Abstract
Baboon reovirus (BRV) is a member of the fusogenic subgroup of orthoreoviruses. Unlike most other members of its genus, BRV lacks S-segment coding sequences for the outer fiber protein that binds to cell surface receptors. It shares this lack with aquareoviruses, which constitute a related genus and are also fusogenic. We used electron cryomicroscopy and three-dimensional image reconstruction to determine the BRV virion structure at 9.0-Å resolution. The results show that BRV lacks a protruding fiber at its icosahedral 5-fold axes or elsewhere. The results also show that BRV is like nonfusogenic mammalian and fusogenic avian orthoreoviruses in having 150 copies of the core clamp protein, not 120 as in aquareoviruses. On the other hand, there are no hub-and-spoke complexes attributable to the outer shell protein in the P2 and P3 solvent channels of BRV, which makes BRV like fusogenic avian orthoreoviruses and aquareoviruses but unlike nonfusogenic mammalian orthoreoviruses. The outermost “flap” domains of the BRV core turret protein appear capable of conformational variability within the virion, a trait previously unseen among other ortho- and aquareoviruses. New cDNA sequence determinations for the BRV L1 and M2 genome segments, encoding the core turret and outer shell proteins, were helpful for interpreting the structural features of those proteins. Based on these findings, we conclude that the evolution of ortho- and aquareoviruses has included a series of discrete gains or losses of particular components, several of which cross taxonomic boundaries. Gain or loss of adhesion fibers is one of several common themes in double-stranded RNA virus evolution.
INTRODUCTION
Baboon reovirus (BRV) is the prototype strain of species Baboon orthoreovirus, one of five species formally recognized to date in the genus Orthoreovirus (subfamily Spinareovirinae, family Reoviridae) of double-stranded RNA (dsRNA) viruses with 10-segment genomes and two-layer, icosahedral capsids. Viruses in the other four species—Mammalian orthoreovirus, Avian orthoreovirus, Nelson Bay orthoreovirus, and Reptilian orthoreovirus—infect mammals, birds, or reptiles (16, 17). Broome virus (BroV), a recently described Australian isolate from an encephalitic fruit bat, is divergent from Nelson Bay virus (NBV) and related strains of bat-borne orthoreoviruses and represents a new, though not yet formally recognized, orthoreovirus species (50). Related 11-segment dsRNA viruses isolated from fish define a distinct phylogenetic cluster and are grouped in the separate genus Aquareovirus (1, 19). Interestingly, a 10-segment fish reovirus (piscine reovirus, or PRV) was recently described and proposed to represent the prototype strain of another new orthoreovirus, not aquareovirus, species (38). The diversity of viruses from these two genera suggests that additional comparative studies are likely to provide important new insights into not only their evolution but also their structural and functional properties.
BRV was first described in 1995 (18), having been isolated in 1993 to 1994 from a single colony of Papio cynocephalus baboons at the Southwest Foundation for Biomedical Research in San Antonio, TX (24). It was recovered from the brains of five juvenile animals that showed clinical signs of progressive meningoencephalomyelitis as well as histopathological evidence for that disease. A causative role for BRV was established by intracranial inoculations of two young baboons, both of which developed the disease, although the source of the virus and the mode of transmission among the baboons remain unknown. The severity of disease in these primates nevertheless raises concern that BRV may represent a group of viruses circulating in nature that has the potential to emerge as a serious human pathogen. In fact, partial sequence evidence for another virus closely related to BRV has been reported, based on samples derived from skunk brain tissue collected in 1974 by the California Department of Public Health (52).
In cultured cells, BRV is fusogenic, inducing formation of multinucleated syncytia by cell-cell fusion (18). This unusual activity for a nonenveloped virus is mediated by a virally encoded nonstructural protein, called the FAST (fusion-associated small transmembrane) protein (46), which is expressed on the surface of infected cells but is not present in mature virions (12, 13). Possession of a FAST protein and associated fusogenicity are shared by avian reovirus (ARV) and reptilian reovirus (RRV) strains (16, 17), but prototypical mammalian reovirus (MRV) strains are nonfusogenic. The only fusogenic mammalian orthoreoviruses in addition to BRV are the bat-borne NBV-like viruses (11, 21) and the divergent bat-borne isolate BroV (50). Notably, several of the NBV-like viruses have been recently associated with human cases of high fever and acute respiratory illness in eastern Asia (21).
Full-length cDNA sequences have been reported to date for the four small (S) genome segments of BRV, and their encoded, σ-class proteins show only low levels of sequence identity with homologous proteins of other representative orthoreoviruses (<35%), consistent with the evolutionary divergence of BRV and its assignment to a separate species (13, 16, 17). One distinctive aspect of the BRV σ proteins is that they do not include a homolog of the outer fiber protein found in most other orthoreoviruses (e.g., ARV σC or MRV σ1) (12, 13, 16) (Table 1). Each of the orthoreovirus fiber proteins, which bind to cell surface receptors (3, 39), is distinguished by heptad repeats of hydrophobic residues in the N-terminal half of the deduced amino acid sequence, consistent with formation of a long, α-helical coiled coil (4, 17, 47). In most other orthoreoviruses, the outer fiber protein is encoded on the same bi- or tricistronic S genome segment as the FAST protein and/or a poorly conserved nonstructural protein of unclear function (4, 17, 47). The S4 genome segment that encodes the FAST protein of BRV is also bicistronic, but the second encoded protein, p16, shows no sequence similarity to the outer fiber protein. Moreover, p16 is a nonstructural protein (12) and is therefore not a functional analog of the fiber protein. Which protein in BRV virions then plays the primary role in adhesion to cell surface receptors remains undefined.
Table 1.
Virion proteins of orthoreoviruses BRV, MRV, and ARV and aquareovirus GCRV
| Protein namea | No. of copies per virionb | Protein homolog |
Protein size (aa) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| BRV | MRV | ARV | GCRV | BRVc | MRVd | ARVe | GCRVf | ||
| Core RdRp | ∼12 | λB | λ3 | λB | VP2 | 1,261 | 1,267 | 1,259 | 1,274 |
| Core NTPase | ∼24 | μA | μ2 | μA | VP5 | 738 | 736 | 732 | 728 |
| Core shell | 120 | λC | λ1 | λA | VP3 | 1,231 | 1,275 | 1,293 | 1,214 |
| Core clamp | 150/120 | σA | σ2 | σA | VP6 | 413 | 418 | 416 | 412 |
| Core turret | 60 | λA | λ2 | λC | VP1 | 1,284 | 1,289 | 1,285 | 1,299 |
| Outer shell | 600 | μB | μ1 | μB | VP4 | 676 | 708 | 676 | 648 |
| Outer clamp | 600 | σB | σ3 | σB | VP7 | 396 | 365 | 367 | 276 |
| Outer fiber | ∼36/0 | σ1 | σC | 455 | 326 | ||||
The names are according to shared location-function in each virus. Proteins are listed in approximate inside to outside position in each virus. Rows for the two main shell proteins (core and outer) are bolded.
Values are for MRV virions, which are probably also correct for ARV and BRV except that BRV lacks an outer fiber protein. The values with approximate signs (∼) are estimates from best available data for MRV. These values are probably also correct for aquareovirus virions, except that aquareoviruses contain only 120 core clamp subunits and also lack an outer fiber protein.
GenBank accession numbers are (top to bottom) HQ847903, HQ847904, HQ847905, HQ847906, HQ847907, AAC18124, and AAC18128.
Values are for MRV-T3D. GenBank accession numbers are (top to bottom) AF129822, J03488, M31058, AF461684, M20161, X01161, L19776, and K02739, in respective order.
Values are for ARV-176. GenBank accession numbers are (top to bottom) ACH72474, ACH72476, ACH72478, AAT52025, AAW78486, AAC18121, AAC18125, and AF218358.
GenBank accession numbers are (top to bottom) AF260511S3, AF260511S2, AF260511S1, AF403391, AF403392, AF403394, and AAM92742.
Even though their genomes comprise 11 dsRNA segments, members of the genus Aquareovirus exhibit clear homologies to orthoreoviruses in all but a few of their proteins, both structural and nonstructural (1, 19, 23) (Table 1). Also, like many orthoreoviruses as noted above, aquareoviruses are fusogenic in cultured cells (43). The virion structures of striped bass reovirus (SBRV) (from species Aquareovirus A) and grass carp reovirus (GCRV) (from species Aquareovirus C) have been determined by transmission electron cryomicroscopy (cryo-TEM) and three-dimensional (3D) image reconstruction (10, 32) and show substantial similarities to those of ARV and MRV virions (15, 58). One notable exception is the absence of an adhesion fiber in the aquareovirus cryo-reconstructions (10, 32). In ARV and MRV virions, the base of the adhesion fiber is anchored to the flattened top of the pentameric core turret protein (ARV λC and MRV λ2) (15, 58). In GCRV and SBRV virions, in contrast, the tops of the core turrets (formed by VP1) adopt a more open arrangement (10, 32), presumably reflecting that the aquareovirus turrets need not be closed since there is no adhesion fiber to be anchored at each icosahedral 5-fold (5f) axis.
Given the several notable characteristics of BRV, as well as its status as the prototype strain of a distinct, fusogenic mammalian orthoreovirus species, we sought to determine its virion structure. In particular, based on the structures of other ortho- and aquareoviruses, we hypothesized that the virions of BRV might differ from those of other orthoreoviruses by not only lacking an adhesion fiber but also having more open core turrets, like those in aquareovirus virions. To address these hypotheses, we determined the structure of BRV virions by cryo-TEM and 3D image reconstruction to an estimated resolution of 9.0 Å. Sequences of the BRV L1 and M2 genome segments, respectively, encoding the core turret protein (BRV λA) and the outer shell protein (BRV μΒ), were also newly determined to aid in interpreting the structure. Results indicate that the evolution of ortho- and aquareoviruses has included a series of discrete gains or losses of particular components, several of which cross taxonomic boundaries.
MATERIALS AND METHODS
Virion and dsRNA purifications.
Vero cells were cultured in roller bottles at 37°C in medium 199 with Hank's salts supplemented with 5% fetal bovine serum (FBS). Subconfluent monolayers (2 × 108 cells per bottle; 10 bottles) were infected with BRV at a multiplicity of infection of 3 to 5 PFU per cell, and infected cultures were incubated at 37°C for 21 h. At that time, syncytium formation was extensive, but monolayers remained intact. Culture medium was removed, and cells were disrupted in TMN buffer (25 mM Tris, pH 7.5, 5 mM MgCl2, 150 mM NaCl) supplemented with 50 μg/ml RNase A, 1% Triton X-100, and protease inhibitors (200 nM aprotinin, 1 μM leupeptin, and 1 μM pepstatin). The inclusion of RNase and Triton was required to remove the nonstructural protein σNS, which tends to associate with BRV virions. Cell debris was removed from the pooled cell lysates (50 ml) by centrifugation at 10,000 × g for 30 min at 4°C, and virus particles were concentrated from the supernatant by centrifugation at 100,000 × g for 60 min at 4°C through a 20% sucrose cushion. The virus pellet was resuspended in TMN buffer, and virus particles were purified on linear CsCl density gradients as previously described (58). Isolated virus particles were dialyzed against TMN buffer and stored at 4°C until analysis. For obtaining genomic dsRNA, purified BRV virions were disrupted by incubation for 30 min at 37°C in the presence of 1% SDS, and the dsRNA was then isolated by phenol-chloroform extraction and ethanol precipitation. Resuspended dsRNA was subjected to centrifugation through a CHROMA SPIN 1000 column (Clontech) to remove small oligonucleotides as a final step in purification.
Cryo-TEM.
Small (3.5-μl) aliquots of purified BRV particles (∼5 × 1012 particles/ml) were vitrified and examined using standard procedures (2). Briefly, this involved applying samples to holey carbon grids that were glow discharged for ∼15 s in an Emitech K350 evaporation unit. Grids were blotted with Whatman filter paper for ∼5 s, plunged into liquid ethane, and transferred into a precooled, FEI Polara multispecimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron-image film at both 300 and 200 keV in an FEI Polara microscope under minimal-dose conditions (ranging from ∼25 to 35 electrons/Å2) at a nominal magnification of 39,000 (Table 2).
Table 2.
Parameters for cryoTEM and 3D image reconstruction of BRV virions
| Samplea | No. of micrographs by instrument typeb |
Pixel size (Å) | Defocus range (μm) | No. of boxed particle images/no. of boxed particle images used for 3D reconstruction | Resolution (Å)c | |||
|---|---|---|---|---|---|---|---|---|
| Polara |
Zeiss | Nikon | ||||||
| 200 keV | 300 keV | |||||||
| BRV | 0 | 91 | 48 | 19 | 1.80d | 0.25–5.09 | 2669/2138 | 9.0 |
| BRV−flap | 30 | 146 | 0 | 176 | 1.63 | 0.41–5.38 | 3778/2993 | 9.2 |
BRV, baboon reovirus with turret flaps; BRV−flap, baboon reovirus without turret flaps.
No. of micrographs obtained or scanned with each indicated instrument. Zeiss, Zeiss SCAI microdensitometer (7.0-μm step size); Nikon, Nikon Super CoolScan 8000 scanner (6.35-μm step size).
Resolution of final 3D reconstruction as estimated by FSC0.5 criterion (51).
Nikon-scanned data were interpolated to 1.80-Å pixels so that they could be merged with Zeiss-scanned data.
3D image reconstruction.
Micrographs were digitized either on a Nikon Super CoolScan 8000 scanner at 6.35-μm intervals (representing 1.63-Å pixels at the specimen) or on a Zeiss SCAI microdensitometer at 7.0-μm intervals (1.80-Å pixels) (Table 2). Only micrographs that exhibited negligible astigmatism and specimen drift or charging were selected for further processing. Estimation of the defocus level of each micrograph was performed using ctffind3 (30). RobEM (http://cryoem.ucsd.edu/programs/robem_usersguide_v4.01.htm) was used to extract individual BRV virion images from the digitized micrographs (Table 2). We used the random model computation method to create an initial, low-resolution model of BRV (53) and then used the AUTO3DEM automated 3D reconstruction package (version 4.01.8) to determine and refine the origin and orientation parameters for each particle (54). The estimated resolution limits of all reconstructed density maps were computed based on a conservative Fourier shell correlation threshold value of 0.5 (FSC0.5) (51). Final density maps used for visualization, analysis, and interpretation were sharpened to enhance high-resolution features by imposing an inverse temperature factor (1/400 Å2) (22).
During the course of the image-processing procedures, we sometimes obtained cryo-reconstructions of the BRV virion that were missing the core turret flap densities. Upon investigating this phenomenon, we determined that it varied from grid to grid. The presence or absence of flaps in the resulting reconstruction was found to be independent of the virion preparation (three independent preparations were tested at different times, and each produced a mixture of reconstructions with or without flaps in a grid-specific manner) and also independent of the three types of support film on which the particles were frozen (home-made continuous, home-made holey, or Quantifoil carbon grids). In addition, the presence or absence of flaps in the resulting reconstruction seemed to reflect an all-or-none phenomenon, whereby all particles processed from any particular grid either contributed flap densities or did not. The images were therefore sorted into two groups to give rise to the cryo-reconstructions of the BRV virion with or without core turret flaps as described below.
First-strand cDNA synthesis, PCR amplification, and sequencing.
To synthesize viral cDNA, purified dsRNA was first denatured in 30 mM methylmercury hydroxide for 10 min at room temperature, followed by addition of a poly(A) tail to both 3′ ends of each dsRNA molecule using a Saccharomyces cerevisiae poly(A) polymerase (Epicentre Technologies). First-strand cDNA synthesis was next conducted using SuperScript III reverse transcriptase (Invitrogen) and an oligo(dT)20 primer with the 5′ 20-nucleotide (nt) extension GCGATAGTCTCCGTTCAGGA, followed by digestion of cRNA with RNase H (New England BioLabs). PCR was then performed at low annealing temperatures (50°C) with iProof DNA polymerase (Bio-Rad) for 35 cycles of amplification, using a primer to the anchor sequence along with one of 28 internal primers based on reported skunk orthoreovirus partial sequences (GenBank accession numbers EU789369, EU789370, and EU789372) (52). Each amplified sample was then sequenced directly using the same internal primer that was used for amplification, and sequences from the targeted BRV genome segments (L1 and M2) were identified by BLAST homology (http://blast.ncbi.nlm.nih.gov/) with previously reported sequences of other orthoreoviruses. These internal sequence toe holds were then used to design new primers for PCR amplifying the remaining portions of BRV L1 and M2, followed again by direct sequencing. In this manner, every nucleotide of the BRV L1 and M2 genome segments was read in both forward and reverse directions, except for about 50 nt at either end of each segment, which were read with high confidence in a single (outward) direction. All primers were obtained from Integrated DNA Technologies.
Sequence alignments.
Pairwise sequence alignments were performed using EMBOSS Stretcher, with default settings except for use of the Blosum 30 matrix, as implemented at http://www.ebi.ac.uk/Tools/psa/emboss_stretcher/. Multiple sequence alignments were performed using the Clustal W2 algorithm as implemented at http://guidance.tau.ac.il/in combination with the GUIDANCE algorithm to calculate guide tree-based alignment confidence scores (40). Confidence scores from 100 bootstraps for the core turret and outer shell protein alignments were 97.4% and 97.2%, respectively, compared with only 45.5% from a parallel analysis of the outer clamp protein. The program PROMALS3D as implemented at http://prodata.swmed.edu/promals3d/promals3d.php and the program MUSCLE, version 3.7, and T-Coffee, version 6.85, as implemented at http://phylogeny.fr/ were used as alternatives for multiple sequence alignments to confirm results. GenBank accession numbers for published sequences used in these analyses were AF378003 (MRV type 1 Lang [T1L]), AF378005 (MRV type 2 Jones [T2J]), EU707937 (ARV-138), EU707938 (ARV-176), GQ258978 (BroV), GU994014 (PRV), AF260511S1 (GCRV), and EF589098 (American grass carp reovirus [AGCRV]) for alignment with BRV core turret protein λA and AF490617 (MRV-T1L), M19355 (MRV-T2J), AY50052 (ARV-138), AY50053 (ARV-176), GQ258981 (BroV), GU994016 (PRV), AF403392 (GCRV), and EF589103 (AGCRV) for alignment with BRV outer shell protein μB.
Fitting MRV crystal structures.
In preparation for model-fitting experiments, the BRV 3D density maps were scaled to the same size as the MRV virion structure from Zhang et al. (57) by using the program UCSF Chimera (http://www.cgl.ucsf.edu/chimera). The entire asymmetric unit containing one or more copies of all five major structural proteins of MRV (two core shell λ1 subunits, two and a half core clamp σ2 subunits, one core turret λ2 subunit, 10 outer shell μ1 subunits, and 10 outer clamp σ3 subunits) was then fitted into each BRV map without any manual adjustments. The resulting, overall fit among all of these structural components was extremely good, as described below. Images of the MRV λ2 and μ1 subunits from this fit were then extracted for depictions in figures (see Fig. 6 and 7).
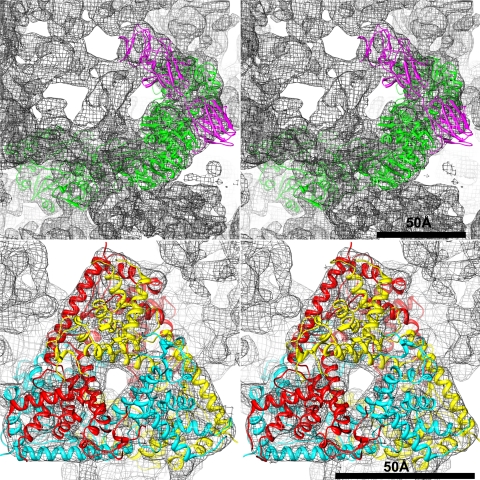
Fig. 6.
Atomic model of the MRV asymmetric unit fitted into the BRV cryo-reconstruction. Excerpted portions are shown in stereo to illustrate goodness of fit for the core turret protein (top) and the outer shell protein (bottom). For the core turret protein, the MRV λ2 model is shown in magenta for the three flap domains and in green for the other domains of one subunit. For the outer shell protein, the MRV μ1 model is shown in a different color (red, yellow, or cyan) for the three subunits within one trimer.
Fig. 7.
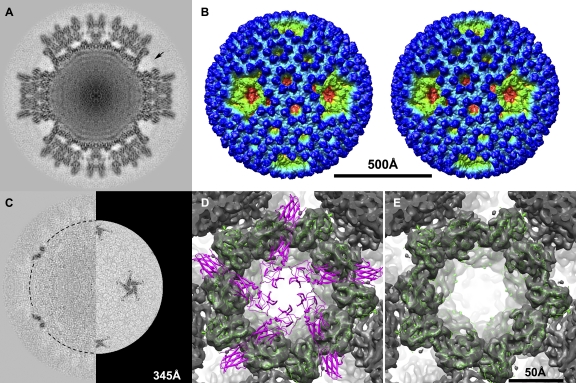
Cryo-reconstruction at 9.2-Å resolution of flapless BRV virions. (A) Density-coded, equatorial 2f-axis section, comparable to the sections shown at top of Fig. 2. The region of the missing flap domain densities of one core turret is indicated by an arrow. (B) Stereo surface representation viewed down an icosahedral 2f axis, comparable to the stereo view of the flap-containing BRV virion shown at the bottom of Fig. 2 with the same radial color map employed. The scale bar applies to panels A, B, and C. (C) Difference map obtained by subtracting the cryo-reconstruction of the BRV virion lacking core turret flaps from that of the BRV virion containing core turret flaps. The left side shows a density-coded, equatorial 2f-axis section of the difference map. The right side shows a radial section at 345 Å through the level of the flap domains in the difference map, as indicated by the dashed half circle on the left side. (D and E) Close-up view down the icosahedral 5f axis of a single core turret from the cryo-reconstruction of flapless BRV virions. In panel D, a pentamer of the atomic model of the full-length MRV core turret protein λ2 is fitted. The flap domains are shown in magenta, and the other domains are shown in green. In panel E, a pentamer of the atomic model of the MRV core turret protein λ2 minus its flap domains is fitted, with the remaining domains shown in green. The scale bar applies to panels D and E.
Phylogenetic analysis.
Phylogenetic analysis (see Fig. 8) was performed using programs implemented at http://phylogeny.fr/, with default settings unless otherwise indicated. Multiple-sequence alignment was performed with the program MUSCLE, version 3.7, without subsequent curation. Maximum-likelihood analysis was then performed with the program PhyML, version 3.0, using the chi-square parametric method of the approximate-likelihood ratio test to generate confidence scores. The tree was drawn with the program TreeDyn, version 198.3, and refined for publication with the program FigTree, version 1.3.1. GenBank accession numbers for published core clamp protein sequences used in this analysis were AAA47239 (MRV-T1L), AAA47248 (MRV-T2J), AAA47261 (MRV type 3 Dearing [MRV-T3D]), AAC18124 (BRV), AAY54282 (BroV), AAC18123 (NBV), AAR13234 (Pulau reovirus), ABM67658 (Melaka reovirus), AAC18122 (ARV-138), AAC18121 (ARV-176), ABK96865 (ARV Muscovy duck S12), ABY57290 (psittacine reovirus), AAM93413 (SBRV), AAM92740 (GCRV), and ABV01047 (AGCRV).
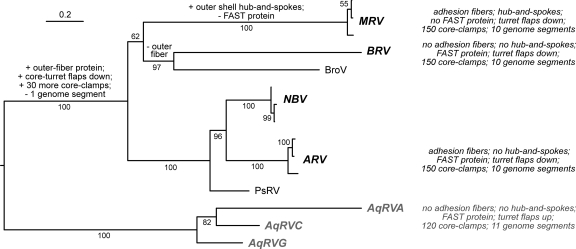
Fig. 8.
Phylogenetic tree and evolution schematic. A maximum-likelihood phylogenetic tree is shown for the core clamp proteins of the indicated ortho- and aquareovirus species or tentative species. Species names are abbreviated as follows: MRV, Mammalian orthoreovirus; BRV, Baboon orthoreovirus; NBV, Nelson Bay orthoreovirus; ARV, Avian orthoreovirus; AqRVA, Aquareovirus A; AqRVC, Aquareovirus C; and AqRVG, Aquareovirus G. For MRV, NBV, and ARV, three representative isolates were used in generating the tree. Tentative new species are represented by Broome virus (BroV) and psittacine reovirus (PsRV) isolates. Branch labels are confidence scores (%). The tree is rooted on the aquareovirus clade. The scale bar indicates horizontal branch lengths in units of substitutions per amino acid position in the multiple-sequence alignment. Highlighted properties of each species for which one or more representative virion structures has been determined (MRV, BRV, ARV, and AqRVA and C) are shown at right. Suggested evolutionary events to explain differences in the highlighted features are labeled on appropriate branches.
Nucleotide sequence accession numbers.
GenBank accession numbers for newly determined BRV sequences reported here are HQ847903 for L1/λA and HQ847907 for M2/μB. GenBank accession numbers for additional new BRV sequences to be reported separately are HQ847904 for L2/λB, HQ847905 for L3/λC, HQ847906 for M1/μA, and HQ847908 for M3/μNS.
RESULTS
Electron cryomicrographs of purified BRV virions.
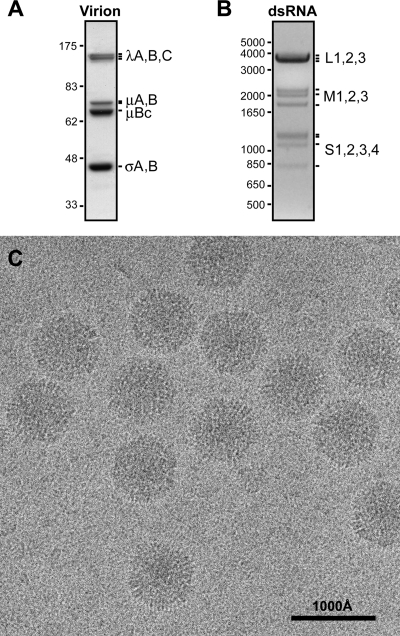
BRV virions were purified from infected Vero cells by differential and density gradient centrifugation. Denaturing gel electrophoresis of the purified virions showed the expected pattern of λ, μ, and σ structural proteins (Fig. 1A) as well as the expected pattern of L, M, and S dsRNA genome segments (Fig. 1B) (16). Transmission electron cryomicrographs were recorded from unstained, vitrified samples of the purified virions and showed fields of regularly sized, intact particles with diameters near 850 Å (Fig. 1C). As evident in the figure, the projected profiles of individual particles appear largely round though some are more angular. There are no evident structures projecting far out from the particle surfaces. Striated surface features viewed in profile in some areas are reminiscent of regularly spaced subunits. Overall, the images are very similar to those of other orthoreoviruses (15, 58).
Fig. 1.
Purified BRV virions. (A) Protein gel lane of purified virions. Positions of the structural proteins are labeled at right. Size marker positions (in kDa) are indicated at left. (B) dsRNA gel lane of purified virions. Positions of the 10 genome segments are labeled at right. Size marker positions (in bp) are indicated at left. (C) Transmission electron cryomicrograph of unstained, vitrified BRV virions, with scale bar as indicated.
Cryo-reconstruction of BRV virions.
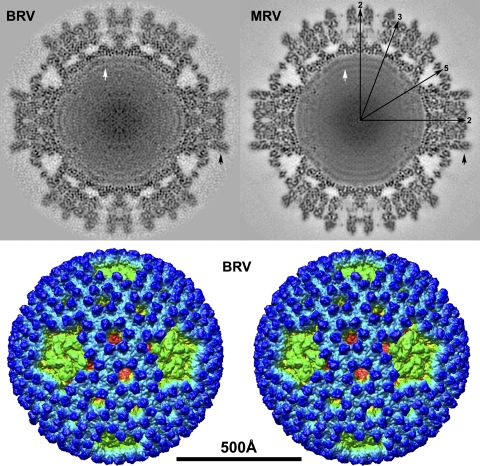
Using current processing procedures (49), we obtained a cryo-reconstruction at 9.0-Å resolution from 2,138 particle images of the BRV virions. A density-coded equatorial section of the cryo-reconstruction highlights the approximately round profile of the virion's outer surface, with flattened areas, or “divots,” around the icosahedral 5f axes (Fig. 2, top left). These divots are explained by absence of the short projections, each ∼40 Å high, that regularly stud the remainder of the surface. The outermost radius approximates 425 Å to the tops of these projections near the icosahedral 2-fold (2f) and 3-fold (3f) axes and 355 Å near the icosahedral 5f axes. A radially color-coded surface view of the BRV virion in stereo (Fig. 2, bottom) clarifies these surface features and is discussed in detail below. The innermost radius of the BRV capsid layers approximates 250 Å near the icosahedral 2f and 3f axes and is less near the 5f axes though its limit is hard to discern at those positions (Fig. 2, top left). Outer and inner capsids are separated by large solvent cavities at many positions in the equatorial section. Around the icosahedral 2f and 3f axes, these cavities appear smaller because “nodules” reach up by about 25 Å at regular intervals from the top of the inner capsid to contact the bottom of the outer capsid. Around each 5f axis, the solvent cavity is larger and heart shaped in profile. Punctate and linear features visible within most capsid regions appear likely to represent secondary-structure elements including α-helices and β-sheets. Two or three regularly spaced rings of density, with an average center-to-center distance of about 30 Å and attributable to genomic dsRNA, are evident in the particle interior (Fig. 2, top left). The RNA rings are interrupted at the icosahedral 5f axes by what appear to be limited regions of protein density that project inward from the bottom of the inner capsid, as also seen in MRV and attributable to the viral transcriptase complex (14, 59). Overall similarities of these BRV features to those of the MRV virion (Fig. 2, top right) are obvious.
Fig. 2.
Cryo-reconstruction of the BRV virion at 9.0-Å resolution. (Top) Density-coded, equatorial 2f-axis sections are shown for BRV and MRV. Highest and lowest densities are depicted in black and white, respectively. The MRV map is that from Zhang et al. (57) recalculated at 9.0 Å. Icosahedral symmetry axes (2f, 3f, and 5f) are labeled for one quadrant of MRV. A difference in the relative erectness of the outer clamp subunits of BRV and MRV is highlighted by black arrows. RNA ring densities in the two viruses are highlighted by white arrows. (Bottom) Stereo surface representation of the BRV cryo-reconstruction viewed along an icosahedral 2f axis with radial color cueing applied to highlight features at different radii (from blue outside to red inside). The scale bar applies to the entire figure.
BRV L1 and M2 sequences.
To assist in further interpreting features of the BRV virion, we determined the full-length cDNA sequences of the genome segments encoding the core turret and outer shell proteins. The full-length cDNA sequence has been previously reported for the outer clamp protein σB (16) but not for these other two proteins that are expected to cover the remaining bulk of the BRV surface. Based on the lengths of the cDNA sequences recently determined for the other four L and M genome segments of BRV (R. P. Goodman., J. Shou., R. Duncan, and M. L. Nibert, unpublished data), the segments encoding the core turret and outer shell proteins were found to be L1 and M2, respectively, and the encoded BRV proteins were therefore designated λA and μB (Table 1). Both are discussed below.
(i) L1/λA.
The L1 genome segment is 3,903 bp long, and its plus strand contains one long open reading frame (ORF) spanning nucleotide positions 18 to 3872. The encoded λA protein comprises 1,284 amino acids (aa), with a calculated molecular mass of 147 kDa. In pairwise sequence alignments, it exhibits homology to the core turret protein of representative MRV and ARV strains (λ2 and λC; 51 to 53% similarity and 26 to 29% identity) as well as to the core turret protein of aquareoviruses (VP1; 49 to 50% similarity and 23 to 25% identity). This protein is also known to mediate three of the four enzymatic reactions during cotranscriptional 5′ capping of orthoreovirus plus-strand RNAs (26, 28, 44). In multiple sequence alignments, the BRV λA protein aligns well—end to end and with only occasional, small internal gaps—with the homologous protein of other ortho- and aquareoviruses. The core turret protein of ortho- and aquareoviruses is known to comprise multiple domains (10, 44), and the end-to-end alignments indicate that the homologous BRV protein contains all of these domains, including the C-terminal flap domains (see below) that are missing from the core turret proteins of certain, more divergent members of the subfamily Spinareovirinae (9, 31). Specific residues of apparent functional importance for the substrate binding or enzymatic activities are conserved in the RNA 5′ guanylyltransferase domain (e.g., Lys171, Lys190, His223, and His232 in MRV-T3D; Lys172, Lys191, His221, and His230 in BRV), in the RNA methyltransferase-1 domain (e.g., Ser518, His521, Tyr552, Asp553, Asp561, Asp577, and Val578 in MRV-T3D; Ser515, His518, Tyr548, Asp549, Asp557, Asp573, Val574, and 570-VYCDVDQV-577 in BRV), and in the RNA methyltransferase-2 domain (e.g., Asn812, Asp827, Gly829, Asp850, Asp871, Tyr872, and Ser891 in MRV-T3D; Asn807, Asp822, Gly824, Asp845, Asp866, Tyr867, and Ser886 in BRV) (5, 26, 28, 41, 44). Two potential integrin-binding motifs conserved in MRV λ2 as noted by Maginnis et al. (29) (882-RGD-884 and 1083-KGE-1085) are not conserved in BRV or in other ortho- or aquareoviruses. A few long regions notable for variability and poor scores in the alignments include BRV aa 693 to 722 (MRV-T3D; aa 696 to 725), which contributes to a small domain that bridges the methyltransferase domains and probably serves as a structural support for them as seen in MRV λ2 (44), and BRV aa 1056 to 1104 (MRV-T3D aa 1060 to 1113), which contributes to the first (N-terminal-most) of the three C-terminal flap domains as seen in MRV λ2 (44).
(ii) M2/μB.
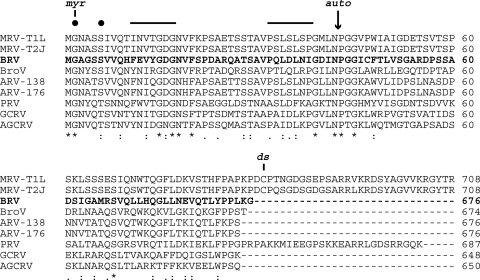
The M2 genome segment is 2,143 bp long, and its plus strand contains one long ORF spanning nt positions 24 to 2054. The encoded μB protein comprises 676 aa, with a calculated molecular mass of 74 kDa. In pairwise sequence alignments, it shows clear homology to the outer shell protein of representative MRV and ARV strains (μ1 and μB; 59 to 64% similarity and 33 to 39% identity) as well as to the outer shell protein of aquareoviruses (VP4; 47 to 48% similarity and 20 to 22% identity). This protein is also known to mediate cellular membrane penetration during cell entry by MRV and presumably other ortho- and aquareoviruses as well (6, 25). In multiple sequence alignments, the BRV μB protein aligns very well with the homologous proteins of other ortho- and aquareoviruses. The alignment includes not only occasional, small, internal gaps scattered throughout the sequences but also an ∼30-aa C-terminal extension present in MRVs and PRV but missing from BRV, BroV, ARVs, and aquareoviruses (Fig. 3). In the MRV virion, this C-terminal extension is known to form the hub-and-spoke complexes visible by cryo-EM within the P2 and P3 channels of the MRV outer capsid (57), structures that are missing from ARV and aquareovirus virions (10, 58). On the other hand, sequences near the N terminus are among the best conserved in the alignment (Fig. 3), including the extreme N-terminal myristoylation motif (1-MGNASS-6 in MRV-T3D; 1-MGAGSS-6 in BRV) (35), a β-hairpin motif implicated in membrane interaction by the μ1N peptide (11-INVTGDGNVFKPSAETSSTAVPSLSLSPG-39 in MRV-T3D [predicted β strands underlined]; 11-FEVYGDGNVFSPDARQATSAVPQLDLNIG-39 in BRV) (56), and the autocleavage motif spanning the μ1N/μ1C junction [42-(N/P)GG-45 in both MRV-T3D and BRV; cleavage between Asn42 and Pro43 as indicated by a slash] (34).
Fig. 3.
Multiple-sequence alignment of ortho- and aquareovirus outer shell proteins including the newly determined sequence from BRV (bold). N-terminal (top) and C-terminal (bottom) portions of the alignment are shown. The alignment was generated using the program Clustal W2 as implemented at http://guidance.tau.ac.il/, using sequences as identified in Materials and Methods. The number at the right end of each line indicates the position of the last amino acid in the sequence. Identities (*), strong similarities (:), and weaker similarities (.) in the aligned sequences are indicated at bottom. Other annotations are for the N-terminal myristoylation site (myr), the conserved myristoylation consensus sequence (filled circles), the putative transmembrane β-strands (solid lines at top) described in the text, the autolytic cleavage site (auto, arrow), and the disulfide-bonding Cys residue of MRVs (ds).
3D structure of the BRV virion.
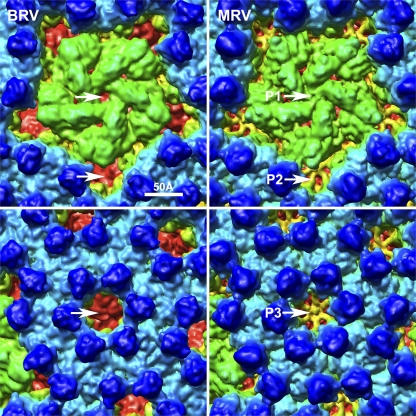
A stereo surface view of the BRV virion (Fig. 2, bottom) reveals many characteristic features such as the radially directed P2 and P3 solvent channels that perforate the outer capsid, the small projections attributable to outer clamp protein σB that cover much of the virion surface, and the extent to which the flattened top of the pentameric core turret is inset (i.e., having divots) relative to other regions of the surface. These and other surface details are quite similar to those of MRV (15, 57, 59) though closer inspection reveals three notable differences, as follows. (i) An open P1 channel is present at the icosahedral 5f axes of BRV, through which underlying portions of the core turret and core shell proteins are visible from outside the virion (Fig. 4, top). This finding is indicative of the fact that BRV lacks an adhesion fiber, the base of which obscures this channel in MRV and ARV virions by inserting into the top of the pentameric core turret at this position (15). (ii) The hub-and-spoke complexes in the P2 and P3 channels of MRV (57) are missing from those channels in BRV, allowing underlying portions of the core clamp and core shell proteins to be visible again from outside the virion (Fig. 4, bottom). BRV is like ARV in this regard (58). The absence of these complexes in BRV is consistent with new sequence evidence above, namely, that the BRV outer shell protein is missing the C-terminal sequences that form these complexes in MRV (Fig. 3). (iii) There are differences in the surface topography of the outer clamp proteins of BRV and MRV, that is, differences in the positions and sizes of various “bumps” on the protein surfaces. In addition, as perhaps best seen in the equatorial sections of the two virions (Fig. 2, top), the outer clamp subunits of BRV are “less erect” than those of MRV. For example, the outer clamp subunits that flank the icosahedral 2f axis are less parallel to that axis in BRV than in MRV. Such differences are consistent with the fact that the outer clamp proteins of BRV and MRV share the lowest level of sequence identity in pairwise alignments (<20%) among all of the structural proteins of these viruses (16; also this study and R. P. Goodman et al., unpublished), especially in the sequences that have been shown to form the upper lobe of the MRV outer clamp protein σ3 (16, 25, 37).
Fig. 4.
Close-up surface views of BRV and MRV virion cryo-reconstructions. The map and color coding of the BRV virion (left) are the same as used as in Fig. 2, but the map is shown in close-up view to highlight the P1 and P2 (top) and P3 (bottom) channel areas (arrows). Matching regions of the MRV virion surface are shown on the right for comparison to BRV. The MRV map is the same as that for Fig. 2, with radial color cueing applied to match that for BRV.
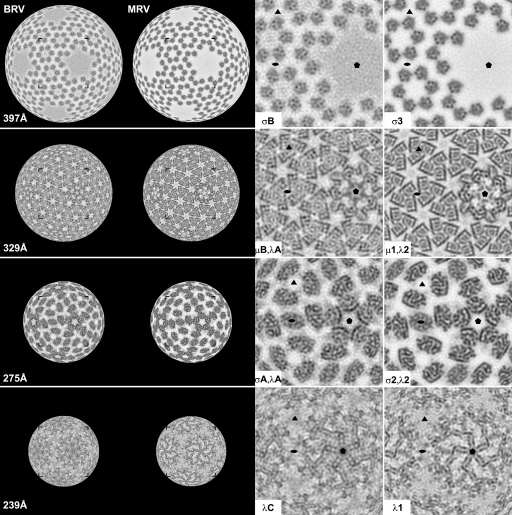
Density-coded radial sectioning of the cryo-reconstructions provided a means to compare the more internal features of the BRV and MRV capsids. Overall features are similar between BRV and MRV at most radii (Fig. 5, left columns). One notable observation is that sections through the core clamp protein, such as at 275 Å (Fig. 5), reveal that both BRV and MRV have 150 of these subunits per virion, which is also like ARV (58) but unlike aquareoviruses, which have only 120 per virion because none are present at the 30 icosahedral 2f-axis positions (10, 32). As one examines more carefully for specific high-density features at particular radii in the BRV and MRV capsids, which likely represent individual α-helices and β-sheets, one again sees a number of similarities between BRV and MRV (Fig. 5, right columns), suggesting that many individual components of their protein folds are conserved. For example, again at radius 275 Å, there is a curving helix in the guanylyltransferase domain at the base of the core turret protein on the perimeter of this domain facing the P1 channel, which is present in both BRV and MRV. In addition, at radius 329 Å in both BRV and MRV, intersecting, straight helices adopt a “backward-L” pattern in each outer shell protein subunit, such that the perimeter of each outer shell trimer is rimmed by these features. Both of these exemplified helical features are indeed present in X-ray crystal structures of the MRV core turret and outer shell proteins (25, 44) (Fig. 6). On the other hand, one can also find many differences in high-density features between BRV and MRV at essentially all capsid radii. Such differences are consistent with the fact that all five major BRV and MRV structural proteins demonstrate ≤41% sequence identity in pairwise alignments: core shell, 41%; core turret, 29%; core clamp, 29%; outer shell, 33%; and outer clamp, 18%) (16; also this study and R. P. Goodman, unpublished).
Fig. 5.
Selected radial sections of BRV and MRV virion cryo-reconstructions. Density-coded radial sections (each 2.3 Å thick) are shown for BRV and MRV (left columns as labeled for each virus). The particle radius at which each section was generated is indicated at left. Corner brackets frame the region in each section that is shown in close-up view in one of the right columns. The proteins for which densities are visible in each close-up view are indicated. The MRV map is the same as that for Fig. 2. Filled ellipse, triangle, and pentagon symbols demarcate the positions of 2f, 3f, and 5f axes in each close-up panel.
Fitting MRV crystal structures into the BRV virion cryo-reconstruction.
To assist with comparisons, we also fitted the atomic model of an asymmetric unit containing the five major capsid proteins of the MRV virion (37, 44, 57) into the BRV cryo-reconstruction. The results corroborated that the capsid positions and overall domain organizations of these proteins are largely the same in the two viruses. For instance, the series of domains in the core turret protein subunit and the trimeric organization of the outer shell protein are readily seen to be shared between MRV and BRV (Fig. 6). For these two proteins, as well as the core shell and core clamp proteins, the goodness of fit extends even to many individual secondary-structure elements within each subunit. As examples, several α-helices in the MRV core turret and outer shell proteins are seen to fit very well into corresponding rod-like densities in the BRV cryo-reconstruction (Fig. 6), including the backward-L pattern of intersecting helices that rim the μ1 trimer, as also highlighted in the radial sections (Fig. 5). On the other hand, the correspondence between individual secondary-structure elements within the outer clamp protein subunits of MRV and BRV is poorer, especially within their outer lobes, consistent with the greater sequence divergence of these proteins.
Cryo-reconstruction and 3D structure of BRV virions missing the λA flap domains.
During the course of this study, we encountered many frozen grids in which the BRV particles, when separately subjected to 3D image reconstruction (3,798 particles; 9.2-Å resolution), were found to be missing the C-terminal flap domains of the core turret protein (Fig. 7A and B). Difference maps between these “flapless” particles and those with flaps showed the missing flap domains to be the only substantive difference (Fig. 7C). The phenomenon was observed with three different preparations of purified BRV virions and moreover was confined to only certain grids; in other words, from the same virion preparation, the particles from some grids did not show flaps in the subsequent reconstruction, whereas the particles from other grids did. SDS-PAGE of one such virion preparation, after the flaps were found to be missing from the cryo-reconstruction, showed that the λ proteins appeared to remain intact, suggesting that the flap had not been removed (e.g., proteolytically) during storage. Instead, the flaps were either removed or exhibited irregular mobility only after being placed on those grids.
By examining the fitted MRV core turret protein structure within the context of the flapless BRV particles, we were able to discern that the cryo-reconstruction of these particles is missing all three of the C-terminal domains (designated A, B, and C) that constitute the flap (MRV λ2 aa 1024 to 1289; BRV λA aa 1020 to 1284 according to our alignment) of each BRV core turret subunit (Fig. 7D and E). Based on these observations, we conclude that the flap domains of the BRV core turret protein are capable of assuming variable positions within the virion, probably consequent to “swiveling” about a hinge near amino acid position 1020 in BRV λA, within a region of sequence that forms an extended chain between the methyltransferase-2 domain and the A (i.e., most N-terminal) flap domain, as seen in MRV λ2 (44). A similar type of flap rearrangement, though involving a different hinge region (see Discussion), has been observed in association with structural transitions between virions and cores during MRV particle disassembly and assembly (7, 15) but not within MRV virions per se. The capacity for such variable arrangement in BRV virions may reflect the absence of an outer fiber protein, which inserts at the icosahedral 5f axes in MRV and ARV virions (15, 58) and thereby stabilizes the flaps of those viruses in the “down” position. Why flap rearrangement occurred irregularly, and only on certain grids, with the BRV virions is difficult to answer but presumably reflects that the potential for movement of these domains was actuated only under certain specific conditions.
DISCUSSION
Because the S genome segments of BRV lack coding capacity for a homolog of the outer fiber protein encoded by most other orthoreoviruses (13, 16), BRV has been thought likely to contain only seven structural proteins, like aquareoviruses (1, 19) but unlike ARV and MRV strains, which have eight (Table 1). Limited densities corresponding to the adhesion fibers in the cryo-reconstructions of ARV and MRV virions (15, 58) are indeed missing from the cryo-reconstruction of the BRV virion shown here, resulting in an open P1 solvent channel at the icosahedral 5f axis. Thus, although it was previously conceivable that an outer fiber protein might be encoded by one of the L or M genome segments of BRV that have yet to be sequenced, the current results suggest that this is unlikely and that BRV does indeed lack an adhesion fiber anchored at the icosahedral 5f axes or elsewhere. Completion of the BRV genome sequences in this and a pending report (R. P. Goodman et al., unpublished) confirm that the BRV L and M segments encode only the expected λ- and μ-class homologs of ARV and MRV proteins and thus lack the coding capacity for an outer fiber protein (Table 1).
The absence of an adhesion fiber raises the question of which other BRV protein might take the place of the fiber in binding to cell surface receptors. One strong candidate is the outer clamp protein σB, which is present in 600 copies per virion and the upper lobes of which are the most dominant feature on the BRV virion surface. Another strong candidate to participate in adhesion to cell surface receptors is the pentameric core turret protein λA, which is present in 60 copies per virion and the tops of which are also well exposed on the BRV virion surface. The homologous MRV protein λ2 has been suggested to play a role in coreceptor (β1 integrin) binding and uptake of virions from the cell surface (29); however, proposed integrin-binding sites in the MRV core turret protein are not conserved in the BRV protein.
Aquareoviruses also lack an outer fiber protein and moreover have an altered conformation of their core turret protein (1, 10, 19, 32). The conformation of the aquareovirus turret protein differs in that the flap domains have pivoted around a hinge point following the first (A) domain so that the second and third (B and C) domains project to higher radii and further away from the icosahedral 5f axes (10), thereby opening a larger P1 channel than do the homologous, down-positioned flap B and C domains in MRV, ARV, and BRV virions. Which proteins function in aquareovirus adhesion to cell surface receptors remains unknown. However, it is intriguing that both aquareovirus and BRV virions display distinct differences in the arrangement or structural plasticity of their core turrets relative to the fiber-containing MRV and ARV virions. It will be interesting to determine whether BroV, another orthoreovirus that lacks an outer fiber protein (50), shares similar core turret features with BRV and aquareoviruses and to compare the strategies for adhesion to, and uptake by, cells of orthoreoviruses lacking the fiber protein.
In both MRV and aquareovirus particles, pivoting of the core turret flap domains occurs during the transitions between virions and cores that accompany particle disassembly and assembly (7, 10, 15, 32). In both cases, the hinge point for these rearrangements appears to be in an extended region of structure between the A and B flap domains in each subunit, such that the A domain remains largely in the same position in virions and cores. The new results for BRV virions, however, point to rearrangements also involving the A domain, with the main hinge region being between the methyltransferase-2 domain and the flap A domain. The flap rearrangements in BRV virions are therefore more comparable to those that occur in MRV cores upon heating to 52°C, in which the A flap domain of each core turret subunit is also involved (27). The results for BRV virions are also distinct in that all three flap domains of each subunit appear to assume variable positions such that they are missing from the cryo-reconstruction of the flapless particles, whereas in MRV and aquareovirus particles, though pivoted, the B and C flap domains of each subunit assume regular positions and remain well visualized in the reconstructions of both virions and cores (7, 10, 15, 32).
BRV virions are additionally seen to be like ARV and aquareovirus virions in lacking the hub-and-spoke complexes that reside within the P2 and P3 solvent channels of the MRV outer capsid. In MRV virions, these unique structures are formed by a 33-aa C-terminal extension of the outer shell protein (25, 57), which is missing from the homologous proteins of BRV, ARV, and aquareoviruses. The significance of these P2/P3 structures in MRV is not fully known, but they appear to play a role in stabilizing the outer capsid, including formation of intersubunit disulfide bonds within this region (36). A reciprocal possibility is that the open P2 and P3 channels, as well as the open P1 channels, in BRV and others may facilitate or otherwise influence conformational changes in the outer capsid that are associated with cell entry (6, 15, 25). Like BRV, ARV, and aquareoviruses, BroV also lacks a C-terminal extension on its outer shell protein, but, interestingly, PRV contains an extension, though not the disulfide-bonding Cys residue found in MRVs (Fig. 3).
The present cryo-reconstruction of BRV highlights structural differences in the ortho- and aquareoviruses that cross taxonomic boundaries. For instance, while BRV resembles ARV and MRV in containing 150 copies of the core clamp protein, rather than 120 copies as in aquareoviruses, BRV resembles aquareoviruses in its lack of an adhesion fiber and dissimilarities in the arrangement or stability of its core turret structure compared to ARV and MRV (10, 15, 32, 58). Some more divergent members of the subfamily Spinareovirinae, cypoviruses and oryzaviruses, whose core turret proteins wholly lack the flap domains, also contain only 120 copies of the core clamp protein (9, 31). In combination, this absence of a correlation between the presence of fibers and corresponding changes in the core turret with the number of copies of the core clamp protein implies that these structural features are not strictly linked in evolutionary terms. The significance of the various differences for particle function or assembly, however, has yet to be deciphered.
The taxon-crossing differences in structural features and coding capacities among ortho- and aquareoviruses have led us to ponder the detailed steps in their evolution. A series of discrete transitions that could account for gain or loss of particular features is diagrammed in Fig. 8. It is important to recognize, however, that because ortho- and aquareoviruses have segmented genomes, evolution can proceed by both drift (random point substitutions) and shift (transfer of an entire genome segment), analogous to influenza virus antigenic variation. Moreover, horizontal transfer of genetic elements between different viruses may well have occurred during the complex evolutionary pathway of the nonenveloped viruses, as suggested by the previously described relationships between orthoreovirus and adenovirus fiber proteins (8, 20, 33). This diagram is therefore clearly speculative and intended solely to help frame certain questions or predictions that may be addressed by future experiments.
We have recently described the structure of a provisional member of family Totiviridae—penaeid shrimp infectious myonecrosis virus (IMNV)—whose virions are adorned at the icosahedral 5f axes by surface fibers, making it unlike other totiviruses (48). Here, we reciprocally describe a member of family Reoviridae, subfamily Spinareovirinae, genus Orthoreovirus—BRV—whose virions lack surface fibers, making it unlike most other orthoreoviruses. These and other examples suggest that gain or loss of adorning fibers is one of several common themes in dsRNA virus evolution. The outer fiber protein VP4 of rotaviruses, as one other example from the family Reoviridae (subfamily Sedoreovirinae, genus Rotavirus), shows little if any sequence similarity to the outer fiber protein of orthoreoviruses and instead anchors its base into the P2 channels of the rotavirus outer capsid (45, 55). This malleability seems likely to reflect the ease of accommodating a surface fiber that simply anchors atop an established capsid architecture. Perhaps the bigger obstacle is how to incorporate fiber-encoding sequences into the viral genome so as to ensure their proper expression. Interestingly, in fusogenic orthoreoviruses, the fiber protein is commonly encoded on the same bi- or tricistronic S genome segment as the FAST protein (17, 47). The evolution of these multicistronic segments was presumably facilitated by the fact that neither the FAST protein nor the fiber protein needs to be expressed at high levels (there are no more than 36 copies of the fiber protein per orthoreovirus virion), allowing expression of these proteins to be mediated by atypical and possibly inefficient translation initiation mechanisms (42). Pressures driving the gain or loss of fiber proteins and FAST proteins, for that matter, are likely based in the ever-continuing battles between viruses and their hosts, relating to such elements as innate and adaptive immune responses, tropism, virulence, transmission, and host range.
ACKNOWLEDGMENTS
We thank Xing Zhang for assistance with MRV maps, Alex Rusnak for exploratory TEM work on BRV, Kelly Dryden for help with the Zeiss SCAI microdensitometer at The Scripps Research Institute, and Kevin Chiang and Debbie Kim for assistance with scanning images.
This work was supported in part by NIH grant R01 AI46440 (to M.L.N.), Canadian Institutes of Health Research grants 13723 and 100584 and Natural Sciences and Engineering (NSERC) grant 0183745 (to R.D.), NIH fellowship F32 AI078624 (to K.N.P.), NIH grants R37 GM-033050 and 1S10 RR-020016, and support from University of California—San Diego and the Agouron Foundation to establish and equip cryo-TEM facilities at USCD (to T.S.B.).
Footnotes
M.L.N., R.D., and T.S.B. are co-senior authors.
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Attoui H., et al. 2002. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J. Gen. Virol. 83:1941–1951 [DOI] [PubMed] [Google Scholar]
- 2. Baker T. S., Olson N. H., Fuller S. D. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barton E. S., et al. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451 [DOI] [PubMed] [Google Scholar]
- 4. Bassel-Duby R., et al. 1985. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. Nature 315:421–423 [DOI] [PubMed] [Google Scholar]
- 5. Breun L. A., et al. 2001. Mammalian reovirus L2 gene and λ2 core spike protein sequences and whole-genome comparisons of reoviruses type 1 Lang, type 2 Jones, and type 3 Dearing. Virology 287:333–348 [DOI] [PubMed] [Google Scholar]
- 6. Chandran K., Farsetta D. L., Nibert M. L. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandran K., et al. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chappell J. D., Prota A. E., Dermody T. S., Stehle T. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng L., et al. 2011. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc. Natl. Acad. Sci. U. S. A. 108:1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng L., et al. 2010. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J. Mol. Biol. 397:852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chua K. B., et al. 2007. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 104:11424–11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dawe S., Boutilier J., Duncan R. 2002. Identification and characterization of a baboon reovirus-specific nonstructural protein encoded by the bicistronic S4 genome segment. Virology 304:44–52 [DOI] [PubMed] [Google Scholar]
- 13. Dawe S., Duncan R. 2002. The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 76:2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dryden K. A., et al. 1998. Internal structures containing transcriptase- related proteins in top component particles of mammalian orthoreovirus. Virology 245:33–46 [DOI] [PubMed] [Google Scholar]
- 15. Dryden K. A., et al. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316–328 [DOI] [PubMed] [Google Scholar]
- 17. Duncan R., Corcoran J., Shou J., Stoltz D. 2004. Reptilian reovirus: a new fusogenic orthoreovirus species. Virology 319:131–140 [DOI] [PubMed] [Google Scholar]
- 18. Duncan R., Murphy F. A., Mirkovic R. R. 1995. Characterization of a novel syncytium-inducing baboon reovirus. Virology 212:752–756 [DOI] [PubMed] [Google Scholar]
- 19. Fang Q., et al. 2000. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (Genus Aquareovirus, family Reoviridae). Biochem. Biophys. Res. Commun. 274:762–766 [DOI] [PubMed] [Google Scholar]
- 20. Furlong D. B., Nibert M. L., Fields B. N. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gard G., Compans R. W. 1970. Structure and cytopathic effects of Nelson Bay virus. J. Virol. 6:100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Havelka W. A., Henderson R., Oesterhelt D. 1995. Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 247:726–738 [DOI] [PubMed] [Google Scholar]
- 23. Kim J., Tao Y., Reinisch K. M., Harrison S. C., Nibert M. L. 2004. Orthoreovirus and Aquareovirus core proteins: conserved enzymatic surfaces, but not protein-protein interfaces. Virus Res. 101:15–28 [DOI] [PubMed] [Google Scholar]
- 24. Leland M. M., Hubbard G. B., Sentmore H. T., III, Soike K. F., Hilliard J. K. 2000. Outbreak of Orthoreovirus-induced meningoencephalomyelitis in baboons. Comp. Med. 50:199–205 [PubMed] [Google Scholar]
- 25. Liemann S., Chandran K., Baker T. S., Nibert M. L., Harrison S. C. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108:283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luongo C. L., Contreras C. M., Farsetta D. L., Nibert M. L. 1998. Binding site for S-adenosyl-l-methionine in a central region of mammalian reovirus λ2 protein. Evidence for activities in mRNA cap methylation. J. Biol. Chem. 273:23773–23780 [DOI] [PubMed] [Google Scholar]
- 27. Luongo C. L., et al. 1997. Localization of a C-terminal region of λ2 protein in reovirus cores. J. Virol. 71:8035–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luongo C. L., Reinisch K. M., Harrison S. C., Nibert M. L. 2000. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein λ2. J. Biol. Chem. 275:2804–2810 [DOI] [PubMed] [Google Scholar]
- 29. Maginnis M. S., et al. 2006. Beta1 integrin mediates internalization of mammalian reovirus. J. Virol. 80:2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mindell J. A., Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 [DOI] [PubMed] [Google Scholar]
- 31. Miyazaki N., et al. 2008. Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell. J. Virol. 82:11344–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nason E. L., Samal S. K., Prasad B. V. V. 2000. Trypsin-induced structural transformation in aquareovirus. J. Virol. 74:6546–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nibert M. L., Dermody T. S., Fields B. N. 1990. Structure of the reovirus cell-attachment protein: a model for the domain organization of σ1. J. Virol. 64:2976–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nibert M. L., Odegard A. L., Agosto M. A., Chandran K., Schiff L. A. 2005. Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345:461–474 [DOI] [PubMed] [Google Scholar]
- 35. Nibert M. L., Schiff L. A., Fields B. N. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Odegard A. L., Chandran K., Liemann S., Harrison S. C., Nibert M. L. 2003. Disulfide bonding among μ1 trimers in mammalian reovirus outer capsid: a late and reversible step in virion morphogenesis. J. Virol. 77:5389–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olland A. M., Jané-Valbuena J., Schiff L. A., Nibert M. L., Harrison S. C. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 20:979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palacios G., et al. 2010. Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PLoS One 5:e11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paul R. W., Choi A. H., Lee P. W. K. 1989. The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology 172:382–385 [DOI] [PubMed] [Google Scholar]
- 40. Penn O., Privman E., Landan G., Graur D., Pupko T. 2010. An alignment confidence score capturing robustness to guide-tree uncertainty. Mol. Biol. Evol. 27:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qiu T., Luongo C. L. 2003. Identification of two histidines necessary for reovirus mRNA guanylyltransferase activity. Virology 316:313–324 [DOI] [PubMed] [Google Scholar]
- 42. Racine T., Duncan R. 2010. Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic Acids Res. 38:7260–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Racine T., et al. 2009. Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site. J. Virol. 83:5951–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reinisch K. M., Nibert M. L., Harrison S. C. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960–967 [DOI] [PubMed] [Google Scholar]
- 45. Settembre E. C., Chen J. Z., Dormitzer P. R., Grigorieff N., Harrison S. C. 2011. Atomic model of an infectious rotavirus particle. EMBO J. 30:408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shmulevitz M., Duncan R. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 19:902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shmulevitz M., et al. 2002. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 76:609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang J., et al. 2008. Infectious myonecrosis virus has a totivirus-like, 120-subunit capsid, but with fiber complexes at the fivefold axes. Proc. Natl. Acad. Sci. U. S. A. 105:17526–17531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang J., et al. 2010. Backbone trace of partitivirus capsid protein from electron cryomicroscopy and homology modeling. Biophys. J. 99:685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thalmann C. M., et al. 2010. Broome virus, a new fusogenic Orthoreovirus species isolated from an Australian fruit bat. Virology 402:26–40 [DOI] [PubMed] [Google Scholar]
- 51. van Heel M., Schatz M. 2005. Fourier shell correlation threshold criteria. J. Struct. Biol. 151:250–262 [DOI] [PubMed] [Google Scholar]
- 52. Victoria J. G., Kapoor A., Dupuis K., Schnurr D. P., Delwart E. L. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yan X., Dryden K. A., Tang J., Baker T. S. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yan X., Sinkovits R. S., Baker T. S. 2007. AUTO3DEM—-an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yeager M., Dryden K. A., Olson N. H., Greenberg H. B., Baker T. S. 1990. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J. Cell Biol. 110:2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang L., et al. 2009. Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J. Virol. 83:7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang X., et al. 2005. Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Å resolution. Structure 13:1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X., et al. 2005. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 343:25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X., Walker S. B., Chipman P. R., Nibert M. L., Baker T. S. 2003. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 10:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]