HCV biology, most notably that of HCVcc based on the Japanese fulminant hepatitis 1 (JFH1) strain, have been created. Nonetheless, the creation of an immunocompetent, breedable, and technically simple small-animal model remains a major goal of HCV research since currently available systems, immunodeficient mice with transplanted human hepatocytes, do not fully meet these criteria (25). It is thought that a number of genetic alterations in host factors involved in all steps of the HCV replication cycle will be required to enable efficient HCV infection in a mouse. At the stage of cell entry, it is thought that provision of human CD81 and OCLN either in addition to or in place of the murine homologues will enable efficient cell entry (29, 31). At the same time, replacing a murine component with a homologue from a different species may perturb its interactions with cellular partner molecules and alter its physiological functions. Hence, it is important to know in quantitative terms whether humanizing a given factor will likely be essential or merely advantageous to the HCV cell entry process. Our data indicate that murine OCLN is 5- to 8-fold less efficient than human OCLN but does have significant residual (co)receptor function. This finding explains the recent observation that an HCV variant selected to utilize murine CD81 with efficiency equal to that of human CD81 was able to enter cells expressing only murine entry factors (4). In line with this, primary hepatocytes of the small mammal Tupaia belangeri were recently reported to be susceptible to HCVcc entry (39). Moreover, the alanine-to-glycine exchanges in positions 223 and 224 account for this difference in coreceptor efficiency, as was predicted by HCVpp assays (29), indicating that despite differences in particle architecture, HCVpp remain a model strongly predictive of the behavior of HCVcc.
The recent breakthrough discovery that two SNPs in the IL-28B gene are strongly predictive of whether individuals will clear infection either spontaneously during the acute phase of hepatitis C or in response to interferon-based treatment during the chronic phase (17, 36, 37), has substantiated the notion that genetic variation in host genes accounts for the variable clinical course of hepatitis C in humans. Genetic variation in viral (co)receptors has a major impact on infection in the case of HIV, in which the CCR5 Δ32 allele is known to be strongly protective (2). No such allele has been described for HCV infection. However, a pedigree from northern Africa that includes individuals homozygous for a null mutation in the CLDN1 coreceptor has been described (19). The HCV statuses of these individuals are unknown, but one may speculate that this very rare genetic variant may confer resistance to HCV infection. The OCLN coreceptor is characterized by high variability with a high number of genetic variants (http://www.ncbi.nlm.nih.gov/snp) as well as splice variants (23). We have assessed the frequencies of the three known coding nonsynonymous SNPs and found that they do not significantly affect the (co)receptor function of OCLN in vitro. Although the in vivo situation may be more complex, we believe that this makes a major clinical impact of these variants unlikely.
We have used the OCLNlow Huh-7.5 subclone to test the involvement of OCLN in cell-to-cell spread. Cell-to-cell spread is the dissemination of virus between adjacent cells without the appearance of a cell-free virion in the extracellular space. In the case of HCV that has the tightly packed liver parenchymal cells as a reservoir and utilizes two tight junction components, CLDN1 and OCLN, as (co)receptors, the direct cell-to-cell route may be advantageous, as it may allow spread without exposure to the broadly neutralizing antibodies that are generated during chronic infection. Whether the mechanisms of cell entry during cell-to-cell spread are distinct from those during cell entry by an extracellular virion is unclear. Most notably, there is conflicting data on whether cell-to-cell spread is CD81 independent (22, 38, 41) or not (7). OCLN knockdown has been reported to reduce cell-to-cell transmission by 2- to 3-fold in an assay in which spread, despite the presence of neutralizing antibodies in the supernatant, was defined as occurring cell to cell (7). We used an agarose overlay to prevent cell-free spread and a HCV-NS3/4A-sensitive fluorescent reporter to monitor spread of infection in living cells and found that the absence of OCLN in target cells abolished cell-to-cell spread, indicating that this route is strictly OCLN dependent. Interestingly, cell-to-cell spread mediated by murine OCLN was somewhat less efficient than that mediated by human OCLN. This difference was not statistically significant, but we cannot rule out that statistical significance would become apparent in a larger-scale assay.
Finally, while many HCV entry factors are known, much remains to be learned regarding their exact functions and interplay during the entry process. To this end, cell lines deficient for individual key factors (8) or expressing functionally relevant variants may be very useful, especially when combined with innovative approaches that make the HCV cell entry process directly observable (10). A better understanding of the entry process may inform both the development of antiviral approaches and the development of better small-animal models.
ACKNOWLEDGMENTS
We thank Charles Rice for providing pseudovirus production vectors, lentiviral OCLN expression vectors, the tagRFP-NLS-IPS reporter, the pLenti-3′-U6-EC-EP7 shRNA vector, and cell lines (Huh-7.5 and 786-O); Jens Bukh for providing the chimeric H77/JFH1 genome; and Eike Steinmann for helpful discussions.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) Emmy Noether program (grant HA 4393/2-1) and by the Hochschulinterne Leistungsförderung (HiLF) program of Medizinische Hochschule Hannover.
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Altshuler D. M., et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arenzana-Seisdedos F., Parmentier M. 2006. Genetics of resistance to HIV infection: role of co-receptors and co-receptor ligands. Semin. Immunol. 18:387–403 [DOI] [PubMed] [Google Scholar]
- 3. Baldick C. J., et al. 2010. A novel small molecule inhibitor of hepatitis C virus entry. PLoS Pathog. 6:e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bitzegeio J., et al. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 6:e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonnafous P., et al. 2010. Characterization of hepatitis C virus pseudoparticles by cryo-transmission electron microscopy using functionalized magnetic nanobeads. J. Gen. Virol. 91:1919–1930 [DOI] [PubMed] [Google Scholar]
- 6. Brazzoli M., et al. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brimacombe C. L., et al. 2011. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J. Virol. 85:596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Catanese M. T., et al. 2010. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J. Virol. 84:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciesek S., et al. 2010. Glucocorticosteroids increase cell entry by hepatitis C virus. Gastroenterology 138:1875–1884 [DOI] [PubMed] [Google Scholar]
- 10. Coller K. E., et al. 2009. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5:e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cormier E. G., et al. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 101:7270–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Demaison C., et al. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of immunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803–813 [DOI] [PubMed] [Google Scholar]
- 13. Evans M. J., et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 14. Farquhar M. J., et al. 2008. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 82:8797–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flint M., Logvinoff C., Rice C. M., McKeating J. A. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flint M., et al. 2006. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 80:11331–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ge D., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- 18. Gottwein J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 19. Hadj-Rabia S., et al. 2004. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology 127:1386–1390 [DOI] [PubMed] [Google Scholar]
- 20. Haid S., Pietschmann T., Pecheur E. I. 2009. Low pH-dependent hepatitis C virus membrane fusion depends on E2 integrity, target lipid composition, and density of virus particles. J. Biol. Chem. 284:17657–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris H. J., et al. 2010. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285:21092–21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones C. T., et al. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28:167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohaar I., et al. 2010. Splicing diversity of the human OCLN gene and its biological significance for hepatitis C virus entry. J. Virol. 84:6987–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koutsoudakis G., et al. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Legrand N., et al. 2009. Humanized mice for modeling human infectious disease: challenges, progress, and outlook. Cell Host Microbe 6:5–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindenbach B. D., Thiel H., Rice C. M. 2006. Flaviviridae: the viruses and their replication, p. 991–1041 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 27. Liu S., et al. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mee C. J., et al. 2010. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology 138:1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michta M. L., et al. 2010. Species-specific regions of occludin required by hepatitis C virus for cell entry. J. Virol. 84:11696–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pileri P., et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 31. Ploss A., et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scarselli E., et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seeff L. B. 2009. The history of the “natural history” of hepatitis C (1968–2009). Liver Int. 29(Suppl. 1):89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shepard C. W., Finelli L., Alter M. J. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 35. Sirven A., et al. 2001. Enhanced transgene expression in cord blood CD34(+)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol. Ther. 3:438–448 [DOI] [PubMed] [Google Scholar]
- 36. Suppiah V., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 [DOI] [PubMed] [Google Scholar]
- 37. Thomas D. L., et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Timpe J. M., et al. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17–24 [DOI] [PubMed] [Google Scholar]
- 39. Tong Y., et al. 2011. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J. Virol. 85:2793–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tscherne D. M., et al. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Witteveldt J., et al. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeisel M. B., et al. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731 [DOI] [PubMed] [Google Scholar]
- 43. Zeuzem S., et al. 2009. Expert opinion on the treatment of patients with chronic hepatitis C. J. Viral Hepat. 16:75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]