Abstract
Human APOBEC3 cytidine deaminases target and edit single-stranded DNA, which can be of viral, mitochondrial, or nuclear origin. Retrovirus genomes, such as human immunodeficiency virus (HIV) genomes deficient in the vif gene and the hepatitis B virus genome, are particularly vulnerable. The genomes of some DNA viruses, such as human papillomaviruses, can be edited in vivo and in transfection experiments. Accordingly, herpesviruses should be no exception. This is indeed the case for herpes simplex virus 1 (HSV-1) in tissue culture, where APOBEC3C (A3C) overexpression can reduce virus titers and the particle/PFU ratio ∼10-fold. Nonetheless, A3A, A3G, and AICDA can edit what is presumably a small fraction of HSV genomes in an experimental setting without seriously impacting the viral titer. Hyperediting was found in HSV genomes recovered from 4/8 uncultured buccal lesions. The phenomenon is not restricted to HSV, since hyperedited Epstein-Barr virus (EBV) genomes were readily recovered from 4/5 established cell lines, indicating that episomes are vulnerable to editing. These findings suggest that the widely expressed A3C cytidine deaminase can function as a restriction factor for some human herpesviruses. That the A3C gene is not induced by type I interferons begs the question whether some herpesviruses encode A3C antagonists.
INTRODUCTION
The seven-gene human APOBEC3 (A3) cytidine deaminase locus came to the fore with the identification of APOBEC3G (A3G) as the interaction partner of the human immunodeficiency virus (HIV) Vif protein (8, 16, 26, 29, 30, 43, 57). These enzymes belong to a larger group that can edit nucleic acids, of which activation-induced cytidine deaminase (AICDA), responsible for class switch recombination and somatic hypermutation of rearranged immunoglobulin V region genes, is perhaps the most widely known (11). All functional A3 enzymes show specificity for single-stranded DNA (ssDNA). Since the reaction product is uridine (dU), A3 activity results in DNA peppered by C → U substitutions, referred to as hypermutants. Editing can range from a few cytidine targets to over 80% (2, 3, 8, 16, 26, 29, 30, 49, 54). To a good first approximation, all A3 enzymes preferentially edit ssDNA when the edited base is 5′ flanked by thymidine or cytidine, i.e., TpC and CpC. In contrast, AICDA prefers GpC and ApC (2, 3, 9, 17, 27, 39, 49, 54).
Since the HIV-encoded Vif protein protects its genome from the mutagenic effects of A3G, HIV hypermutants are associated with a defective or deleterious vif background (43). In contrast, hepatitis B virus (HBV) DNA is particularly susceptible to genetic editing by at least two A3 enzymes in vivo (54), perhaps because its small genome does not encode interferon or A3 antagonists. Double-stranded DNA (dsDNA) is prone to editing during replication or transcription when it is partially single-stranded. Human papillomavirus genomes are vulnerable to APOBEC3 editing in vivo and in transfection experiments (53). In contrast, vaccinia virus, which replicates in the cytoplasm, is apparently resistant to A3G (22).
Given their very large genomes, between 124 and 241 kb (32), herpesviruses, which replicate in the nucleus, might be particularly sensitive to A3 deamination, since even low levels of deamination, say, <0.1%, would introduce several hundred mutations per genome. The seven A3 genes are expressed in a very wide variety of cell types, with some of the genes, notably human APOBEC3A and APOBEC3G (A3A and A3G), being strongly upregulated by type I interferons (IFNs) (5, 21, 40, 44, 55). Yet since herpes simplex virus (HSV) replication is comparatively resistant to IFN signaling and IFN-mediated responses in tissue culture (13, 34, 35, 41), they may not function as restriction factors. In contrast, APOBEC3C (A3C) is not only the most abundantly expressed of all A3 genes across a wide range of tissues and cells but also is insensitive to IFN (19). It can edit transfected human papillomavirus (HPV) DNA and mitochondrial DNA (mtDNA) (46, 53). Hence, it is plausible that A3C has posed a particular problem for primate herpesviruses. Here it is shown that HSV-1 is particularly vulnerable to the editing effects of APOBEC3C both in tissue culture and in vivo. Equally, Epstein-Barr virus (EBV) genomes in EBV-transformed oligoclonal B-cell lines can be edited by at least one APOBEC3 enzyme.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (PAA) at 37°C in 5% CO2. The EBV cell lines were maintained in RPMI supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (PAA). QT6 cells were maintained in Ham's medium supplemented with 100 U/ml penicillin, 2 mM glutamine, 5% tryptose phosphate, 1% chicken serum, and 10% fetal calf serum. HSV-1 strain 17 was grown in HeLa cells. Two days postinfection, supernatants were recovered and treated with 40 U of Turbo DNase (Ambion) for 30 min at 37°C and frozen at −80°C. Infections were carried out on 6 × 105 QT6 cells in 6-well plates at a multiplicity of infection (MOI) of 1. QT6 supernatants were treated with 40 U of Turbo DNase as described above.
For transfection, 5 × 104 HeLa cells were seeded in 24-well tissue culture plates and incubated for 24 h. Transfections were performed using Lipofectamine 2000 (Invitrogen) or jetPRIME. Briefly, HeLa cells were transfected with equal amounts (2.5 μg) of the individual expression plasmids in duplicate. Controls were performed in parallel without APOBECs. An enhanced green fluorescent protein (EGFP) expression plasmid was transfected in parallel, and the transfection efficiency was determined via flow cytometry: transfection efficiencies were ∼70%. Transfection medium was changed after 3 h, and transfected cells were incubated for 24 h and infected with HSV-1 (MOI = 1) for 90 min. After 48 h of incubation, the virus-containing supernatants were collected after centrifugation and stored at −80°C. Western blotting was performed as described previously (46).
Plaque assay.
Vero cells (7.5 × 104) were seeded in 24-well tissue culture plates and incubated for 24 h. The confluent cell monolayer was inoculated with serial virus dilutions in DMEM for 90 min and then overlaid with methylcellulose, incubated for 48 h, and washed with phosphate-buffered saline solution (PBS). Plaques were counted manually after fixing and staining with crystal violet.
Particle/PFU ratios.
Particle counts of viral supernatants were determined by mixing the supernatants with a preparation of 250-nm-diameter biotin-conjugated latex beads (Sigma) of known concentration. The mixture was then adsorbed onto glass coverslips for 1 h at room temperature and fixed with methanol. Samples were labeled using the mouse VP5-specific monoclonal antibody (DM165) (31) to label capsids or the rabbit polyclonal PTNC antibody (37) to label whole virions and L particles. Secondary antibodies used were goat anti-mouse Alexa 488 and goat anti-rabbit Alexa 568 conjugated antibodies (Molecular Probes). Latex beads were stained using Alexa 633-conjugated streptavidin (Molecular Probes). The relative number of virus particles was estimated according to the number of counted latex beads.
PCR.
DNAs were extracted using an Epicentre kit. The HSV-1 ICP22 gene primers were as follows: 5′ OUT, CGACGCGGGCCCGAGCRTATRCTYYAT; 3′ OUT, GGAAATGGCGGACACCTTCCTGGAYAYYAT; 5′ IN, CTCGTAGTAGACCCRAATCTCCACATT; 3′ IN, GCCGACGTACGCGATGAGATYAAT.
The outer and inner fragments were 880 and 461 bp, respectively. The first reaction involved standard amplification. Reaction parameters were as follows: 95°C for 7 min, followed by 42 cycles (each consisting of 95°C for 1 min, 60°C for 30 s, and 72°C for 3 min), and finally 20 min at 72°C. Differential amplification occurred in the second round (using 1 μl of the first-round reaction as input) by using an Eppendorf gradient Mastercycler S thermal cycler programmed to generate a 6°C gradient in the denaturation temperature. The reaction parameters were 89 to 95°C for 5 min, followed by 42 cycles (each consisting of 89 to 95°C for 1 min, 55°C for 30 s, and 72°C for 2 min), and finally 10 min at 72°C.
For HSV ICP0, the primers were as follows: 5′ OUT and IN, 5′ TTGCGCAATTGCATCCARRTTTTCAT; 3′ OUT, 5′ GAGGGGGAACTCGTGGGTGYTGATT; 3′ IN, 5′ GGACAGCACGGACAYGGAAYTGTT.
The outer and inner fragments were, respectively, 420 bp and 217 bp without primers. The conditions were as for ICP22, except for a 63°C annealing step for first-round PCR and an 8°C gradient at the denaturation temperature. The differential DNA denaturation PCR (3DPCR) reaction parameters were 87 to 95°C for 5 min, followed by 42 cycles (each consisting of 87 to 95°C for 1 min, 63°C for 30 s, and 72°C for 2 min), and finally 10 min at 72°C.
For HSV ICP8, the primers were as follows: 5′ OUT, 5′ CAAAGCCCAAGACGGCAACCACCATCAA; 3′ OUT, 5′ CTGGCTGGCTTCGAAGGCCGTGAAYGTA; 5′ IN, 5′ CACCTGGACCCCAGCACCCAGRCCCCAA; 3′ IN, 5′ GCTAAAATCCGGCATGAACAGCTGYAA.
The outer and inner fragments were, respectively, 810 bp and 263 bp without primers. The conditions were as for ICP22 except for a 65°C annealing step for first-round PCR and an 8°C gradient in the denaturation temperature. The reaction parameters were 87 to 95°C for 5 min, followed by 42 cycles (each consisting of 87 to 95°C for 1 min, 62°C for 30 s, and 72°C for 2 min), and finally 10 min at 72°C.
The EBV EBNA-1 gene primers were as follows: 5′ OUT, GTAGCATCTCTGTCTGGTGACCTTGAA; 3′ OUT, TTTTGGGGTCTCCGGACACCATCTCTA; 5′ IN, AGGCCTGGCTTGAGGCTCAGGACGCAA; 3′ IN, GACATGATTCACACTAAAAGAGATCAA.
The outer and inner fragments were 567 and 254 bp, respectively. The first reaction involved standard amplification. The reaction parameters were 95°C for 7 min, followed by 42 cycles (each consisting of 95°C for 1 min, 60°C for 30 s, and 72°C for 3 min), and finally 20 min at 72°C. Differential amplification occurred in the second round (using 1 μl from the first-round reaction as input) by using a 10°C gradient in the denaturation temperature. The reaction parameters were 80 to 90°C for 5 min, followed by 42 cycles (each consisting of 80 to 90°C for 1 min, 60°C for 30 s, and 72°C for 2 min), and finally 10 min at 72°C.
The EBV EBNA-2 gene primers were as follows: 5′ OUT, 5′ TAACGTGCAAGACGCTAAACTTAACCAA; 3′ OUT, 5′ AGCCTCGGTTGTGACAGAGGTGACAA; 5′ IN, 5′ TGTGTTTTGCTTTATCTGCCGCCATCA; 3′ IN, 5′ CGTCATATCCTAGCGGATCCCTATCAA.
The outer and inner fragments were, respectively, 907 bp and 345 bp without primers. The conditions were as for EBNA-1 except for a 62°C annealing step for first-round PCR. For the 3DPCR, reaction parameters were 80 to 90°C for 5 min, followed by 42 cycles (each consisting of 80 to 90°C for 1 min, 63°C for 30 s, and 72°C for 2 min), and finally 10 min at 72°C. PCR products were cloned into the pCR2.1 Topo cloning vector (Invitrogen). Sequencing was outsourced to GATC.
RESULTS
A3C restricts HSV replication.
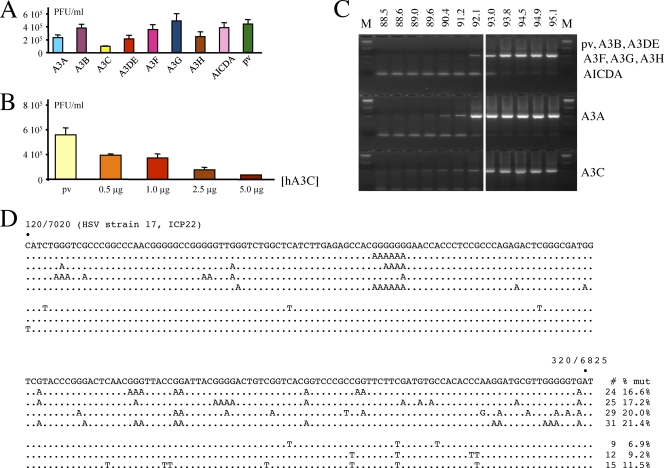
To explore the hypothesis that A3 enzymes may impact HSV-1 replication, HeLa cells were transfected by a variety of human cytidine deaminases, including AICDA. Twenty-four hours posttransfection, the cells were infected with HSV-1 at a MOI of 1 and allowed to grow for a further 48 h, after which titers of virus supernatants were determined on Vero cells. A3C reduced HSV titers by ∼4-fold (Fig. 1A), in keeping with transfection frequencies of ∼70%. The other deaminases had no significant impact compared to controls. A titration was performed with increasing amounts of A3C, using a plasmid vector to provide for a constant DNA concentration. A dose-response relationship was obtained (Fig. 1B).
Fig. 1.
Several human A3 enzymes may restrict HSV-1 replication and edit the genome. (A) Transfection of HeLa cells by 2.5 μg of plasmid DNA followed by HSV-1 infection. pv, empty vector. Titers are the means for triplicate experiments. (B) Dose-response relationship of supernatant titers as a function of A3C DNA concentration. The DNA concentration was maintained constant with the plasmid vector. (C) 3DPCR of HSV-1 DNA for all DNA transfections. The denaturation temperature (Td) is given across the top; M, molecular weight markers. The HSV-1 sequence between the primers is 410 bp in length. The white vertical bar indicates the minimal Td (93 to 93.8°C) for cloned wild-type HSV-1 DNA. Bands to lower temperature can be taken as prima facie evidence of cytidine deamination. (D) A selection of A3-edited HSV-1 genomes from the HeLa + pv transfection. For clarity, only 200 bp of the 410-bp ICP22 sequence are shown, while differences are scored with respect to the reference sequence (G → A, n = 25, 10,250 bp; C → T, n = 9, 3,690 bp). The positions in the HSV-1 strain 17 sequence are also given. To the right of each sequence is the total number of C → T transitions on the plus or minus (shown as G → A transitions) strand.
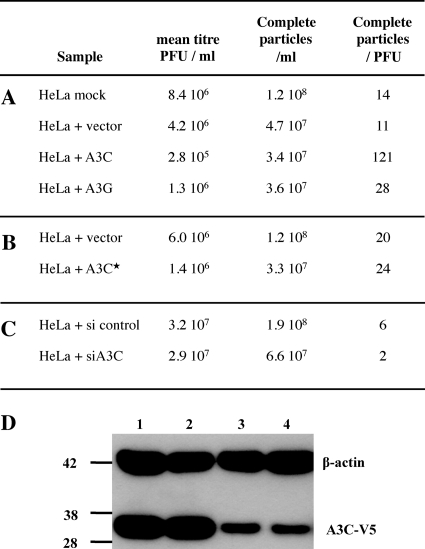
Another measure of HSV infectivity is the particle/PFU ratio, which is typically of the order of 10 to 100 for herpesviruses (12). When HSV was grown on A3C-transfected HeLa cells the ratio was increased ∼10-fold (Fig. 2A), indicating a higher number of defective particles in this population. In contrast, A3G failed to seriously impact the ratio. The vector transfection control yielded a titer comparable to that of virus from mock-transfected HeLa cells. An A3C catalytic mutant (C97S) yielded a particle/PFU ratio comparable to that of the plasmid vector control (Fig. 2B), while transfection of a small interfering RNA (siRNA) to A3C lowered the particle/PFU ratio compared to that of the siRNA control (Fig. 2C). Since the majority of cells were lysed at 48 h, the efficiency of siRNA knockdown was determined with uninfected cells. At 48 h, there was substantial knockdown of A3C-V5 as shown by Western blotting (Fig. 2D).
Fig. 2.
A3C levels can impact particle/PFU ratios for HSV-1. (A) Impact of active A3 constructs. (B) Impact of the A3C C957S inactive mutant. (C) Impact of A3C siRNA. (D) Western blot of V5-tagged A3C and β-actin loading control for uninfected HeLa cells at 48 h; lanes 1 and 2, 400 ng A3C-V5 tag plus 1 μg siRNA control; lanes 3 and 4, 400 ng A3C-V5 tag plus 1 μg siA3C RNA. Molecular mass markers (in kDa) are shown to the left.
Total DNA was extracted and analyzed by 3DPCR, which is a derivative of PCR that allows selective amplification of AT-rich DNA amid excess normal DNA (48). It exploits the lower denaturation temperature (Td) of AT-rich DNA by carrying out PCR with a Td gradient. If the lowest positive Td at which DNA is recovered is lower than that of the control, this can be considered prima facie evidence of recovery of AT-rich variants. Since the product of APOBEC3 editing of ssDNA is uridine, which base pairs as thymidine and is readily copied by Taq polymerase, it is understandable that 3DPCR has proven immensely useful in analyzing A3 editing (5, 36, 38, 44, 46–48, 51, 53, 54).
We chose the ICP22 immediate-early gene, which encodes a transcription factor involved in regulation of the viral cycle. As can be seen from Fig. 1C, HSV-1 DNA was recovered at denaturation temperatures down to 88.5°C for A3C and 89.6°C for A3A. Since the lowest denaturation temperature for cloned reference HSV-1 is ∼93 to 93.8°C (indicated by a white vertical line in Fig. 1C), recovery of HSV-1 DNA at lower temperatures is evidence of AT-rich variants. However, hyperedited HSV-1 DNA was obtained even for the HeLa plasmid vector (pv) transfection control (Fig. 1C). A sequence analysis of 3DPCR products derived from the pv sample with a Td of 92.1°C revealed hyperedited HSV genomes (Fig. 1D), with both DNA strands being vulnerable. The mean level of editing was 13% per clone, with a range of 1 to 23% (n = 25).
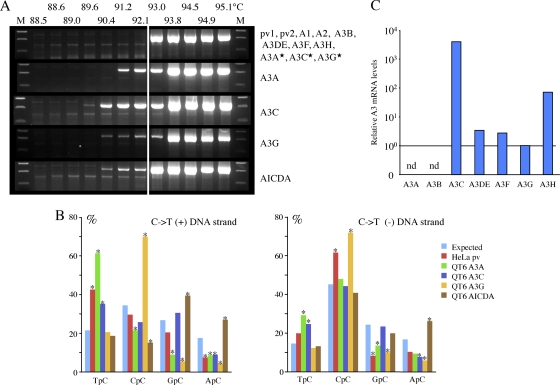
To overcome this endogenous editing background from HeLa cells, HSV-1 was passaged four times on the quail QT6 cell line (HSV-1/QT6). The avian lineage does not encode any A3 genes and has been shown not to give an endogenous editing background when stocks of HIV or HBV are made following transfection (17, 38, 54). DNase treatment of the supernatant was performed each time in order to reduce passive transfer of contaminating edited DNA. As expected, HSV-1/QT6 stock virus failed to give a background signal for editing (Fig. 3A, top). Accordingly, QT6 cells were transfected by human cytidine deaminases and subsequently infected by HSV-1/QT6 virus for 48 h.
Fig. 3.
Four human cytidine deaminases can hyperedit HSV-1 genomes. (A) Agarose gels of ICP22 3DPCR DNA products from HSV-1-infected quail QT6 cells expressing various A3 genes. The annotation is as for Fig. 1C. Stars indicate A3A, A3C, and A3G catalytic mutants. (B) 5′ dinucleotide context associated with editing on the plus and minus strands. Asterisks denote significant deviations from the expected values (χ2 test, P < 0.05) (QT6 + A3A, G → A, n = 24, 9,840 bp; C → T, n = 31, 12,710 bp; QT6 + A3C, G → A, n = 50, 20,500 bp; C → T, n = 78, 31,980 bp; QT6 + A3G, G → A, n = 32, 13,120 bp; C → T, n = 8, 3,280 bp; QT6 + AID, G → A, n = 9, 3,690 bp; C → T, n = 24, 9,840 bp). (C) TaqMan transcriptome analysis of the seven A3 genes from HeLa cells. The levels have been normalized to those of A3G (horizontal bar). nd, not detected.
When total cell DNAs were examined by 3DPCR, AT-rich DNA was identified from the AICDA, A3A, A3C, and A3G transfections but not from the others (Fig. 3A). Not surprisingly, three A3 catalytic site mutants (A3A C105S, A3C C97S, and A3G C281S) failed to edit HSV-1 genomes. Judging by the lowest denaturation temperature (89.6°C) and band intensities, A3C impacted the HSV-1 genome more severely than the others (Fig. 3A). Sequence analysis of cloned 3DPCR products revealed extensive editing, with mean mutation frequencies of ∼23% for the plus strand and between 23 and 42% for the minus strand. For A3A, A3C, and A3G, the minus strand was systematically more heavily edited than the plus strand, whereas for AICDA, the means were comparable (Table 1). Dinucleotide context analysis showed that editing was biased in favor of TpC for A3A and A3C, CpC for A3G, and GpC and ApC for AICDA (Fig. 3B), all of which have been previously noted for other virus genomes (2, 3, 7, 10, 14, 17, 28, 45, 51–54, 56). There was good concordance between the editing biases on both HSV-1 DNA strands (Fig. 3B). With these reference sets, the hyperedited sequences derived from the HeLa stock virus could be examined (Fig. 3B). While the overlap wasn't perfect, there was similarity with the patterns for both A3A and A3C editing. A TaqMan transcriptome analysis (40) of the 7 A3 genes in HeLa cells was performed. Since A3A levels were not detected while A3C levels were far higher than those for any other A3 gene (Fig. 3C), it is most likely that HSV-1 editing is due predominantly to A3C. To establish an overall hypermutant editing frequency, we performed a limiting dilution (cf. reference 47) of the first-round products, followed by PCR at 95°C and in-parallel 3DPCR at 92.1°C. This yielded a differential hypermutant frequency of ∼10−3. Since 3DPCR tends to underestimate lightly edited DNA, this represents an underestimation (54).
Table 1.
Essential statistics for HSV-1 and EBV edited genomes
| Virus (Td, °C) | Deaminase, cell line, and/or sample | Hyperediting | No. of sequences | Mean mut./seq.a | % GC → AT | % other |
|---|---|---|---|---|---|---|
| HSV-1 (91.1–92.1) | A3A/QT6 | C → T | 31 | 30 | 93.7 | 6.3 |
| G → A | 24 | 61 | 96.7 | 3.3 | ||
| A3C/QT6 | C → T | 78 | 32 | 96.2 | 3.8 | |
| G → A | 50 | 46 | 96.9 | 3.1 | ||
| A3G/QT6 | C → T | 8 | 32 | 95.1 | 4.9 | |
| G → A | 32 | 56 | 96.4 | 3.6 | ||
| AICDA/QT6 | C → T | 24 | 32 | 95.6 | 4.4 | |
| G → A | 10 | 34 | 92.9 | 7.1 | ||
| HSV-1 (92.1) | HeLa | C → T | 9 | 14 | 93.4 | 6.6 |
| G → A | 26 | 22 | 94.5 | 5.5 | ||
| HSV-1 (90.4–92.1) | P9 | C → T | 21 | 25 | 91.2 | 8.8 |
| P9 | G → A | 94 | 36 | 96.3 | 3.7 | |
| HSV-1 (93.8–94.9) | P1 | 39 | 2 | 36.1 | 63.9 | |
| P5 | 250 | 3 | 64.7 | 35.3 | ||
| P6 | 186 | 2 | 64.2 | 35.8 | ||
| P8 | 29 | 4 | 64.2 | 35.8 | ||
| P9 | 134 | 2 | 50.2 | 49.8 | ||
| P11 | 30 | 3 | 29.8 | 70.2 | ||
| P13 | 29 | 5 | 70.5 | 29.5 | ||
| P14 | 28 | 6 | 77.6 | 22.4 | ||
| EBV (82.5–89.8) | EBV-blast P1 | G → A | 54 | 17.5 | 96.8 | 3.2 |
| EBV-blast P2 | G → A | 31 | 21.6 | 97.9 | 2.1 | |
| EBV-blast P3 | G → A | 32 | 22.1 | 97.8 | 2.2 | |
| EBV-blast Z | G → A | 99 | 19.2 | 97.5 | 2.5 |
Mean number of mutations per sequence.
Is A3 editing of the HSV-1 genome physiologically relevant?
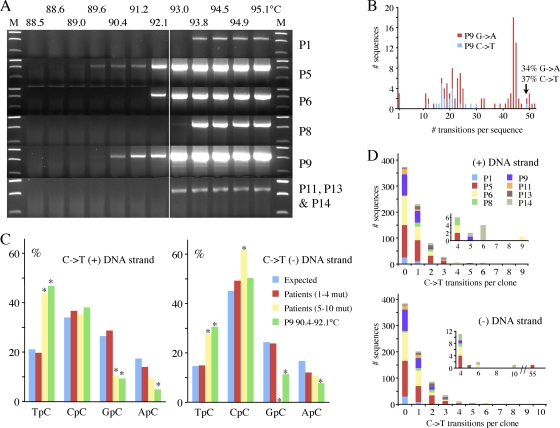
To address this question, we analyzed total DNA extracted directly from several HSV-1-associated lesions, notably pharyngeal washes and labial swabs. Fourteen samples were analyzed, the underlying pathologies ranging from prior liver transplantation to cancer. Using a nested PCR/3DPCR approach, eight samples scored positive for HSV-1 ICP22 DNA. The results were variable, with the last denaturation temperature ranging from 89.6 to 93.8°C (Fig. 4A). Since the lowest Td for cloned HSV-1 from QT6 cells is ∼93 to 93.8°C, samples P5, P6, and P9 apparently include hyperedited HSV-1 genomes. Sequencing of cloned 3DPCR DNA revealed extensively hypermutated sequences for P5 and P9, although only for P9 was an extensive group of hypermutated sequences recovered, with up to 52 targets edited on both strands (Fig. 4B). Dinucleotide contexts associated with P9 editing, from greatest to least association, were TpC, CpC, and RpC (Fig. 4C), typical of editing by A3 deaminases and highly comparable to that observed for HeLa derived stock (Fig. 3B). Limiting dilution of first-round products for patient 9 yielded a hypermutant frequency of ∼10−4.
Fig. 4.
HSV-1 genomes can be edited in vivo. (A) Agarose gels of ICP22 3DPCR DNA products from total DNA derived from pharyngeal washes or labial swabs. The annotation is as for Fig. 1C. (B) Frequency distribution of A3-edited HSV-1 sequences from patient P9. Since the cytidine composition of the two strands varies (32% plus strand; 35% minus strand), the % editing differs slightly (P9 G → A, n = 88, 36,080 bp; C → T, n = 21, 8,610 bp). (C) 5′ dinucleotide context associated with editing on the plus and minus strands for the collection of sequences from patient P9 (Td = 90.4 to 92.1°C). Asterisks denote significant deviations form the expected values (χ2 test, P < 0.05). Also shown are sequences derived at a higher Td (93.8 to 94.5°C) with 1 to 4 C → T transitions or 5 to 10 transitions pooled from all patients [Patients (1–4 mut.), n = 713, 292,330 bp; Patients (5–10 mut.), n = 10, 4,100 bp; P9, G → A, n = 94, 38,540 bp; C → T, n = 21, 8,610 bp]. (D) Frequency distribution of sequences derived at 93.8 to 94.5°C from all eight patients for both strands. The insets expand the region covering 4 to 55 bp.
To see if there were lightly edited HSV-1 sequences, 3DPCR products in the 93 to 94.5°C temperature range were cloned and sequenced. Numerous sequences encoded 1 to 10 transitions (Fig. 4D). Those with 1 to 4 C → T transitions showed no dinucleotide bias and probably reflect the AT-rich end of the HSV mutant spectrum (Fig. 4C). Furthermore, since the dominant PCR-related mutations are AT → GC, the majority cannot be ascribed to PCR error (15, 33). In contrast, those with 5 or more monotonous C → T transitions in either strand showed the same dinucleotide biases as hypermutated genomes from P9 (Fig. 4C), indicating that they do indeed reflect A3 editing. Accordingly, A3-edited HSV genomes were recovered from 4/8 samples (P5, P6, P9, and P14), albeit to different degrees. Finally, to ascertain whether other regions of the HSV-1 genome could be edited in vivo, the ICP0 and ICP8 genes were analyzed from patient 5 and 9 DNA, respectively. Hypermutated sequences were readily recovered (see Fig. S1A and B in the supplemental material), indicating that most probably all parts of the HSV-1 genome are vulnerable to editing.
Since hypermutated HSV genomes are physiologically relevant, we were interested in whether other herpesvirus genomes were also susceptible to A3 editing. Since the A3C gene is highly expressed in leukocytes (40, 46), we tested whether Epstein-Barr virus (EBV) genomes from transformed peripheral blood mononuclear cell lines were vulnerable. In such cell lines, EBV is found in its latent form with little transcription, although EBNA-1 is transcribed (18). Using a nested PCR/3DPCR approach, we amplified part of the EBNA-1 gene from total DNA. Four of five EBV+ cell lines (P1, P2, P3, and Z) (Fig. 5A) proved positive for edited EBV DNA given that the reference denaturation temperature for the segment is 86.7°C, far lower than that for HSV-1, reflecting a lower GC content (EBV EBNA-1, 54%; HSV-1 ICP22, 67%). Cloning and sequencing of the 3DPCR products revealed extensive cytidine editing (Fig. 5B), ranging from 10 to 53% (Fig. 5C). To ascertain whether other regions of the EBV genome could be edited in vivo, the EBNA-2 gene from patient Z was analyzed. Hypermutated sequences were readily recovered (see Fig. S1C in the supplemental material), indicating that most probably all parts of the EBV genome are vulnerable to editing.
Fig. 5.
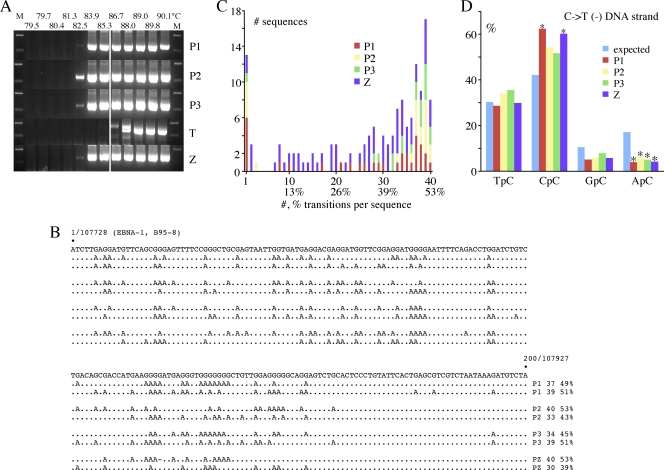
Hyperediting of EBV genomes in EBV-immortalized human B cell lines. (A) Agarose gels of EBNA-1 3DPCR DNA products from total DNA derived from five EBV-immortalized human B cell lines. The annotation is as for Fig. 1C. The P1 to -3 lines are all ung−/−; Z is AICDA−/−, while T is an EBV-cell line from a normal patient without any known leukocyte defect. (B) A selection of hyperedited sequences from P1 to -3 and Z. To the right are the number and % of cytidine residues edited. Plus-strand hypermutants were not always recovered and hence were excluded from analyses. (C) Frequency distribution of A3-edited EBV sequences from four EBV-transformed cell lines. Only minus-strand editing was identified (P1, n = 32, 6,400 bp; P2, n = 21, 4,200 bp; P3, n = 19, 3,800 bp; Z, n = 72, 14,400 bp). (D) 5′ dinucleotide context associated with minus-strand editing. Asterisks denote significant deviations from the expected values (χ2 test, P < 0.05).
The dinucleotide context associated with editing, in order of greatest to least, was CpC, TpC, and RpC, typical of A3G editing (Fig. 5D). A PCR/3DPCR limiting dilution of analysis of Z DNA yielded a hypermutant frequency of ∼10−3 (95°C versus 85.3°C). An APOBEC3 transcriptome analysis of these cell lines showed that A3C was the most abundantly expressed gene, almost a log more than A3G, with A3A being the least expressed (46).
DISCUSSION
Like other viral DNA genomes, those of some human herpesviruses are vulnerable to APOBEC3 editing. For HSV-1, A3C appears to be an important restriction factor and can impact both the titer and particle/PFU ratio. The relevance of the experimental findings is confirmed by the recovery of A3-edited genomes in uncultured samples (Fig. 4). HSV-1 is relatively insensitive to type I interferons, which should help protect it from the A3 enzymes whose genes are upregulated by them. However, since A3C expression is essentially insensitive to alpha IFN (IFN-α), the present findings raise the question as to whether HSV-1 encodes an A3C antagonist, given that it is sensitive to overexpression of A3C (Fig. 1A and B), or whether productive replication occurs in A3Cneg or A3Clow cells. Certainly, A3C siRNA knockdown shows that replication is sensitive to the expression levels in HeLa cells (Fig. 2C). In short, does HSV editing in vivo reflect A3 restriction of genomes defective for the antagonist, analogous to HIV/simian immunodeficiency virus (SIV) hypermutants on a Δvif background (4, 8, 16, 26, 29, 30, 42, 43, 57), or replication in A3Cneg/low cells, analogous to HBV infection in cirrhosis, since this virus does not encode any known IFN-α or A3 antagonist (54)? With this in mind, it would be particularly interesting to determine A3C expression in sensory neurons, particularly since A3 levels are generally low in uninfected brain (40). However, since B cells express high levels of A3 genes yet are propitious for EBV replication, simple correlations with bulk transcription levels may not be reliable. Given that A3C is expressed in a wide range of different cells and tissues and is invariably the most highly expressed of all the seven human A3 genes, variability in expression levels might perhaps account for part of the variability in plaque titers and particle/PFU ratios as a function of different cell lines. For example, varicella-zoster virus grown on a human melanoma cell line shows one of the highest particle/PFU ratios for any virus (6). However, for herpesviruses other factors can impinge on the particle/PFU ratio (1).
Highly edited genomes are clearly defective. Indeed, if A3 editing of ICP22 is representative of the whole genome, ∼40% editing of cytidine targets in the plus or minus strands translates into >20,000 C → T transitions per genome. Even lightly edited ICP22 DNA with 5 edits/410 bp translates into ∼1,860 substitutions per genome, which is far above the average mutation rate for DNA genomes, and so these too must be defective. Indeed, even a log lower amount of editing would also result in defective genomes. Presumably the vast majority of A3-edited HSV-1 genomes must be defective. Given this and a 68.3% GC content (20), it is unlikely that A3 editing contributes to HSV evolution. In a comparison of HSV-1 genomes, GC → AT transitions slightly outnumbered AT → GC transitions (50). However, when normalized to base composition, AT → GC transitions were more frequent, which again suggests that A3C activity does not impact HSV genome evolution. A3C presumably accesses replicating HSV DNA in the nucleus. That it impacts the particle/PFU ratio suggests that the lesions are in the packaged genomes, whether as dG-dU base pairs or as incorrectly repaired derivatives.
An interesting feature of the A3 editing described here is that while both strands are edited, we invariably recover more minus-strand than plus-strand hypermutants (G → A as opposed to C → T) (Table 1). Furthermore, the degree of editing was usually greater for the minus-strand than for the plus-strand hypermutants. This feature also shows up for mtDNA and nuclear DNA editing and for the A3-edited HPV hypermutants (46, 53). The variety among such genomes and amplification primers suggests that it is not an artifact but reflects some feature associated with A3 editing. As the minus strand is transcribed, some degree of protection by the transcription complex is clearly irrelevant.
Since A3 editing of the herpesvirus genomes occurs for HSV-1 and EBV genomes in their lytic and latent forms, respectively, it is possible that some of the other six human herpesviruses may be vulnerable too. The three EBV cell lines P1, P2, and P3 were deficient in the human uracil N-glycosylase (UNG), which we have shown to be an important component in the dynamics of A3-initiated catabolism of DNA: suppression of UNG activity resulted in higher frequencies of edited DNA (46). Yet in the present context, UNG was not crucial to detection of A3-edited EBV genomes since recovery of such genomes from the EBV+ Z cell line (ung+ yet AICDA−/−) was comparable, while the EBV+ line “T,” an EBV cell line from a donor without any known genetic defect, failed to yield hyperedited EBV DNA (Fig. 5A). Both HSV-1 and EBV encode divergent UNG enzymes that are able to excise uridine from DNA and are found at the replication fork. They are clearly orthologous to mammalian UNGs (23). Whether these viral UNG enzymes have any impact on detection of A3-edited genomes needs to be established.
The A3 gene locus appears with the emergence of placental mammals, and while it is generally bounded by the cbx6 and cbx7 genes, there is considerable variation (24). For example, the rat and mouse genomes encode a single A3 gene, the cow genome two, cats three/four, and horses five/six, while the primate lineage encodes seven A3 genes, six of which are functional (24, 25). Since many of these genes are phylogenically grouped with A3C, it is possible that they could represent yet another cross-species barrier, particularly in the sense of herpesvirus transmission to an animal with a more complex A3 locus. Since herpesviruses may go back 300 million to 400 million years, while the initial single gene A3 locus arose ∼125 million years ago, cytidine deamination would seem to be a relatively recent restriction factor.
In conclusion, at least two human herpesviruses are vulnerable to A3 editing, a physiologically relevant observation. The findings extend the list of genomes which are vulnerable: retroviral (3, 10, 14, 16, 26, 28–30, 43), adeno-associated (7), human papillomavirus (53), human mitochondrial, and nuclear genomes (46), as well as transfected plasmid DNA (44). It appears a plausible working hypothesis that yet more DNA viruses are restricted by A3 enzymes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martina Fuss for excellent technical assistance and Frazer Rixon, MRC Virology Unit, Glasgow, United Kingdom, for the DM165 antibody.
R.S. was supported by an EMBO long-term fellowship and a postdoctoral fellowship from l'Association pour la Recherche sur le Cancer (ARC). M.-M.A. is supported by a graduate fellowship from La Ligue contre le Cancer. This work was supported by grants from the Institut Pasteur, CNRS, and HOMFOR and the Spanish Ministry of Science and Innovation (SAF2010-21336). The Molecular Retrovirology Unit is an Equipe Labelise par la Ligue contre le Cancer.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Ace C. I., McKee T. A., Ryan J. M., Cameron J. M., Preston C. M. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beale R. C., et al. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596 [DOI] [PubMed] [Google Scholar]
- 3. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 4. Bogerd H. P., Doehle B. P., Wiegand H. L., Cullen B. R. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U. S. A. 101:3770–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonvin M., et al. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364–1374 [DOI] [PubMed] [Google Scholar]
- 6. Carpenter J. E., Henderson E. P., Grose C. 2009. Enumeration of an extremely high particle-to-PFU ratio for varicella-zoster virus. J. Virol. 83:6917–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H., et al. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480–485 [DOI] [PubMed] [Google Scholar]
- 8. Conticello S. G., Harris R. S., Neuberger M. S. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009–2013 [DOI] [PubMed] [Google Scholar]
- 9. Conticello S. G., Thomas C. J., Petersen-Mahrt S. K., Neuberger M. S. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol. Biol. Evol. 22:367–377 [DOI] [PubMed] [Google Scholar]
- 10. Delebecque F., et al. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Noia J. M., Neuberger M. S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 [DOI] [PubMed] [Google Scholar]
- 12. Dohner K., Radtke K., Schmidt S., Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eidson K. M., Hobbs W. E., Manning B. J., Carlson P., DeLuca N. A. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esnault C., et al. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433 [DOI] [PubMed] [Google Scholar]
- 15. Goodenow M., et al. 1989. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J. Acquir. Immune Defic. Syndr. 2:344–352 [PubMed] [Google Scholar]
- 16. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 17. Henry M., et al. 2009. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One 4:e4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imai K., et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028 [DOI] [PubMed] [Google Scholar]
- 19. Jarmuz A., et al. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296 [DOI] [PubMed] [Google Scholar]
- 20. Karlin S., Mocarski E. S., Schachtel G. A. 1994. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J. Virol. 68:1886–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koning F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kremer M., et al. 2006. Vaccinia virus replication is not affected by APOBEC3 family members. Virol. J. 3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krusong K., Carpenter E. P., Bellamy S. R., Savva R., Baldwin G. S. 2006. A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J. Biol. Chem. 281:4983–4992 [DOI] [PubMed] [Google Scholar]
- 24. LaRue R. S., et al. 2009. Guidelines for naming non-primate APOBEC3 genes and proteins. J. Virol. 83:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LaRue R. S., et al. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 27. Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 [DOI] [PubMed] [Google Scholar]
- 28. Mahieux R., et al. 2005. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 86:2489–2494 [DOI] [PubMed] [Google Scholar]
- 29. Mangeat B., et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 30. Mariani R., et al. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 31. McClelland D. A., et al. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGeoch D. J., Rixon F. J., Davison A. J. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 33. Meyerhans A., et al. 1989. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell 58:901–910 [DOI] [PubMed] [Google Scholar]
- 34. Mossman K. L., Saffran H. A., Smiley J. R. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nicholl M. J., Robinson L. H., Preston C. M. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215–2218 [DOI] [PubMed] [Google Scholar]
- 36. Noguchi C., et al. 2009. G-to-A hypermutation in hepatitis B virus (HBV) and clinical course of patients with chronic HBV infection. J. Infect. Dis. 199:1599–1607 [DOI] [PubMed] [Google Scholar]
- 37. Pasdeloup D., Blondel D., Isidro A. L., Rixon F. J. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petit V., et al. 2009. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J. Mol. Biol. 385:65–78 [DOI] [PubMed] [Google Scholar]
- 39. Pham P., Bransteitter R., Petruska J., Goodman M. F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103–107 [DOI] [PubMed] [Google Scholar]
- 40. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sainz B., Jr., Halford W. P. 2002. Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J. Virol. 76:11541–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schrofelbauer B., Chen D., Landau N. R. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. U. S. A. 101:3927–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 44. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stenglein M. D., Harris R. S. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837–16841 [DOI] [PubMed] [Google Scholar]
- 46. Suspène R., et al. 2011. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl. Acad. Sci. U. S. A. 108:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suspène R., et al. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suspène R., Henry M., Guillot S., Wain-Hobson S., Vartanian J. P. 2005. Recovery of APOBEC3-edited human immunodeficiency virus G → A hypermutants by differential DNA denaturation PCR. J. Gen. Virol. 86:125–129 [DOI] [PubMed] [Google Scholar]
- 49. Suspène R., et al. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Szpara M. L., Parsons L., Enquist L. W. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J. Virol. 84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsuge M., et al. 2010. G to A hypermutation of TT virus. Virus Res. 149:211–216 [DOI] [PubMed] [Google Scholar]
- 52. Turelli P., Mangeat B., Jost S., Vianin S., Trono D. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 53. Vartanian J. P., Guetard D., Henry M., Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233 [DOI] [PubMed] [Google Scholar]
- 54. Vartanian J. P., et al. 2010. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 6:e1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang F. X., Huang J., Zhang H., Ma X. 2008. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J. Gen. Virol. 89:722–730 [DOI] [PubMed] [Google Scholar]
- 56. Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang H., et al. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.