Abstract
The herpes simplex virus (HSV) UL17 and UL25 minor capsid proteins are essential for DNA packaging. They are thought to comprise a molecule arrayed in five copies around each of the capsid vertices. This molecule was initially termed the “C-capsid-specific component” (CCSC) (B. L. Trus et al., Mol. Cell 26:479-489, 2007), but as we have subsequently observed this feature on reconstructions of A, B, and C capsids, we now refer to it more generally as the “capsid vertex-specific component” (CVSC) (S. K. Cockrell et al., J. Virol. 85:4875-4887, 2011). We previously confirmed that UL25 occupies the vertex-distal region of the CVSC density by visualizing a large UL25-specific tag in reconstructions calculated from cryo-electron microscopy (cryo-EM) images. We have pursued the same strategy to determine the capsid location of the UL17 protein. Recombinant viruses were generated that contained either a small tandem affinity purification (TAP) tag or the green fluorescent protein (GFP) attached to the C terminus of UL17. Purification of the TAP-tagged UL17 or a similarly TAP-tagged UL25 protein clearly demonstrated that the two proteins interact. A cryo-EM reconstruction of capsids containing the UL17-GFP protein reveals that UL17 is the second component of the CVSC and suggests that UL17 interfaces with the other CVSC component, UL25, through its C terminus. The portion of UL17 nearest the vertex appears to be poorly constrained, which may provide flexibility in interacting with tegument proteins or the DNA-packaging machinery at the portal vertex. The exposed locations of the UL17 and UL25 proteins on the HSV-1 capsid exterior suggest that they may be attractive targets for highly specific antivirals.
INTRODUCTION
Herpesviruses are incurable human and animal pathogens that generally infect their hosts for life, escaping immune surveillance and producing recurrent infections between periods of latency. They have been shown recently to share functional and structural characteristics with double-stranded DNA (dsDNA) bacteriophages, and the recognition of common features in the capsid assembly pathways has been (and remains) instrumental in identifying roles for herpesvirus proteins that are less easily studied and manipulated than their phage analogs. These common features include (i) initial assembly into an icosahedral procapsid, (ii) maturational proteolysis of structural proteins, (iii) ATP-driven packaging of the dsDNA chromosome through a specialized capsid vertex complex called the “portal,” (iv) maturation of the procapsid during packaging, and (v) capsid stabilization effected by binding accessory proteins or by the formation of intersubunit bonds (reviewed in reference 5). While much of the herpesvirus capsid structure has been detailed, particularly by cryo-electron microscopy (cryo-EM), a number of minor proteins that interact with the capsid during assembly are still understood mostly by analogy with identifiable counterparts in phages. Several of these proteins are essential for packaging and retaining the viral DNA and are potentially valuable targets for interfering with herpesvirus replication.
The herpesvirus virion, ∼200 nm in diameter, consists of an icosahedral capsid of 125 nm in diameter enclosing the dsDNA chromosome and an amorphous layer of tegument proteins linking the capsid to an exterior lipid envelope in which different viral glycoproteins are embedded (5, 7, 12, 25). The herpes simplex virus 1 (HSV-1) capsid is composed primarily of the major capsid protein, VP5, organized as hexameric and pentameric capsomers that are termed “hexons” and “pentons,” respectively (21, 40). Hexons form the faces and edges of the icosahedron, and pentons occupy the vertices, arranged on an icosahedral lattice with triangulation number (T) = 16. Positioned between capsomers on the capsid exterior are protein complexes called “triplexes,” heterotrimers of proteins VP23 and VP19C that serve to stabilize the procapsid and capsid (34). The tips of hexons bind the small VP26 protein that appears to be dispensable for assembly (1, 35, 40). Located at one vertex, in place of a regular penton, is the portal complex, a dodecameric ring of protein UL6 through which genomic DNA is pumped into the preformed procapsid (20). Six minor proteins, UL15, UL17, UL25, UL28, UL32, and UL33, are essential for DNA packaging and retention, although only two of these—UL17 and UL25—are found in the mature virion (16, 18, 26).
Four herpesvirus capsid forms have been isolated from infected-cell nuclei: the on-pathway procapsid and DNA-filled C capsid, as well as two dead-end particles—the A capsid that is essentially empty of protein and DNA and the B capsid that contains a remnant of the cleaved scaffolding protein, VP22a (5, 9). Cryo-EM reconstructions of wild-type C capsids have revealed elongated, astral-like densities on the capsid exterior surrounding the vertices. This density was initially termed the “C-capsid-specific component” (CCSC) (36)—but as we have subsequently observed this feature on reconstructions of A, B, and C capsids, we now refer to it more generally as the “capsid vertex-specific component” (CVSC) (2). Two proteins essential for stable DNA packaging, UL17 and UL25, have been implicated as subunits of the CVSC (30, 36), and we have localized the UL25 protein to the penton-distal portion of the molecule by visualizing bulk tags inserted into the UL25 sequence with cryo-EM-based reconstructions (4). However, the remaining CVSC subunit or subunits have not been identified, nor has the UL17 protein been precisely located on the capsid surface (30). Here we confirm by tandem affinity purification (TAP) tagging and pulldown assays that UL25 and UL17 proteins interact (27), and we localize the UL17 protein through a bulk tag appended to its C terminus and visualized in cryo-EM reconstructions. We find that the UL17 protein occupies the CVSC density as a heterodimer with UL25 and that it is located in the region proximal to the capsid vertex. We further propose an approximate orientation of UL17 within the CVSC molecule that bears on its binding to the capsid and its likely functions at the portal and penton vertices.
MATERIALS AND METHODS
Cell and viruses.
African green monkey kidney cells (Vero) and the UL25 (8-1) and UL17 (G5) complementary cell lines were cultured as described previously (3, 8). The HSV-1 wild-type KOS strain, vNTAP-UL25, and the UL17 deletion mutant, ΔUL17 (26), which was grown on G5 cells, were previously described (3, 31, 32). The vUL17-CTAP and vUL17-GFP mutant viruses were generated by recombination of a KOS genome contained in a bacterial artificial chromosome (BAC) (10), as previously described (3). The following primers were used to amplify the green fluorescent protein (GFP) gene from peGFP-in (31, 32) and the TAP gene from p-EP-TAP-in (obtained from P. Kinchington, University of Pittsburgh): UL17-GFP (5′-CTT AGG TTT TGT CGC AAG GTG TCG TCC GGG AAC GGC CGG TCT CGC ATG GTG AGC AAG GGC GAG GA-3′ and 5′-GGG GAG GAG TGG ATG GGC GAG GTG GCC GGG GGA AGG CGC CCG CTA CTT GTA CAG CTC GTC CAT GCC G-3′) and UL17-CTAP (5′-CTT AGG TTT TGT CGC AAG GTG TCG TCC GGG AAC GGC CGG TCT CGC ATG AAG CGA CGA TGG AAA AAG-3′ and 5′-GGG GAG GAG TGG ATG GGC GAG GTG GCC GGG GGA AGG CGC CCG CTA GCC CAG CTT GCA GCC GCC GGA-3′).
To recover the recombinant viruses, BAC DNA was transfected into Vero cells, and recombinant viruses harvested from the transfected cell lysates were plaque purified on Vero cells.
Capsid purification, SDS-PAGE, and Western blot analysis.
Intranuclear capsids were purified from Vero cells (1.5 × 108) infected overnight (18 h at 37°C) at a multiplicity of infection (MOI) of 5 PFU/cell by sucrose density gradient as previously described (3). The positions of A, B, or C capsids were observed as light-scattering bands with A capsids found highest (least dense) on the gradient and C capsids found lowest (dense fractions). Fractions were collected, and protein was precipitated by adding an equal volume of 16% trichloroacetic acid. Pellets were resuspended in 35 μl of 2× PAGE loading buffer (Invitrogen) supplemented with 0.4 M Tris base. Gradient fractions containing either A, B, or C capsids were run on 4 to 12% SDS-PAGE, and the gels were either stained with Imperial blue (Pierce) to visualize capsid proteins or analyzed by immunoblotting as previously described (3), using UL25 mouse monoclonal antibody, 25E10, a UL17 chicken polyclonal antibody (37), a calmodulin-conjugated rabbit monoclonal antibody (Millipore; catalog no. 05-932), or a rabbit polyclonal GFP antibody, A11122, from Molecular Probes (Eugene, OR) diluted at 1:5,000, 1:25,000, 1:5,000, and 1:1,000, respectively. The diluted antibodies were reacted with the blocked nitrocellulose for 2 h at room temperature, washed five times in Tris-buffered saline (TBS) with 0.5% Tween 20, and incubated with IRdye800-conjugated secondary antibodies diluted 1:15,000 in Rockland near-infrared blocking buffer (Rockland Immunochemicals, Gilbertsville, PA) with 0.1% Tween 20, goat anti-mouse (UL25), donkey anti-chicken (UL17), or goat anti-rabbit (calmodulin and GFP) obtained from LiCor (Lincoln, NE). The blots were washed and scanned using an Odyssey system (LiCor, Lincoln, NE).
TAP.
The TAP protocol was performed by infecting 5 × 108 Vero cells with KOS, vNTAP-UL25, or vUL17-CTAP virus at an MOI of 5 PFU/cell. The infection was allowed to proceed for 18 h at 37°C, after which the cells were harvested and pelleted via centrifugation at 5,000 rpm for 5 min at 4°C. All remaining steps were done at 4°C. The cells were washed in a total volume of 50 ml 1× PBS, and the final cell pellet was resuspended in 24 ml of streptavidin binding buffer (SBB) (300 mM KCl, 40 mM Tris-HCl [pH 7.5], 2 mM EDTA, 0.1% NP-40 substitute, and 5 mM β-mercaptoethanol containing protease inhibitors [Roche 1 697 498]). The cells were lysed by sonication using a probe sonicator. The cell suspension was sonicated 4 times for 10 s each at an output of 6 W with chilling on ice between each sonication step. Benzonase (1,500 U; Novagen catalog no. 70664-3) was added to the samples and left at 4°C for 30 min. The extract was then clarified via centrifugation at 12,000 rpm in a Sorvall SS-34 rotor for 20 min at 4°C. The supernatant was transferred to a new tube, 1.5 ml of streptavidin resin (0.75-ml packed volume) (Pierce; catalog no. 53117) was added, and the samples were rotated at 4°C for 2 h. The resin was pelleted by centrifugation at 3,500 rpm for 5 min, and the supernatant was removed. The resin was washed three times by being resuspended in 5 ml of SBB followed by centrifugation at 3,500 rpm for 5 min. Protein was eluted from the resin by adding 3 ml of streptavidin elution buffer (SEB) (300 mM KCl, 40 mM Tris-HCl [pH 7.5], 2 mM EDTA, 0.1% NP-40 substitute, 5 mM β-mercaptoethanol, 2 mM d-biotin [Sigma 47868]) containing Roche protease inhibitors, and the sample was rotated at 4°C for 30 min. The protein-resin mixture was spun down at 7,000 rpm in a microcentrifuge for 2 min, and the supernatant was collected as the streptavidin eluate. Ten milliliters of calmodulin binding buffer (CBB) [150 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM (CH3COO)2Mg, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40 substitute, 10 mM β-mercaptoethanol containing Roche protease inhibitors] was added to the streptavidin eluate. An additional 10 μl of 1 M CaCl2 was added to the mixture along with 0.6 ml packed calmodulin resin (Agilent Technologies 214303-52), and samples were rotated at 4°C for 2 h. The resin was pelleted by centrifugation at 3,500 rpm for 5 min, and the supernatant was removed. The resin was washed three times by resuspension in 5 ml of CBB followed by centrifugation at 3,500 rpm for 5 min. Protein was eluted from the resin by adding 3 ml of calmodulin elution buffer (CEB) [150 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM (CH3COO)2Mg, 1 mM imidazole, 0.1% NP-40 substitute, 10 mM β-mercaptoethanol, 4 mM EGTA], and the sample was rotated at 4°C for 30 min. The protein-resin mixture was spun down at 7,000 rpm in a microcentrifuge for 2 min, and the supernatant was collected as the final (calmodulin) eluate.
Cryo-EM and image reconstruction of purified capsids.
Three microliters of purified UL17-GFP-tagged C capsids and wild-type C capsids was pipetted onto glow-discharged holey carbon Quantifoil electron microscopy (EM) grids and plunge frozen into liquid ethane-propane (50:50 mix) cooled in a liquid nitrogen bath (33), using an FEI Vitrobot mark III (FEI, Hillsboro, OR) (85% humidity, 5- to 7.5-s blot). The grids were transferred to a Gatan 626 cryoholder and inserted into an FEI Tecnai T20 FEG microscope operating at 200 kV, maintaining liquid nitrogen temperatures throughout. Images were collected at a 29,000× magnification on Kodak SO-163 film (Kodak, Rochester, NY). Micrographs were digitized on a Nikon Super CoolScan 9000 scanner (Nikon, Tokyo) at a sampling rate of 6.35 μm/pixel, corresponding to 2.12 Å/pixel at the sample. C capsids were selected using x3dpreprocess software (6), and defocus/astigmatism values were estimated manually using BSOFT (14). An icosahedrally averaged map of each sample was calculated using AUTO3DEM (39) by refining an initial model generated from the images using the random model procedure (38). The refined maps were calculated from 3,984 and 3,973 particles to resolutions of ∼17.5 and ∼16.5 Å for the tagged and wild-type C capsids, respectively. USCF Chimera was used to visualize the density maps (23). Estimation of occupancy of the CVSC and GFP density on capsids was done from map sections, by comparison of the peak density value within each region to the capsid (100% occupancy) and background (0% occupancy) densities (4, 36). For the CVSC, separate measurements were taken for the vertex-distal and -proximal domains. Density maps have been deposited in the EMDB repository with ID codes 1902, 1903, and 1904.
RESULTS
Analysis of HSV vUL17-GFP and vUL17-CTAP viruses.
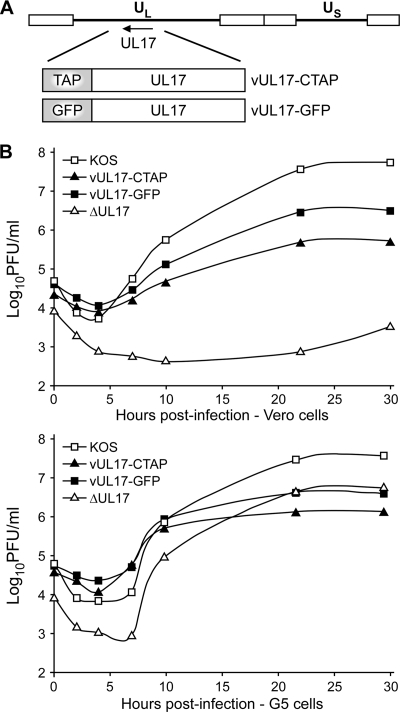
The HSV-1 vUL17-GFP and vUL17-CTAP viruses encode UL17-GFP or UL17-CTAP fusion proteins, respectively, where the 726-bp GFP gene or the 234-bp TAP gene was inserted in frame at the end of the UL17 open reading frame (ORF) (Fig. 1A). The two fusion proteins were introduced into the viral genome through genetic manipulation of an HSV-1 (KOS) genome maintained in a recombinant bacterial artificial chromosome (BAC). The UL17-GFP and UL17-CTAP BACs were transfected onto Vero cells, and the recovered virus was plaque purified on Vero cells and titrated on Vero cells and a UL17-complementing cell line, G5 cells. Both the vUL17-GFP and vUL17-CTAP mutants yielded identical titers on Vero cells and G5 cells (Table 1). Intracellular replication of the UL17 mutants was examined by establishing single-step growth curves in Vero and G5 cells. Wild-type KOS and the UL17-null virus, ΔUL17, were included in these studies. Cells were infected at an MOI of 3 with each virus, and cells were harvested at 0, 2, 4, 7, 10, 22, and 30 h postinfection (hpi) and assayed for infectious virus by plaque assay on G5 cells (Fig. 1B). Both the vUL17-GFP and vUL17-CTAP viruses exhibited growth defects on Vero cells, with titers reduced by approximately 1 to 2 logs compared to KOS. As expected, the UL17-null (ΔUL17) virus failed to produce any virus on Vero cells. When grown on G5 cells, the UL17-GFP and UL17-CTAP mutants along with the UL17-null virus showed increased virus production, but the amount of virus produced was reduced compared to that of KOS (Fig. 1B). Taken together, these results demonstrate that the vUL17-GFP and vUL17-CTAP can grow on noncomplementing cells, but the fusion protein appears to reduce virus yields.
Fig. 1.
UL17-CTAP and UL17-GFP virus constructs and single-step growth curves. (A) The HSV-1 genome is represented at the top. UL and US represent the long and short unique region sequences, respectively. The region of the HSV-1 genome that contains the UL17 gene is expanded below. The UL17-GFP and UL17-CTAP fusion proteins that are expressed from the vUL17-GFP and vUL17-CTAP viruses are shown. (B) Vero or G5 cells were infected with either KOS, vUL17-GFP, vUL17-CTAP, or the UL17-null (ΔUL17) virus at an MOI of 3 at 4°C for 1 h and incubated at 37°C. The cultures were harvested at the indicated times postinfection and freeze-thawed three times, and the yield of virus at each time point was determined by plaque titer determination on G5 cells.
Table 1.
Virus growth assay
| Virus | Virus plating efficiency (PFU/ml) in: |
|
|---|---|---|
| Vero cells | G5 cells | |
| KOS | 2 × 108 | 2 × 108 |
| vUL17-CTAP | 5 × 108 | 3 × 108 |
| vUL17-GFP | 4 × 108 | 5 × 108 |
| ΔUL17 mutant | 1 × 104 | 3 × 109 |
Detection of UL17-GFP and UL17-CTAP proteins on capsids.
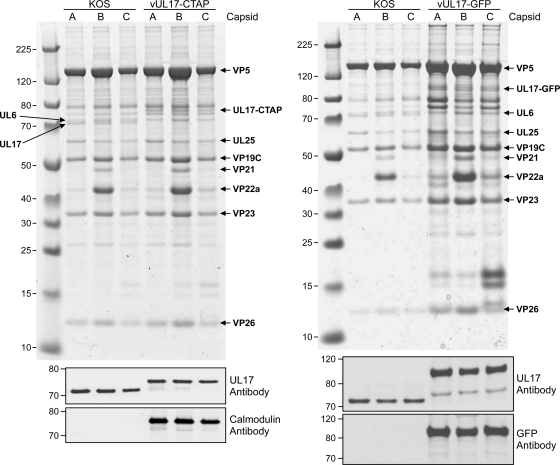
Intranuclear capsids were purified from Vero cells infected with either KOS, vUL17-GFP, or vUL17-CTAP and the presence of the UL17 protein in A, B, and C capsids was examined by SDS-PAGE (Coomassie stained) and Western blot analysis (Fig. 2). A, B, and C capsids were separated by SDS-PAGE, and the capsid protein composition was examined on the stained gels. The five proteins located in the capsid, VP5, VP19C, VP23, VP26, and UL6, and the scaffold protein found only with B capsids, VP22a, are indicated. The CVSC protein UL25 is evident in all three capsid forms, while the wild-type UL17 is seen only with KOS capsids. However, all three capsid forms for the UL17-GFP- and UL17-CTAP-expressing viruses contain bands corresponding to proteins of the correct size for either the UL17-GFP or UL17-CTAP fusion proteins, respectively. Western blot analyses confirmed these were indeed the UL17 fusion proteins, as identical blots were probed with either the chicken UL17 polyclonal antibody, a rabbit GFP antibody, or a rabbit calmodulin monoclonal antibody (Fig. 2B). The UL17-GFP fusion protein, which is approximately 25 kDa larger than the wild-type UL17 protein, was detected in A-, B-, and C-capsids with both the UL17 and GFP antibodies. The UL17-CTAP fusion protein, approximately 9 kDa larger than the wild type, was detected in all three capsids forms with both the UL17 and calmodulin antibodies.
Fig. 2.
Analysis of capsid-bound UL17-GFP and UL17-CTAP. UL17 content in capsids isolated from cells infected with KOS, vUL17-GFP, or vUL17-CTAP. (Top) Equivalent amounts of C, B, or A capsids were separated by SDS-PAGE, and the proteins were detected by Coomassie blue staining. The positions of the capsid proteins (major capsid protein VP5, triplex proteins VP19C and VP23, small capsid protein VP26, the scaffold protein VP22a, and the portal protein UL6) and the two CVSC proteins UL25 and UL17 along with the UL17-GFP and UL17-CTAP fusion proteins are indicated. Molecular mass standards are labeled on the left in kDa. (Bottom) Immunoblot analysis of KOS, vUL17-GFP, or vUL17-CTAP C, B, or A capsids with UL17, GFP, and calmodulin antibodies.
Tandem affinity purification of the UL17-CTAP and UL25-NTAP fusion proteins.
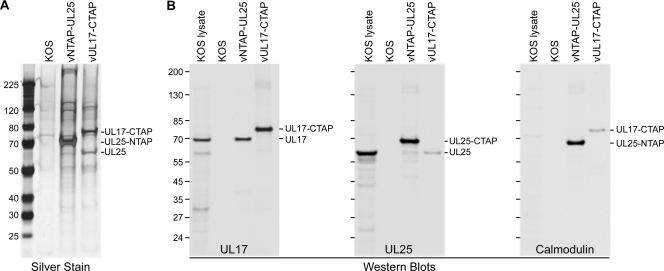
The tandem affinity purification (TAP) method allows for rapid purification of protein complexes under native conditions and here was used to determine what viral and cell proteins UL25 and its putative CVSC binding partner, UL17, interact with. The TAP method requires fusion of a TAP tag to either the N or C terminus of the target protein, resulting in a larger fusion protein that can also be used in cryo-EM to identify and localize minor capsid components such as UL25 and UL17 on purified capsids, as we have recently demonstrated using a virus expressing a UL25-NTAP fusion protein (2). The TAP tag used for these studies consisted of a streptavidin binding peptide and a calmodulin binding peptide (Stratagene-Interplay). This tag was fused to the N terminus of the UL25 protein and to the C terminus of the UL17 protein. Vero cells were infected with KOS or with viruses expressing the UL25-NTAP or UL17-CTAP protein, and the TAP purification procedure was applied to infected cell extracts (Fig. 3). The extracts were applied to a streptavidin resin and eluted from the column with biotin and further purified on a calmodulin resin, with elution by EGTA (Fig. 3A). The UL25-NTAP and UL17-CTAP proteins appear as the major silver-stained band following the last step (calmodulin column) in the TAP procedure. Along with the TAP-tagged protein, several additional proteins were present for each sample and represent proteins that copurify with the TAP-tagged protein. These were identified by Western blot analysis (Fig. 3B), which demonstrated that the wild-type UL25 protein copurifies with UL17-CTAP and the wild-type UL17 protein with UL25-NTAP (Fig. 3B). Note that the wild-type UL25 protein was also observed in the silver-stained gel of the vUL17-CTAP sample (Fig. 3A). The wild-type UL17 protein is not readily apparent in the stained gel since it is approximately the same size as the UL25-NTAP protein.
Fig. 3.
TAP purification of UL25-NTAP and UL17-CTAP. Vero cells were infected with HSV-1 recombinant viruses expressing the indicated TAP-tagged protein or with wild-type (KOS) virus. (A) Following TAP purification, SDS-PAGE analysis (silver stain) was done on the proteins eluted from the KOS, UL25-NTAP, and UL17-CTAP calmodulin column. (B) Western blots (antibodies used listed below each blot) demonstrating the presence of the TAP-tagged proteins along with proteins that copurify with the TAP-tagged proteins. Protein standards (kDa) are shown on the left of each gel or blot.
Location and conformation of the UL17-GFP fusion protein on HSV-1 C capsids.
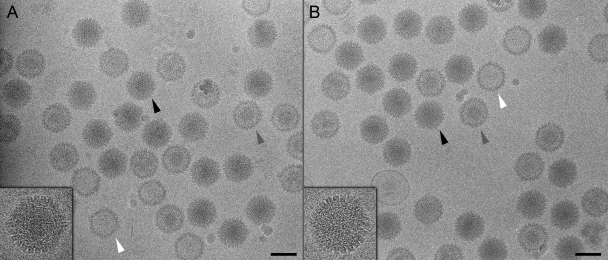
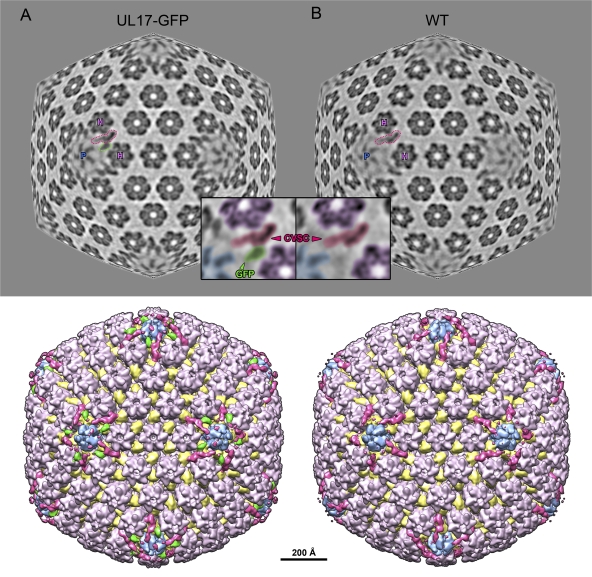
To locate UL17 on C capsids, we compared density maps calculated from purified UL17-GFP C capsids and from wild-type KOS C capsids imaged by cryo-EM. DNA-filled C capsids are the most abundant particle in micrographs of each sample and are easily distinguished from the few contaminating empty capsids and B capsids (Fig. 4). Approximately 4,000 images of each type of C capsid particle were selected and reconstructed to resolutions of ∼17 Å with icosahedral symmetry imposed throughout. The density maps reveal strong density extending from the CVSC region of the UL17-GFP-tagged capsid (Fig. 5A) that is absent in the wild-type capsid (Fig. 5B). This extra density can be seen in a close-up color-coded view of the region surrounding a single CVSC molecule (Fig. 5, inset) and in a close-up surface view of a 5-fold axis (Fig. 6A, black arrows; cf. Fig. 6B). The shape and volume of the extra density snugly fit the GFP crystal structure (Fig. 6C), and the density values show an occupancy level of 68% (compared to capsid density) that is similar to the adjacent CVSC occupancy of 70%, indicating a 1:1 stoichiometry that is consistent with a tagged protein. The same occupancy for the CVSC is also seen on wild-type capsids (70%), in agreement with the SDS-PAGE gels (Fig. 2), showing that the GFP tag does not affect the ability of the labeled protein to incorporate onto the capsid. We attribute the density extension to the CVSC molecule as representing the GFP tag that is fused to the C terminus of the UL17 protein and conclude that UL17 interacts with UL25 to form the CVSC density found on capsids.
Fig. 4.
Cryo-electron microscopy of tagged capsids. Shown are micrographs of UL17-GFP (A) and wild-type (B) capsids. DNA-filled C capsids (black arrowheads and inset close-ups) are darker than, and easily distinguishable from, subset populations of A and B capsids (white and gray arrowheads, respectively). C capsids selected from each data set were subject to the three-dimensional reconstruction procedure.
Fig. 5.
Visualizing the GFP tag on UL17-GFP capsids. Density maps calculated from C capsids with GFP-labeled UL17 (A) and wild-type C capsids with unlabeled UL17 (B). Radial sections partway between spherical and icosahedral (70%) through each map, at a radius of 565 Å at the icosahedral 3-fold axis, are shown at the top, with surface-rendered views beneath. Views are along the icosahedral 2-fold axis. In the sections, a penton and its two adjacent hexons are labeled with “P” and “H,” respectively, and a CVSC density is outlined in magenta. In the UL17-GFP map section, additional density is seen coming off the CVSC and is outlined in green. Insets show close-up views of the CVSC outlined in the sections. The densities have been shaded so that the penton is blue, hexons are purple, the CVSC is magenta, and extra density in the tagged map, which we attribute to GFP, is green. Surface views of the capsid beneath the sections have been color coded as described above, with triplexes in yellow.
Fig. 6.
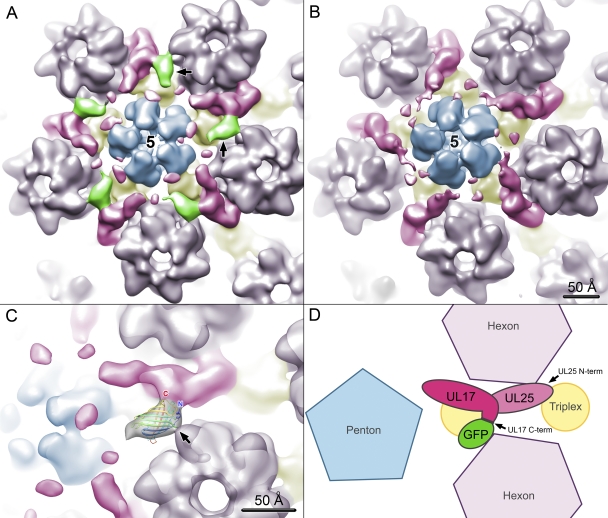
Close-up views of the UL17-GFP density. A penton flanked by five CVSC densities is shown for the UL17-GFP (A) and wild-type (B) density maps. Capsid subunit boundaries have been estimated and colored as for Fig. 5. The GFP density is in green and labeled with black arrows in panel A. Into this density, the crystal structure of GFP (Protein Data Bank ID 1EMA) (22) has been docked in panel C. The capsid density is semitransparent, and the GFP coordinates are in a ribbon representation, rainbow colored, with C and N termini labeled (C and N, respectively). There is a broad contact region between GFP and the CVSC, implying interactions between the GFP moiety and UL17, in addition to the covalent link, and perhaps also with UL25. GFP also contacts an adjacent hexon (black arrow). As demonstrated schematically (D), the location of GFP places protein UL17 in the CVSC density, adjacent to UL25 and in the CVSC region closest to the penton. The UL17 C terminus, tagged by GFP, appears in close proximity to UL25.
The contact area between the GFP tag and CVSC density is broad, indicating GFP makes a number of interactions with UL17/UL25 in addition to the covalent one linking it to the C terminus of UL17. A second set of interactions appears to be made between GFP and a neighboring hexon (arrow in Fig. 6C). Freedom of movement of the GFP tag is therefore rather restricted, sufficient to prevent delocalization of the tag density but without abrogating formation of the CVSC heterodimer. Our previous localization of UL25 penton-distal region of the CVSC and the position of the UL17 C-terminal tag in the midriff of CVSC suggest that UL17 occupies the penton-proximal region (Fig. 6D). The different tagging experiments also suggest that the UL17 C terminal forms the main interface with UL25, possibly in a C-terminal-to-C-terminal manner (Fig. 7), although it is not yet possible to precisely delineate UL25 and UL17 density within the CVSC.
Fig. 7.
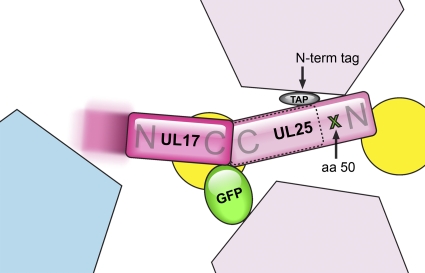
Summary of CVSC subunit tagging. Results of successful attempts to localize and orient CVSC subunits by visualizing bulk tags on HSV-1 capsids are combined in this schematic representation. Color coding of subunits is as for Fig. 5 and 6. Previously we labeled UL25 with GFP at amino acid 50 (aa 50) (4), and the location of this residue is indicated with a green cross. We also labeled the N terminus with a TAP tag (gray oval). The locations of these two tags place UL25 in the distal-most region of the CVSC relative to the penton. The tagging data also support fitting of the crystal structure of UL25 (residues 134 to 580) into the middle part of the CVSC (dashed outline). Tagging of UL17 at the C terminus with GFP localizes UL17 to the CVSC, into the penton-proximal domain. A portion of UL17 nearest to the penton may be flexible (indicated by the motion-blurred extension of UL17). Localization of the UL17 and UL25 tags allows us to predict each protein's orientation within the CVSC (as labeled by “N” and “C,” referring to the N- and C-terminal domains, respectively) with confidence for the C terminus of UL17 and N terminus of UL25 and by inference vice versa. The UL17 and UL25 proteins appear likely to be organized in a C-terminal-to-C-terminal manner.
We also attempted to localize density of the TAP tag from reconstructions of capsids isolated from cells infected with the vUL17-CTAP virus. However, the CVSC density on UL17-CTAP-containing capsids was indistinguishable from that of wild-type CVSC. We suppose that, in contrast to the GFP tag, the TAP tag was not readily visible because it is both smaller than GFP and was less stabilized by serendipitous contact with an adjacent capsid element and thus free to occupy a range of positions. In hindsight, the GFP protein inserted into the UL25 sequence (4) and appended to the UL17 sequence has proven to be valuable for localizing the host protein on the molecular scale (i.e., on capsid reconstructions) as well as on the cellular scale with optical imaging.
A portion of the UL17 protein may be flexible.
The crystal structure of ∼75% of UL25 is available, and docking results combined with tagging results place it toward the middle portion of the CVSC (dashed outline in Fig. 7). The unfilled CVSC density includes a region for the N-terminal 133 residues of UL25 in the vertex-distal-most region of CVSC and the vertex-proximal region, which is insufficient to fully accommodate the 75-kDa UL17 protein. It therefore appears likely that a portion of UL17 is flexible and only poorly visualized in reconstructions. We observe weak densities in the vicinity of pentons (Fig. 6B) in both our density maps for wild-type and tagged C capsids, and these make connections with the penton-proximal portion of the CVSC that we propose contains UL17, although only at low contour levels. These weak densities may represent flexible portions of UL17 that are not stabilized by contact with pentons and so become smeared out with averaging.
Less UL17 protein appears present on capsids than UL25.
Having established that UL17 and UL25 occupy the penton-near and -far domains of the CVSC, respectively, we measured peak density values of these two domains relative to the capsid and background to estimate occupancies. For wild-type capsids, we found 80% occupancy for UL25 and 70% occupancy for UL17, while for the UL17-GFP capsids, we saw a higher occupancy level of 94% for UL25 but 70% for UL17. Generally, the CVSC density has not proven difficult to observe once its presence was established, although herpesvirus capsid reconstructions before 2007 did not include this density. Presumably it is normally present but easily removed during purification, leaving variable amounts on the capsid. Our result also suggests that some UL25 may bind to capsids in the absence of UL17 or that UL25 might remain bound even if UL17 is subsequently lost.
DISCUSSION
The UL25 and UL17 proteins are minor components of the HSV-1 capsid but play essential roles in packaging and retaining the viral genome. The UL25-null mutant appears to package DNA only transiently, yielding empty capsids and near-unit-length genomes, whereas the UL17-null mutant completely abolishes DNA packaging (3, 18, 26, 36). These two proteins, being essential and highly specific to herpesviruses, are therefore of great interest as structural drug targets for new classes of antivirals. We have pursued biochemical and structural characterization of these proteins, aiming to discover their location and conformation on the capsid as well as their binding partners, interfaces, and accessible surfaces. We have previously identified regions of the UL25 N terminus that are essential for binding to capsids (2, 3) and confirmed by cryo-EM that the UL25 protein occupies a portion of the CVSC density that is arrayed around the capsid vertices (4). Its location within the CVSC molecule was constrained to the penton-distal portion by visualization of a GFP tag within the N-terminal portion of the UL25 sequence (Fig. 7). The UL17 protein is proposed to partner with UL25 in comprising the CVSC molecule, presumably occupying the remaining portion of CVSC that is closest to the penton (36). We have now confirmed this by several approaches involving TAP- and GFP-tagged mutants that remain viable on noncomplementing cells, although at yields lower than that of wild-type KOS virus. In particular, we have demonstrated that the tagged UL17 and UL25 proteins are present on A, B, and C capsids and that they copurify in TAP pulldown assays. Further, cryo-EM-based reconstructions of the UL17-GFP mutant virus definitively localize UL17 in the penton-proximal density of the CVSC molecule, in an approximate 1:1 ratio with the UL25 protein and with its C terminus abutting the UL25-containing portion.
The UL17-tagged mutants produce an abundance of empty capsids, as does the UL25-null mutant, and presumably are impaired in the packaging reaction. The evident roles for the UL17 and UL25 proteins in stably packaging DNA would seem at odds with their localization on the capsid exterior around penton vertices, from where packaged viral DNA is presumably inaccessible. Given the imposition of icosahedral symmetry in the density maps calculated for this study, the disposition of CVSC molecules around the unique portal vertex remains unknown, but it is reasonable to assume their presence at this site where the DNA enters the capsid and likely their activity in packaging and maintaining the DNA. However, as is common for virus structural subunits, these proteins also have additional functions. UL25 is implicated in signaling nuclear exit for the DNA-filled mature capsid (15) and in docking at nuclear pores during infection (24). UL17 has been proposed to stabilize the procapsid and prevent premature angularization that would be detrimental to DNA packaging (30). Together, these functions appear consistent with the global coverage of the capsid by the CVSC molecule observed in cryo-EM studies, including its presence on A and B capsids, as well as the mature, DNA-filled C capsid. Detection of the CVSC on procapsids would further support the structural roles in capsid stabilization proposed.
The cryo-EM density maps allow assignment of the UL17 and UL25 proteins to distinct regions of the CVSC molecule as well as their approximate orientation within this molecule and the probable capsid proteins with which they interact. Localization of the UL17 C-terminal GFP tag and UL25 N-terminal GFP tag supports the UL17 and UL25 proteins binding primarily in a C-terminal-to-C-terminal fashion and with these domains of both proteins contacting the underlying penton-proximal triplex (Fig. 7). UL25 extends away from the penton, bridging the gap to the neighboring triplex and contacting one of the adjacent hexons. The density ascribed to UL17 weakens as it extends toward the vertex, suggesting that it is not stabilized by contact with the penton but is instead flexible—although contact at the portal vertex cannot be determined. Nonetheless, occupancy of CVSC density by UL17 is fairly constant at ∼70% for wild-type and UL17-GFP capsids, while the UL25 protein ranges from 80% to 94%. Given the more extensive contacts with the capsid by the UL25 protein, we suspect that the single-triplex and UL25 binding site employed by the UL17 protein may result in greater loss during capsid purification, and indeed the CVSC molecule as a whole appears to be easily dislodged since it is absent from the early structural cryo-EM studies on herpesvirus capsids. While UL17 has been proposed to aid in recruiting UL25 to the capsid, and indeed UL25 does not bind efficiently to capsids in the absence of UL17 (30), UL17 would subsequently appear to be lost from the capsid independently of UL25. However, despite immunodetection of UL25 on capsid preparations of the UL17-null mutant, no CVSC density has been detected in cryo-EM reconstructions, suggesting that the occupancy of the CVSC site is low or that some UL25 may bind to non-CVSC sites on the capsid surface.
The cryo-EM density map indicates that while UL17 is present at relatively high levels in the CVSC, the putative N-terminal penton-proximal domain is smeared out as if it were flexible. This may have functional significance around the portal where the DNA-terminase complex binds and together function as an ATP-driven motor to pump the viral DNA into the capsid and where UL17 may function to support postpackaging cleavage of the DNA. The mechanism of action for such a motor has been modeled for bacteriophages in several different ways, generally involving relative rotation between a capsid-supported stator element and a rotor, and which may exploit the symmetry mismatch between the icosahedral 5-fold vertex and dodecahedral portal complex (13, 28), although alternatives have been proposed (29). Recognition of a terminal DNA sequence or stalling of the motor may separately or together signal cleavage, and the flexibility of the UL17 N-terminal domain may be essential to enable cleavage no matter what part of its cycle the motor and DNA substrate are in. Low-symmetry data analyses will need to target the portal vertex specifically to distinguish it on capsids from the similarly sized penton in order to resolve the conformation of the vertex-proximal CVSC density. In the vicinity of penton vertices, the flexibility of UL17 may have a role in binding the first layers of the irregular tegument and, in particular, the UL37 and UL36 (VP1/2) proteins (11, 17). Nuclear-purified capsids do not address this situation, for which capsids from partially stripped virions (19), or extranuclear capsids, will be needed.
While localization of the UL17 protein on the herpesvirus capsid is an important step in understanding the capsid assembly and DNA packaging processes, specific investigation of its conformation at the portal vertex is a crucial sequel that we are currently pursuing. Nonetheless, identification of the UL17 C termini as accessible and the likely C-terminal-to-C-terminal domain organization with the UL25 protein offer several bases for exploitation in drug targeting. Abolishment of capsid association of either subunit, such as by tightly binding a highly specific molecule or poisoning the CVSC molecule with a defective subunit, would be expected to arrest the formation of C capsids and potentially neutralize viral propagation. In addition to further characterization of CVSC structure and function, we will further identify essential regions for UL17-UL25 dimer formation and for binding to the herpesvirus capsid. This will allow better understanding of interfaces that may potentially be disrupted as well as defining exposed regions that may be targeted for additional labeling experiments that will more comprehensively detail subunit organization.
ACKNOWLEDGMENTS
We gratefully acknowledge expert technical support for electron microscopy from Alexander Makhov. The TAP gene from p-EP-TAP-in was kindly provided by Paul Kinchington (University of Pittsburgh School of Medicine). The UL17-null virus and the UL17 antibody were kindly provided by Joel Baines (Cornell University). The G5 cell line was a gift from Stanley Person and Prashant Desai (Johns Hopkins Medical School).
This work was supported by Public Health Service grant AI060836 from the National Institutes of Health to F.L.H. and by the Commonwealth of Pennsylvania grant SAP 4100031302 to J.F.C.
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Booy F. P., et al. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. U. S. A. 91:5652–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cockrell S. K., Huffman J. B., Toropova K., Conway J. F., Homa F. L. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J. Virol. 85:4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cockrell S. K., Sanchez M. E., Erazo A., Homa F. L. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conway J. F., et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conway J. F., Homa F. L. 2011. Nucleocapsid structure, assembly and DNA packaging of herpes simplex virus, p. 175–193 In Weller S. K. (ed.), Alphaheresviruses. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 6. Conway J. F., Steven A. C. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106–118 [DOI] [PubMed] [Google Scholar]
- 7. Dargan D. J. 1986. The structure and assembly of herpesviruses, vol. 5 Academic Press, London, United Kingdom [Google Scholar]
- 8. Desai P., DeLuca N. A., Glorioso J. C., Person S. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibson W., Roizman B. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gierasch W. W., et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 11. Granzow H., Klupp B. G., Mettenleiter T. C. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grünewald K., et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 13. Hendrix R. W. 1978. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 75:4779–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heymann J. B., Belnap D. M. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 [DOI] [PubMed] [Google Scholar]
- 15. Klupp B. G., Granzow H., Keil G. M., Mettenleiter T. C. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loret S., Guay G., Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luxton G. W., et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McNab A. R., et al. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newcomb W. W., Brown J. C. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newcomb W. W., et al. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newcomb W. W., et al. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499–511 [DOI] [PubMed] [Google Scholar]
- 22. Ormo M., et al. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392–1395 [DOI] [PubMed] [Google Scholar]
- 23. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Preston V. G., Murray J., Preston C. M., McDougall I. M., Stow N. D. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roizman B. 1990. Herpesviridae: a brief introduction, p. 1787–1794 In Fields B. N., Knipe D. M. (ed.), Fields virology, 2nd ed. Raven Press, New York, NY [Google Scholar]
- 26. Salmon B., Cunningham C., Davison A. J., Harris W. J., Baines J. D. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scholtes L., Baines J. D. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pUL17/pUL25 complex of herpes simplex virus 1. J. Virol. 83:12725–12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Simpson A. A., et al. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun S., et al. 2008. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135:1251–1262 [DOI] [PubMed] [Google Scholar]
- 30. Thurlow J. K., Murphy M., Stow N. D., Preston V. G. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tischer B. K., Smith G. A., Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 32. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 33. Tivol W. F., Briegel A., Jensen G. J. 2008. An improved cryogen for plunge freezing. Microsc. Microanal. 14:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trus B. L., et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 35. Trus B. L., et al. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trus B. L., et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wills E., Scholtes L., Baines J. D. 2006. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J. Virol. 80:10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan X., Dryden K. A., Tang J., Baker T. S. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan X., Sinkovits R. S., Baker T. S. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Z. H., et al. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026–1030 [DOI] [PubMed] [Google Scholar]