Abstract
The retinoic acid-inducible gene I (RIG-I)-like receptors (RLR) comprise three homologues: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). They activate the host interferon (IFN) system upon recognition of viral RNA pathogen-associated molecular patterns (PAMPs) in the cytoplasm. Bioinformatic analysis of the sequenced vertebrate genomes suggests that the cytosolic surveillance system is conserved in lower vertebrates, and recent functional studies have confirmed that RIG-I is important to fish antiviral immunity. In this study, we have identified MDA5 and LGP2 homologues from rainbow trout Oncorhynchus mykiss and an additional LGP2 variant with an incomplete C-terminal domain of RIG-I. Trout MDA5 and LGP2 were constitutively produced in fibroblast and macrophage cell lines and upregulated by poly(I:C), recombinant IFN, or infection by RNA viruses (viral hemorrhagic septicemia virus and salmon alphavirus) with a single-stranded positive or negative genome. Overexpression of MDA5 and LGP2 but not of the LGP2 variant resulted in significant accumulation of Mx transcripts in cultured cells, which correlated with a marked enhancement of protection against viral infection. These results demonstrate that both MDA5 and LGP2 are important RLRs in host surveillance against infection of both negative and positive viruses and that the LGP2 variant with a deletion of 54 amino acids at the C terminus acts as a negative regulator for LGP2-elicited antiviral signaling by competing for the viral RNA PAMPs. Interestingly, MDA5 expression was not affected by overexpressed LGP2 in transfected cells and vice versa, suggesting that they likely act in parallel as positive regulators for IFN production.
INTRODUCTION
Toll-like receptors (TLR) and retinoic acid-inducible gene I (RIG-I)-like helicases (RLH) are two major families of pattern recognition receptors (PRRs) that recognize cytosolic viral RNA pathogen-associated molecular patterns (PAMPs) in vertebrates (11, 22). For example, TLR-3 and -7 are endosome-associated cellular PRRs recognizing viral PAMPs released from the uncoating process. In contrast, RIG-I-like helicases sense virus-derived RNA molecules in other regions of the cytoplasm. Upon activation, TLRs and RLHs trigger production of type I interferons (IFNs), leading to an enhanced antiviral state of the host cells.
The RLH family contains three members, RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), all of which share homologous core structural domains, including a DExD/H box helicase domain, helicase C-terminal domain, and C-terminal domain (CTD) of RIG-I (11, 22). The central helicase domain contains six conserved DExD/H helicase motifs and is involved in translocation, ATP hydrolysis, and RNA binding (36). The CTD, also referred to as the repressor domain or carboxy-terminal regulatory domain (RD), has proven to contain multiple diverse functions, such as signaling repression, RNA recognition, and protein dimerization (5, 25). RIG-I and MDA5 have additional caspase activation and recruitment domains (CARDs) at the N terminus that are well conserved in proteins involved in immune signaling, cell differentiation, and apoptosis (1, 18). Activated RIG-I and MDA5 act on the mitochondria via interaction of CARDs between RIG-I/MDA5 and mitochondrial antiviral signaling protein (MAVS), a mitochondrial receptor which also contains a CARD, facilitating phosphorylation of interferon regulatory factors (IRF) 3 and 7 which are key transcription factors involved in triggering IFN production (12, 26, 35).
While it is certain that RIG-I and MDA5 serve as PRRs for viral PAMPs, the role of LGP2 in mediating an antiviral response is contradictory. Since LGP2 lacks CARDs at the N-terminal region, it is believed that it is unable to interact with MAVS and hence may serve as a negative mediator for RIG-I/MDA5-activated antiviral signaling (24, 31, 35). Overexpression of LGP2 was shown to result in decreased IFN production in cells following transfection with poly(I:C) or viral infection. Furthermore, LGP2-deficient mice exhibited enhanced resistance to viral infection, and embryonic fibroblasts isolated from these mice displayed enhanced IFN expression in response to synthetic double-stranded RNA. However, positive regulatory roles of LGP2 in RIG-I/MDA5-activated antiviral signaling have also been reported in LGP2-deficient mice, which lost the ability to synthesize type I IFNs and were unable to mount efficient antiviral responses against infection with encephalomyocarditis virus (32). A more recent study has provided further evidence that the ATPase domain of LGP2 is required for the LGP2-positive regulatory role in mediating RIG-I/MDA5-dependent antiviral responses (26).
In teleost fish, orthologs of human TLRs which recognize viruses have been reported in zebrafish (4, 16, 21), rainbow trout (19, 23), Atlantic salmon (28), grass carp (29), and gilthead sea bream (6). Concerning RLRs, recent studies by our group and others have demonstrated that fish also possess RLR molecules, among which RIG-I and MAVS have been shown to activate the interferon response via a mitochondrion-associated signaling pathway (2, 14, 36). Overexpression of fish RIG-I and MAVS in cultured cells upregulates expression of IFN and IFN-stimulated genes and enhances resistance against not only RNA viruses but also DNA viruses (2, 14). As in mammals, the CARDs at the N-terminal region in both RIG-I and MAVS and the CTD in RIG-I are essential for their antiviral activities. In the present study, we identified two other members of the RLR family, MDA5 and LGP2, known in mammals to be involved in mediating antiviral immunity, from rainbow trout (Oncorhynchus mykiss) and demonstrated that they were induced at the transcriptional level by viral infection, exogenous cytosolic double-stranded RNA (dsRNA), and stimulation with type I IFN. Induction of MDA5 and LGP2 expression was confirmed at the protein level by Western blot analysis using polyclonal antibodies produced against MDA5 and LGP2 peptides. To further clarify the role of trout MDA5 and LGP2, stable trout cell lines overexpressing these RLR molecules were generated and shown to exhibit enhanced resistance to infection against an RNA virus (viral hemorrhagic septicemia virus). Strikingly, we found that a novel LGP2 variant, containing a shortened CTD with a deletion of 54 amino acid (aa) residues at the C terminus, was unable to trigger antiviral responses and downregulated LGP2-induced Mx gene expression. Furthermore, overexpression of MDA5 had no effect on LGP2 in transfected cells and vice versa. Taken together, our data indicate that MDA5 and LGP2 act as independent positive regulators of the IFN response in fish cells, while the LGP2 variant antagonizes LGP2 function.
MATERIALS AND METHODS
Cells and viruses.
The RTG-2 cells were maintained in L-15 medium supplemented with 10% fetal bovine serum (FBS) (Sigma), 100 U/ml penicillin (P), and 100 μg/ml streptomycin (S) at 20°C. The RTS-11 cells were maintained in L-15 medium supplemented with 30% FBS and P-S at 20°C. Salmon alphavirus (SAV) (isolated from a field sample from Inch Kenneth, Scotland [unpublished]) was propagated in CHSE-214 cells for 7 days as described previously (15). Viral hemorrhagic septicemia virus (VHSV; isolate DK-F1) was propagated at 15°C on 90% confluent monolayers of BF-2 cells (bluegill fry; ATCC no. CCL-91) and cultured in Eagle's minimum essential medium (EMEM) supplemented with 5% FBS. Once a full cytopathic effect (CPE) was observed, the virus (50% tissue culture infective dose [TCID50]) was titrated using BF-2 cells by endpoint dilution and stored at −80°C in aliquots.
Cloning and sequencing of trout MDA5 and LGP2.
To search for salmonid MDA5 and LGP2 homologues, expressed sequence tag (EST) sequences in the NCBI database were searched using fish MDA5 and LGP2 protein sequences (37) as bait sequences for BLASTN analysis. Several candidate ESTs were found with high sequence homologies to known MDA5s, among which EST sequences GE782837 (Salmo salar) and GE823089 (Oncorhynchus mykiss) aligned well with the N- and C-terminal sequences of MDA5, respectively. Forward (MDA5-F1) and reverse (MDA5-R1 and MDA5-R2) primers were designed in the 5′- and 3′-end untranslated region (UTR) to amplify the full-length coding sequence of trout MDA5 by seminested reverse transcription (RT)-PCR. For LGP2, the full-length sequence of S. salar was compiled using the CAP3 sequence assembly program (http://pbil.univ-lyon1.fr/cap3.php) with the GE781785, DW582483, DW538655, CX355611, DN165806, DW567942, and CK880416 ESTs, and primers LGP2-F1 and LGP2-R1 located in the 5′- and 3′-end UTR were used for RT-PCR amplification of the full-length coding sequence of trout LGP2. Hot-start PCR was performed for amplification of both molecules under the following conditions: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 25 s, 57°C for 25 s, and 72°C for 2 min; and 1 cycle of 72°C for 10 min. The cDNA template was generated from RTS-11 cells stimulated with 50 μg/ml poly(I:C) for 4 h. The PCR fragments were cloned into pGEM T Easy vector (Promega) and subsequently sequenced by Eurofins MWG Operon.
RNA extraction, cDNA synthesis, and real-time PCR analysis.
Total RNA was extracted using the RNASTAT reagent (AMS Biotechnology [Europe] Ltd.) and treated with RNase-free DNase I (Fermentas Life Sciences, Germany) and RiboLockTM RNase inhibitor (Fermentas Life Sciences, Germany) according to the manufacturer's instructions. The DNase I-treated RNA was reverse transcribed into cDNA using a first-strand cDNA synthesis kit (Fermentas Life Sciences, Germany) and kept at −20°C for real-time PCR analysis.
Primers used for real-time PCR analysis are listed in Table 1. Real-time PCR was performed on a Roche LightCycler 480 using the following conditions: 1 cycle of 95°C for 10 min and 45 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 40 s. Each sample was run in triplicate in a 96-well plate and the mean value recorded. The relative expression of target genes was then normalized to the expression of elongation factor 1α (EF-1α) and expressed as arbitrary units. Fold changes were calculated by comparison to the corresponding controls. Three independent experiments were conducted for statistical analysis.
Construction of MDA5 and LGP2 plasmids.
For expression of recombinant proteins, the coding regions of MDA5, LGP2a, and LGP2b were amplified by PCR using pQE30MDA5F/pQE30MDA5R, pQE30LGP2F/pQE30LGP2aR, and pQE30LGP2F/pQE30LGP2bR (Table 1), respectively. The PCR products of LGP2a and LGP2b were digested with BamHI/KpnI and ligated into digested pQE30 vector, giving rise to plasmids pQE30-LGP2a and pQE30-LGP2b. Similarly, the amplified MDA5 fragment was digested with SacI/HindIII and cloned into pQE30, generating pQE30-MDA5.
Table 1.
Primer sequences used in this study
| Primer name | Sequence | Application |
|---|---|---|
| MDA5-F1 | TGAAAGTGCTTGCATTGACGGGA | Gene cloning |
| MDA5-R1 | CACACTGCATCAATGACGGCTTTC | Gene cloning |
| MDA5-R2 | CTACCACAGATACACGGAGCTG | Gene cloning |
| LGP2-F1 | GTGGACCAATAGACCGGTAGGA | Gene cloning |
| LGP2-R1 | CATAGGACTATGACACACTTCAGGA | Gene cloning |
| PQE30MDA5F | GTGGAGCTCATGGCCGCTGACAAAGATAAC | pQE30-MDA5 |
| PQE30MDA5R | GACAAGCTTTTACATTGATTCTTCATCCTC | pQE30-MDA5 |
| PQE30LGP2F | GTGGGATCCATGACAGACTTTGGGCTG | pQE30-LGP2a pQE30-LGP2b |
| PQE30LGP2aR | GACGGTACCTCAATCCAACATGTTAGGG | pQE30-LGP2a pQE30-LGP2b |
| PQE20LGP2bR | GACGGTACCCTAGAGATTAAAGGTCAAC | pQE30-LGP2a pQE30-LGP2b |
| FlagMDA5F | GTGAGATCTGATGGCCGCTGACAAAG | pMDA5-FLAG |
| FlagMDA5R | TTGGGTACCATTGATTCTTCATCCTCTG | pMDA5-FLAG |
| ptGFPMDAF | GTGAGATCTGCGATGGCCGCTGACAAAG | pTurbo-MDA5-GFP |
| GFPMDAR1 | GACGGTACCATTGATTCTTCATCCTCTG | pTurbo-MDA5-GFP |
| ptGFPLGP2F | GTGAGATCTACGATGACAGACTTTGGGCTG | pTurbo-LGP2a-GFP pTurbo-LGP2b-GFP |
| GFPLGP2aR | GACGGTACCCAATCCAACATGTTAGGGAA | pTurbo-LGP2a-GFP pTurbo-LGP2b-GFP |
| GFPLGP2bR | GACGGTACCAGATTAAAGGTCAACCTCACC | pTurbo-LGP2a-GFP pTurbo-LGP2b-GFP |
| ptGFPMDAF | GTGAGATCTGCGATGGCCGCTGACAAAG | ptGFP1-MDA5 |
| ptGFPMDAR | GACGGTACCTTACATTGATTCTTCATCCTC | ptGFP1-MDA5 |
| ptGFPLGP2F | GTGAGATCTACGATGACAGACTTTGGGCTG | ptGFP1-LGP2a ptGFP1-LGP2b |
| ptGFPLGP2R | GACGGTACCTCAATCCAACATGTTAGGG | ptGFP1-LGP2a ptGFP1-LGP2b |
| MDA5F | AGAGCCCGTCCAAAGTGAAGT | Real-time PCR |
| MDA5R | GTTCAGCATAGTCAAAGGCAGGTA | Real-time PCR |
| LGP2aF | ACACCTGCTCTTTCCGTCAC | Real-time PCR |
| LGP2aR | GTTGGCTGGATGTCCTTTGG | Real-time PCR |
| LGP2bF | GTGGCAGGCAATGGGGAATG | Real-time PCR |
| LGP2bR | CCTCCAGTGTAATAGCGTATCAATCC | Real-time PCR |
| EF-1αF | CAAGGATATCCGTCGTGGCA | Real-time PCR |
| EF-1αR | ACAGCGAAACGACCAAGAGG | Real-time PCR |
| MDA5Rexo | GAGCCGCGGTACCTTACATTG | Real-time PCR |
| LGP2Rexo | GAGCCGCGGTACCTCAATCCAAC | Real-time PCR |
| LGP2bFexo | AGCACCTTTCGCCAGAGTAGCC | Real-time PCR |
| LGP2aFall | TGGCAGGCAATGGGGAATG | Real-time PCR |
| LGP2aRall | TAAACTCCTCCACTGTAAACTCAAC | Real-time PCR |
To generate expression vectors of MDA5-green fluorescent protein (GFP) and LGP2-GFP fusion proteins for cellular localization studies, the full-length coding sequences of MDA5, LGP2a, and LGP2b were amplified with primer pairs ptGFPMDAF/GFPMDAR1, ptGFPLGP2F/GFPLGP2aR, and ptGFPLGP2F/GFPLGP2bR (Table 1), respectively, and inserted into the BglII and KpnI sites of the pTurboGFP-N vector (Evrogen) to construct pTurbo-MDA5-GFP, pTurbo-LGP2a-GFP, and pTurbo-LGP2b-GFP. For overexpression studies of MDA5 and LGP2, expression vector ptGFP1 was modified from the pTurboGFP-N vector and contained two sets of CMV promoter and SV40 3′UTR to drive expression of the target gene product and GFP as separate proteins rather than as a fusion protein. The transfected cells expressing exogenous MDA5 and LGP2 were GFP positive. For antiviral assays, the full-length coding sequences of MDA5, LGP2a, and LGP2b were amplified with primer pairs ptGFPMDAF/ptGFPMDAR (for MDA5) and ptGFPLGP2F/ptGFPLGP2R (for LGP2a and LGP2b) and inserted into the BglII and KpnI sites of the ptGFP1 vector to make constructs ptGFP1-MDA5, ptGFP1-LGP2a, and ptGFP1-LGP2b for overexpression in stably transfected RTG-2 cells. Lastly, the coding region of MDA5 was amplified with primers FlagMDA5F/FlagMDA5R and inserted into the BglII/KpnI sites of p3xFLAG-CMV-14 expression vector (Sigma) to construct plasmid pMDA5-FLAG.
Expression analysis of MDA5 and LGP2 in RTG-2 and RTS-11 cells by real-time PCR.
The cells were passaged into fresh flasks 2 days before being stimulated for 4 h and 24 h with either 50 μg/ml poly(I:C) (Sigma-Aldrich) or 20 ng/ml recombinant trout type I IFN2 protein (38). For poly(I:C) transfection, the RTG-2 cells (5 × 106 cells in 100 μl Nucleofector solution) were electroporated with 5 μg poly(I:C) using an Amaxa Nucleofector II transfection system (Lonza) under Programme T20, washed immediately with 5 ml Hanks balanced salt solution (HBSS) buffer, and cultured at 20°C for 4 h or 24 h. The cells were collected for RNA extraction and real-time PCR analysis as described previously (3). The relative expression of target genes was normalized to the expression of EF-1α and expressed as arbitrary units or fold change relative to the corresponding control group. The means of results from three independent experiments were used for statistical analysis, as described below.
Expression analysis of MDA5 and LGP2 in RTG-2 cells by Western blot analysis.
Polyclonal antibodies against trout MDA5 and LGP2 were generated in rabbits by Genscript using standard procedures with synthetic peptides for MDA5-1 (CEHLDSRRKEGRPGK), LGP2-1 (PRKRFDIVDRRPQDC), and LGP2-2 (CETPEGRKLAKKWKN). Other antibodies included mouse monoclonal anti-Arabidopsis actin (Thermo Scientific), rabbit polyclonal anti- TurboGFP (Evrogen), and horseradish peroxidase-conjugated goat anti-mouse IgG (Thermo Scientific).
Recombinant proteins of trout MDA5 and LGP2 were produced to verify the polyclonal antibodies generated against the synthetic peptides. For this, the full-length cDNA fragments of MDA5, LGP2a, and LGP2b were amplified using the primers listed in Table 1, inserted into the pQE30 vector (Qiagen), and expressed in Escherichia coli M15 cells (Qiagen). The recombinant MDA5 and LGP2 proteins were produced as previously described and purified under denaturing conditions (38). The recombinant proteins were separated on a 4 to 12% precast SDS-PAGE gel (Invitrogen) and transferred onto a polyvinylidene difluoride membrane. Western blotting was performed using a WesternBreeze chemiluminescent Western blot immunodetection kit (Invitrogen). The primary antibodies were diluted 400-fold.
To examine the expression of MDA5 and LGP2 at the protein level in RTG-2 cells, the cells were passaged into 25-cm2 flasks and cultured at 20°C until they reached 80% confluence (2 days), the culture media were replaced by fresh media, and equal volumes of PBS or poly(I:C) (50 or 100 μg/ml) were added. After 24 h, the cells were harvested and resuspended in 1× SDS-PAGE sample buffer. Cell lysates were boiled at 95°C for 5 min, centrifuged at 13,000 × g for 1 min, and loaded immediately onto the SDS-PAGE gels. After gel electrophoresis, a Western blot analysis was performed as described previously by using polyclonal antibodies against MDA5 and LGP2. Actin protein was detected with mouse monoclonal antiactin antibody (1:1,000, vol/vol) and the secondary antibody (horseradish peroxidase-conjugated goat anti-mouse IgG [1:2,000, vol/vol]) as an internal control to normalize the amount of protein loaded onto the gels.
Kinetics of gene expression in head kidney of fish infected with VHSV.
VHSV has a single-stranded negative RNA genome and can infect a wide range of marine and freshwater fish species, including salmonids. In this study, fish (approximately 15 g) were reared in the Marine Scotland Science Marine Laboratory in Aberdeen, Scotland, and acclimated for 14 days prior to challenge. Fish were stocked in 30-liter tanks of freshwater maintained at 10°C, with a flow rate of 50 liters/h with aeration. Fish were starved for 24 h prior to the experiment, during which they were maintained on a commercial diet. Fish in duplicate tanks were anesthetized with methane tricaine sulfonate (MS222; Sigma; 0.1 mg/ml) and injected intraperitoneally (i.p.) with 100 μl of the isolate DK-F1 (1 × 107 TCID50 per fish) or mock infected with control medium. Head kidney tissues were sampled for preparation of total RNA for real-time PCR analysis.
Kinetics of gene expression in TO cells infected with SAV.
SAV, containing a single-stranded positive RNA genome, is known to cause sleeping pancreatic disease in salmonids and has recently been shown to upregulate IFN expression (3). To determine whether MDA5 and LGP2 are involved in antiviral responses against SAV, we assessed gene expression of MDA5 and LGP2 in Atlantic salmon TO cells infected with SAV. Since the salmon MDA5 and LGP2 (compiled with EST sequences) share significant nucleotide sequence identities in the coding regions (>95.0% for both genes) and the regions used for primer design for trout MDA5 and LGP2 expression are almost identical to the corresponding sequences of the Atlantic salmon genes, the trout primers were tested to confirm that they worked for real-time PCR analysis of these Atlantic salmon genes. Atlantic salmon TO cells (passage no. P95) (33) were propagated in L-15 medium supplemented with 5% FBS, 2 mM l-glutamine, and 1% nonessential amino acids. Stock cultures were incubated at 20°C for 7 to 21 days. Cells were passaged at 50 to 60% confluence and cultured for 24 to 48 h before viral challenge. Triplicate cultures of TO cells at approximately 80% confluence were infected with SAV (F93-125 isolate [subtype 1]) (7, 34) at a multiplicity of infection (MOI) of 0.1 in 6-well plates at 15°C. Parallel uninfected cells were set up in the same format under the same conditions. At days 1, 2, 3, 4, 6, and 8, the cells were detached with trypsin-EDTA (Invitrogen) and centrifuged at 400 × g for 5 min. The cell pellets were drained and kept at −80°C. RNA was extracted from the infected cell cultures using the Allprep RNA/DNA/protein kit (Qiagen) according to the manufacturer's instructions. The RNA was eluted in 50 μl RNase-free distilled water (dH2O). RNA was reverse transcribed to cDNA using the TaqMan reverse transcription reagent kit (ABI) with oligo(dT)16 as follows: 9.63 μl of total RNA (approximately 0.5 μg) and 1.25 μl 50 μM oligo(dT)16 were mixed and heated to 70°C for 10 min and chilled on ice. The final volume was adjusted to 25 μl by adding master mix comprised of the following: 1× RT buffer (25 mM Tris-HCl [pH 8.3], 37.5 mM KCl, 5.5 mM MgCl2), and 0.5 mM each deoxynucleoside triphosphate (dNTP), 0.4 U RNase inhibitor, and 1.25 U Multiscribe reverse transcriptase. The reaction mixtures were incubated at 48°C for 90 min, heat inactivated at 95°C for 5 min, and stored at −80°C until use. Real-time PCR analysis was carried out as outlined above.
Localization of MDA5 and LGP2 in RTG-2 cells.
The RTG-2 cells were passaged into fresh flasks and cultured for 24 h. Two micrograms of plasmid constructs (pTurboGFP-N, pTurbo-MDA5-GFP, pTurbo-LGP2a-GFP, or pTurbo-LGP2b-GFP) was transfected into 5 × 106 cells using the Amaxa Nucleofector II transfection system (Lonza) under Programme T20. The cells were transferred into 6-well plates and cultured at 20°C with 3 ml L-15 medium supplemented with 10% FBS and P-S. Approximately 24 h posttransfection, the cells were collected for confirmation of LGP2a-GFP and LGP2b-GFP fusion proteins by Western blotting. Subsequently, the cells were examined under a fluorescent microscope and photographed.
Antiviral activities of MDA5, LGP2a, and LGP2b.
The RTG-2 cells were transfected with ptGFP1, ptGFP1-MDA5, ptGFP1-LGP2a, or ptGFP1-LGP2b as described above. After 36 h, the cells were cultured in medium containing 800 μg/ml G-418 for 2 weeks to enrich for transfected cells and then maintained in the medium containing 200 μg/ml G-418. The cells were checked under a fluorescent microscope every 2 weeks, and when a minimum of ∼80% of cells were GFP positive, they were deemed suitable for assessment of their antiviral state. For antiviral assays, the transfected cells were seeded into a 96-well plate (Nunc) at approximately 80% confluence. The cells were left to attach overnight in 100 μl per well L-15 medium (Invitrogen), 10% FBS (Nalgene), and 200 μg/ml G-418 at 23°C and acclimatized at 15°C for 24 h prior to viral infection. Ten microliters of supernatant containing VHSV (isolate DKF3592), 1× TCID50, previously adapted to RTG-2 cells, was added to column 1 and serially diluted 10-fold in the following columns. The plates were incubated at 15°C for 2 weeks, drained, fixed in 10% formalin for 10 min at room temperature, and then stained with 0.05% (wt/vol) crystal violet (Sigma) for 30 min. The plates were photographed under a light box. To quantify stained cells, the crystal violet was dissolved in 100 μl 1% SDS solution for 5 min in an orbital shaker at 150 rpm. The absorbance was read at a 562-nm wavelength using a Bio Lab-Tek plate reader. The relative OD was calculated by dividing the average OD of the infected wells (n = 6) with that of the uninfected control (n = 24). In addition, in parallel uninfected cultures, the cells were also collected for examining gene expression of MDA5, LGP2, and Mx by real-time PCR.
Inhibitory activity of LGP2b on LGP2a elicited antiviral responses.
RTG-2 cells (5 × 106 cells) were cotransfected with 5 μg poly(I:C) and ptGFP1 (2 μg), ptGFP1-LGP2a (2 μg), ptGFP1-LGP2b (2 μg), or ptGFP1-LGP2a (2 μg) plus ptGFP1-LGP2b (2 μg). The cells were washed immediately with culture medium and incubated at 20°C for 6 h. Cells were harvested for extraction of total RNA and real-time PCR analysis of gene expression.
Interaction of MDA5 and LGP2 in RTG-2 cells.
To study whether overexpression of MDA5 affects LGP2 expression, 5 × 106 RTG-2 cells were transfected with 2 μg of p3xFLAG-CMV-14 vector (Sigma), pMDA5-Flag, ptGFP1, or ptGFP1-LGP2a plasmid as described above. The cells were immediately washed with HBSS buffer, cultured in 10% FBS containing L-15 medium at 20°C for 48 h, and collected for protein extraction using cell extraction buffer (Invitrogen). Western blotting was performed to determine trout MDA5 and LGP2 proteins as described above.
Binding affinity of MDA5 and LGP2 with poly(I:C).
Poly(C)-conjugated agarose beads (Sigma) and poly(I) were resuspended in buffer containing 50 mM Tris (pH 7.0) and 150 mM NaCl to a final concentration of 2 mg/ml and mixed at a ratio of 1:2 (vol/vol). The mixture was incubated at 4°C overnight, centrifuged at 1,000 × g for 1 min, and washed once. The beads were resuspended in the same buffer as a 50% slurry and stored at 4°C for use.
Poly(I:C) pulldown assays were performed as described by Sumpter et al. (30). The poly(I:C)-coated beads were equilibrated in binding buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 1% NP-40 and resuspended as a 10% slurry. The beads were then incubated with the purified recombinant MDA5, LGP2a, or LGP2b at 4°C for 1 h, centrifuged at 1,000 × g, rinsed three times with binding buffer, resuspended in three volumes of 1× SDS-PAGE sample buffer, and boiled for 3 min before being loaded onto an SDS-PAGE gel for Western blotting. For competition experiments, the poly(I:C)-coated beads were incubated with recombinant proteins and 50 μg/ml of poly(I:C).
Statistical analysis.
The relative expression of target genes was normalized to the expression of EF-1α and expressed as arbitrary units or fold change relative to the corresponding control group. The means of results from at least three independent experiments were used for statistical analysis using the paired Student t test. P values of <0.05 were considered to be significantly different.
RESULTS
Sequence analysis of trout MDA5 and LGP2.
The cloned trout MDA5 cDNA contains a 3,009-nucleotide (nt) open reading frame (ORF) (GenBank accession no. FN396357), encoding a protein of 1,002 aa with a predicted molecular mass of 113.2 kDa and a pI of 5.93. Sequence analysis of the MDA5 protein in the Pfam HMM database (Pfam version 24.0; http://pfam.sanger.ac.uk) identified one conserved N-terminal CARD (aa 125 to 197) with an E value of 0.00063, followed by a DExD/H box helicase domain (aa 306 to 496), a helicase C-terminal domain (aa 712 to 796), and a RIG-I CTD (C-terminal domain of RIG-I, aa 872 to 992) (see Fig. S1A in the supplemental material). Although it was not detected by the Pfam program, a distantly related CARD located near the N terminus (aa 12 to 98) was noticeable when the protein was aligned with the human MDA5. The two CARDs, the DExD/H box domain, helicase C-terminal domain, and CTD of trout MDA5 showed similarities of 58.3%, 77.0%, 80.0%, and 69.7% with the corresponding domains of human MDA5 and of 64.2 to 66.3%, 85.9 to 89.0%, 81.2 to 89.4%, and 77.0 to 86.1% with the corresponding domains of fish MDA5, respectively. Furthermore, other conserved features include an ATP binding motif (PTGSGKT), an ATPase motif (DECH), and an RNA unwinding motif (TAS) located within the DExD/H domain of trout MDA5. An RNA binding motif (QARGRGRA) was found within the helicase C domain of trout MDA5.
The identified trout LGP2 cDNA (named LGP2a) is 2,194 bp in length, which encodes a protein of 678 aa (GenBank accession no. FN396358). An alternatively spliced form of 3,230 bp (named LGP2b; GenBank accession no. FN396359) was also sequenced, with an intron of 1,040 bp retained at the 3′-end region of the ORF, resulting in early termination of translation. As a result, LGP2b is 54 aa shorter than LGP2a (see Fig. S1B in the supplemental material). TMpred analysis showed that the LGP2a and LGP2b proteins contained a transmembrane helix at aa 549 to 568. Pfam HMM analysis predicted a typical type III restriction enzyme Res subunit domain (aa 3 to 176) at the N terminus, which is highly homologous to the DExD/H helicase domain of MDA5. The helicase C-terminal domain (aa 396 to 477) and the RIG-I CTD (aa 552 to 672) were also detected both in LGP2a and LGP2b (see Fig. S1B). The type III restriction enzyme subunit, conserved C-terminal helicase domain, and CTD of trout LGP2a/LGP2b have similarities of 73.0%, 74.4%, and 63.1% with the corresponding domain of human LGP2 and of 62.6 to 83.9%, 83.1 to 92.8%, and 63.6 to 74.8% with the corresponding domain of fish LGP2. In addition, a conserved ATP binding motif (PTGGGKT), an ATPase motif (DECH), an RNA unwinding motif (TAS), and an RNA binding motif (QASGRARA) were predicted in the trout LGP2a and LGP2b proteins.
Expression of trout MDA5 and LGP2 after stimulation with poly(I:C) or type I IFN.
Constitutive expression of trout MDA5, LGP2a, and LGP2b was detected in fibroblast-like RTG-2 and monocyte/macrophage-like RTS-11 cells by real-time PCR. The expression level of trout MDA5, LGP2a, and LGP2b in RTG-2 cells was higher than that in RTS-11 cells. In RTG-2 cells, the MDA5 transcript level was the lowest among the 3 genes; however, it was higher relative to LGP2a/LGP2b expression in the RTS-11 cells (Fig. 1A).
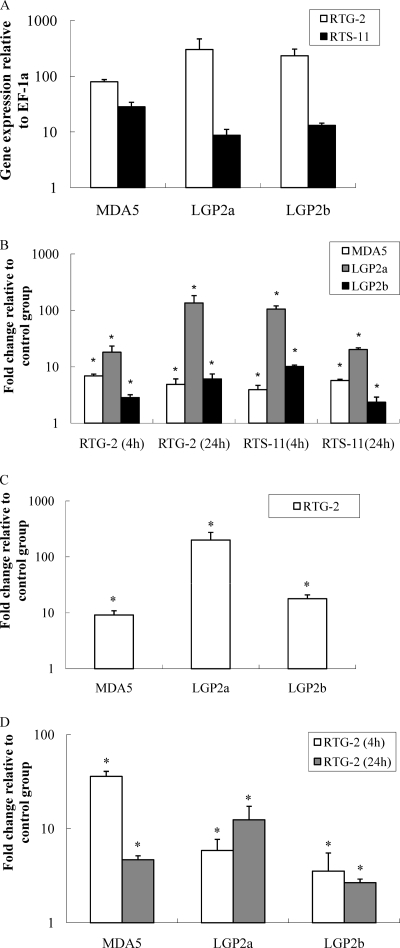
Fig. 1.
Trout MDA5 and LGP2 are upregulated at the transcript level in cultured cells in response to intracellular/extracellular poly(I:C) and type I IFN. (A) The expression of trout MDA5 and LGP2 in normal RTG-2 and RTS-11 cell lines; (B) the effect of extracellular poly(I:C) stimulation on trout MDA5 and LGP2 expression in RTG-2 and RTS-11 cell lines; (C) the effect of transfected poly(I:C) on trout MDA5 and LGP2 expression in RTG-2 cells; (D) the effect of recombinant trout IFN2 protein stimulation on trout MDA5 and LGP2 expression in RTG-2 cells. The RTG-2 and RTS-11 cells were stimulated for 4 h with either 50 μg/ml poly(I:C) or 20 ng/ml trout type I rIFN2 protein. In addition, the RTG-2 cells (5 × 106 cells) were electroporated with 5 μg poly(I:C), washed, and cultured for 24 h. The cells were collected and used for RNA extraction and real-time PCR analysis. The relative expression of target genes was normalized to the expression of EF-1α and expressed as arbitrary units or fold changes relative to the appropriate control group. The means of results from three independent experiments are shown, and bars indicate the standard errors of the means (SEMs). Asterisks indicate where the increase of gene expression is significant (P < 0.05).
To study the effect of the double-stranded synthetic RNA, poly(I:C), a known activator of MDA5 and LGP2 in mammals, the trout RTG-2 and RTS-11 cells were stimulated by the addition of poly(I:C) (50 μg/ml) into the culture medium for 4 h and 24 h. At 4 h poststimulation, poly(I:C) stimulation resulted in approximately 7-, 18-, and 3-fold increases of transcript levels for MDA5, LGP2a, and LGP2b in RTG-2 cells and approximately 4-, 105-, and 10-fold increases in RTS-11 cells, respectively (Fig. 1B). The stimulatory effects were sustained at 24 h for all three genes in both cell types. It seemed apparent that LGP2 was most inducible in response to poly(I:C) stimulation. To examine the effect of intracellular poly(I:C) on MDA5 and LGP2 gene expression, the RTG-2 cells were electroporated with poly(I:C), washed with PBS immediately after transfection to remove poly(I:C) from the culture medium, and incubated for 24 h. MDA5, LGP2a, and LGP2b were all induced, with approximately 9-, 200-, and 18-fold increases of expression levels, respectively (Fig. 1C).
In mammals, activation of RIG-I/MDA5 by viral RNA PAMPs induces IFN expression, and these surveillance mechanisms are enhanced by the synthesized IFNs via a positive feedback. In this study, the RTG-2 cells stimulated with 20 ng/ml rIFN2 for 4 h and 24 h exhibited increased expression for MDA5, LGP2a, and LGP2b (Fig. 1D). Notably, a 36-fold increase in MDA5 transcript level was detected at 4 h, while the increase was reduced to 5-fold at 24 h.
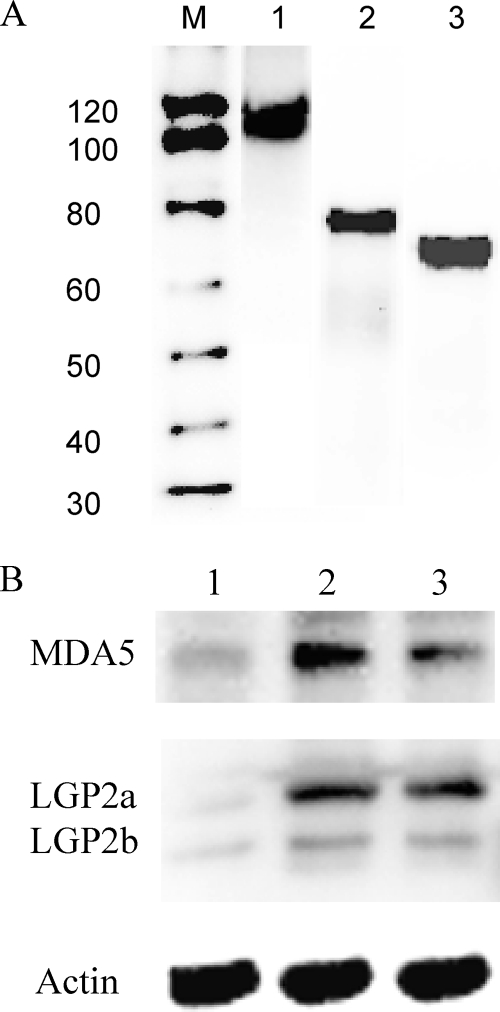
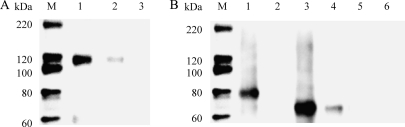
To confirm induction of MDA5 and LGP2 detected by real-time PCR, polyclonal antibodies were raised against synthetic MDA5 and LGP2 peptides for Western blot analysis. The polyclonal antibodies reacted well with the recombinant proteins produced in bacteria (Fig. 2 A). These antibodies were used to analyze MDA5 and LGP2 protein expression in RTG-2 cells after poly(I:C) stimulation by Western blotting. In control cells, a low level of constitutive expression was detected for MDA5 and the two isoforms of LGP2. Induction of all three proteins after 24 h stimulation with either 50 or 100 μg/ml poly(I:C) was apparent (Fig. 2B). These data confirmed the transcriptional results obtained by real-time PCR.
Fig. 2.
Trout MDA5 and LGP2 are upregulated at the protein level in RTG-2 cells after stimulation with poly(I:C). (A) Verification of rabbit polyclonal antibodies generated against trout MDA5, LGP2a, and LGP2b by Western blotting. Four hundred nanograms of purified recombinant trout MDA5 (lane 1), LGP2a (lane 2), and LGP2b (lane 3) was applied, and bands of the correct sizes were visualized after blotting. Lane M, molecular weight markers. (B) Protein expression of MDA5, LGP2a, LGP2b, and actin in unstimulated cells (lane 1) and cells stimulated with 50 μg/ml (lane 2) or 100 μg/ml (lane 3) of poly(I:C) for 24 h. Note that the actin bands are similar between lanes.
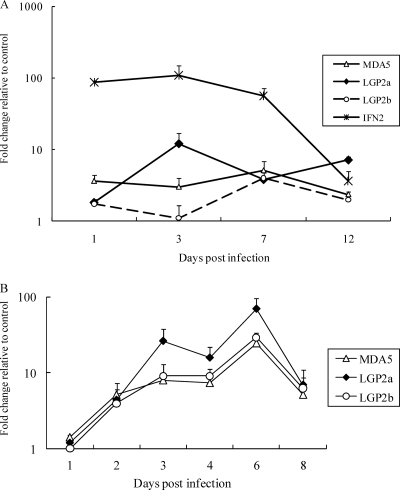
Kinetics of trout MDA5 and LGP2 expression in response to VHSV and salmon alphavirus (SAV) infection.
Expression of MDA5 and LGP2 was studied in trout after infection with viral hemorrhagic septicemia virus (VHSV), a virus with a single-stranded negative RNA genome (Fig. 3A). At day one postinfection, increased transcript levels of MDA5, LGP2a, and LGP2b were detected. MDA5 expression appeared to stay at a constant level during the 12-day sampling period, while induction of LGP2a peaked at day 3. Compared with MDA5 and LGP2a, a much bigger increase of gene expression was observed for IFN2.
Fig. 3.
Trout MDA5 and LGP2 expression is induced by infection with negative and positive single-stranded RNA viruses. (A) Rainbow trout were challenged intraperitoneally with VHSV or medium (mock infected), and head kidney tissues were sampled at days 1, 3, 7, and 12 for gene expression analysis. (B) Atlantic salmon TO cells were infected with salmon alphavirus (SAV) or medium (mock infected) and collected at days 1, 2, 3, 4, 6, and 8 for gene expression analysis. Gene expression of trout MDA5, LGP2a, LGP2b, and IFN2 (panel A only) was analyzed by real-time PCR, and their expression levels were normalized to the expression of EF-1α. Fold change relative to the appropriate control group is presented. The means of results from three independent experiments are shown, and bars indicate the SEMs.
SAV is a single-stranded positive RNA virus which infects salmonid species. Recent studies have shown that SAV infection led to elevation of IFN and Mx expression in TO cells (3, 8). In the present work, the expression of MDA5 and LGP2 was analyzed to establish whether these cytosolic PRRs were also involved in antiviral responses against infection of RNA viruses with a positive genome. During the 8-day infection period, expression of MDA5, LGP2a, and LGP2b was induced from day 2 and peaked at day 6. Induction patterns were similar for all 3 molecules, but the magnitude of trout LGP2a induction was higher than that of MDA5 or LGP2b (Fig. 3B).
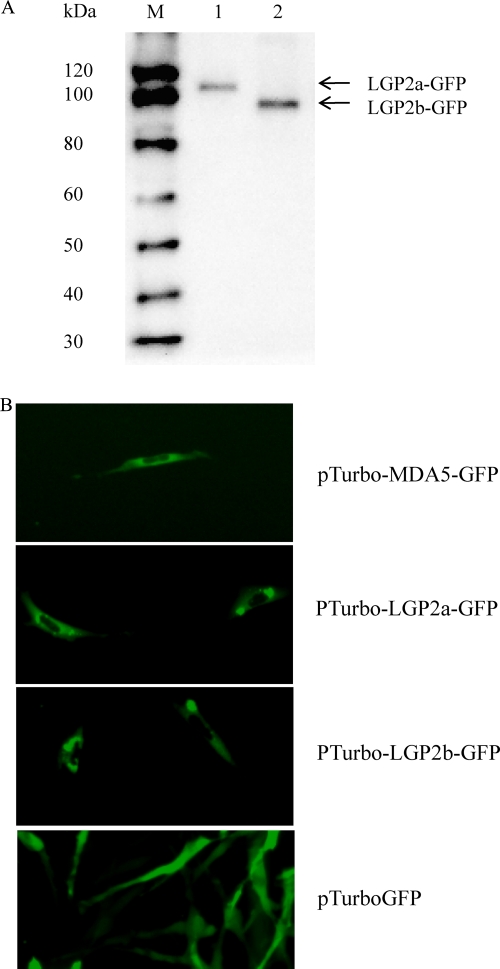
Localization of trout MDA5 and LGP2 within cells.
To determine their localization within cells, the ORFs of trout MDA5, LGP2a, and LGP2b were inserted into the pTurboGFP-N expression vector, to facilitate overproduction of fusion proteins in transfected cells. The RTG-2 cells transfected with plasmids pTurbo-GFP (vector), pTurbo-MDA5-GFP, pTurbo-LGP2a-GFP, or pTurbo-LGP2b-GFP were tested by Western blot analysis for confirmation of expression of fusion proteins and examined under a fluorescent microscope 24 h after transfection. The presence of LGP2a-GFP and LGP2b-GFP proteins was confirmed using a polyclonal antibody against the GFP protein (Fig. 4A). A global cytosolic distribution was seen for all three GFP fusion proteins (Fig. 4B). However, marked intensified green spots were apparent for the two LGP2 isoforms, most of which were located in the region surrounding the nucleus, possibly the nuclear envelope and endoplasmic reticulum. Control cells transfected with empty vector pTurbo-GFP showed a global cytosolic localization.
Fig. 4.
Trout MDA5 and LGP2 are localized in the cytoplasmic region of RTG-2 cells. The RTG-2 cells were transiently transfected with pTurboGFP, pTurbo-MDA5-GFP, pTurbo-LGP2a-GFP, or pTurbo-LGP2b-GFP. After 24 h, cells were collected for analysis. (A) Western blot confirmation of expression of fusion proteins. Lane 1, cell lysate transfected with pTurbo-LGP2a-GFP; lane 2, cell lysate transfected with pTurbo-LGP2b-GFP. (B) Cellular localization of MDA5, LGP2a, and LGP2b in transfected cells under a fluorescent microscope. Note the cytoplasmic expression of MDA5 and LGP2.
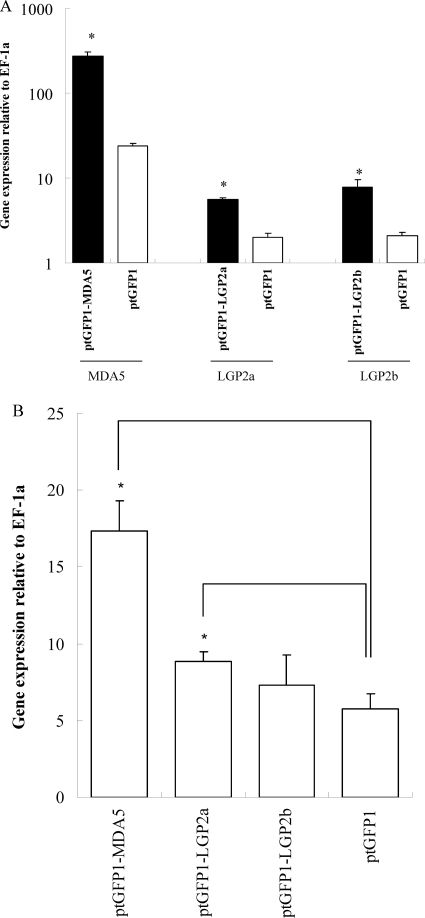
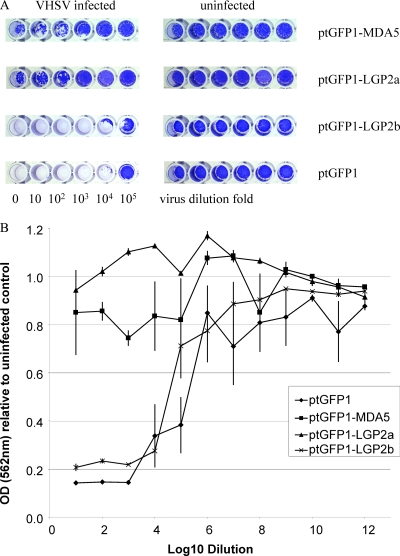
Antiviral activity of trout MDA5 and LGP2 in RTG-2 cells.
The RTG-2 cells were transfected with ptGFP1-MDA5, ptGFP1-LGP2a, and ptGFP1-LGP-2b and stable cell lines established by selection with G-418. Constitutive overexpression of transfected genes was apparent, with a significant increase of the transcript levels compared with that of untransfected cells: 12-fold for MDA5, 3-fold for LGP2a, and 4-fold for LGP2b (Fig. 5A). In each case, the expression of Mx was significantly increased in MDA5- and LGP2a-transfected cells (P < 0.5) but not in LGP2b-transfected cells (Fig. 5B). These cells were also examined for their resistance against VHSV infection. The cells transfected with MDA5 and LGP2a were more resistant to VHSV infection than the control cells transfected with empty vector plasmid (Fig. 6). Overexpression of LGP2b had little impact on the antiviral state of the cells.
Fig. 5.
Overexpression of trout MDA5 and LGP2 enhances Mx gene expression in RTG-2 cells. The RTG-2 cells were transfected with ptGFP1, ptGFP1-MDA5, ptGFP1-LGP2a, or ptGFP1-LGP2b. Gene expression of MDA5, LGP2a, and LGP2b (A) and Mx (B) was determined in stably transfected cells. The means of results from three independent experiments are shown, and bars indicate the SEMs. Asterisks indicate where the increase of gene expression is significant relative to ptGFP1-transfected cells (P < 0.05).
Fig. 6.
Overexpression of MDA5 and LGP2a but not LGP2b in RTG-2 cells enhances protection against VHSV infection. The RTG-2 cells stably transfected with ptGFP1, ptGFP1-MDA5, ptGFP1-LGP2a, or ptGFP1-LGP2b were infected with 10-fold-diluted VHSV for 2 weeks and stained with crystal violet. The plates were photographed, and one row of cells from the 96-well plates is shown (n = 6 wells) (A). Stained cells were subsequently dissolved in 1% SDS solution, and optical density at 562 nm (OD562) was measured. The relative OD was calculated by comparing the average OD of the infected wells (n = 6) with that of the uninfected control (n = 24 wells) (B).
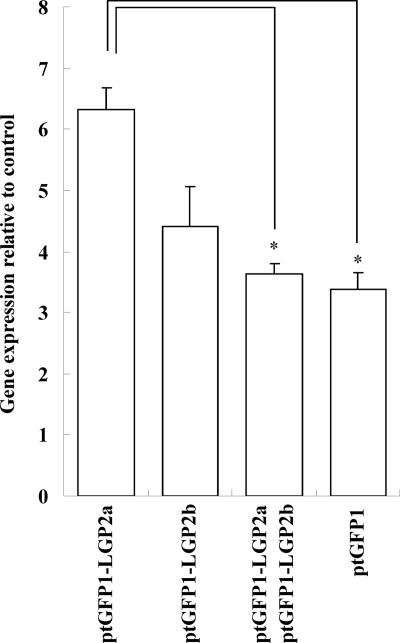
Inhibitory activity of LGP2b on LGP2a-activated antiviral response.
To further investigate whether LGP2b has any effect on the LGP2a-elicited antiviral responses, RTG-2 cells were cotransfected with 5 μg poly(I:C) and ptGFP1 (2 μg), ptGFP1-LGP2a (2 μg), ptGFP1-LGP2b (2 μg), or ptGFP1-LGP2a (2 μg) plus ptGFP1-LGP2b (2 μg) and cultured for 6 h. The cells cotransfected with LGP2a and LGP2b exhibited decreased Mx expression compared with that of cells transfected with LGP2a alone (Fig. 7) (P < 0.05), demonstrating that LGP2b suppresses the LGP2a-induced antiviral responses.
Fig. 7.
The LGP2 variant lacking a 54-aa C-terminal region (LGP2b) suppresses LGP2-induced gene expression of antiviral factors. The RTG-2 cells were transiently cotransfected with poly(I:C) and ptGFP1, ptGFP1-LGP2a, ptGFP1-LGP2b, or ptGFP1-LGP2a plus ptGFP1-LGP2b, washed, and cultured for 6 h before cell collection for RNA extraction. The means of results from three independent experiments are shown, and bars indicate the SEMs. Asterisks indicate where the increase of gene expression is significant (P < 0.05).
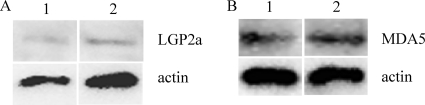
Interaction of MDA5 and LGP2 in RTG-2 cells.
It has been demonstrated recently that LGP2 is involved in the primary recognition of encephalomyocarditis virus upstream of MDA5 during signaling (26). To study whether LGP2 regulates MDA5 expression, MDA5 was overexpressed in transfected RTG-2 cells, where LGP2 expression was analyzed by Western blotting using polyclonal antibodies. The MDA5 protein was apparently not affected by LGP2 (Fig. 8). Similarly, overexpression of MDA5 did not result in any change of LGP2 at the protein level. These data suggest that MDA5 and LGP2 may serve as parallel positive regulators to activate the IFN system.
Fig. 8.
Overexpression of MDA5 and LGP2 in RTG-2 cells does not affect gene expression of either molecule at the protein level. (A) The RTG-2 cells were transfected with plasmid p3xFLAG-CMV-14 vector (lane 1) or pMDA5-Flag (lane 2) and analyzed for LGP2 expression using rabbit polyclonal anti-LGP2 antibody. (B) The RTG-2 cells were transfected with ptGFP1 (lane 1) or ptGFP1-LGP2a (lane 2) and analyzed for MDA5 expression using rabbit polyclonal anti-MDA5 antibody.
Binding affinity of MDA5 and LGP2 with poly(I:C).
To determine whether trout MDA5, LGP2a, and LGP2b bind to synthetic poly(I:C), a poly(I:C) pulldown assay was performed using poly(I:C)-conjugated agarose. All three proteins bound to poly(I:C) as shown in Fig. 9, and the binding was inhibited by coincubation with soluble poly(I:C). In the case of MDA5 and LGP2b, inhibition by poly(I:C) was incomplete, as a small amount of proteins was still able to bind to the poly(I:C) agarose. No proteins were detected from poly(C)-conjugated agarose for all three proteins.
Fig. 9.
Binding affinity of MDA5 and LGP2 with poly(I:C). Purified recombinant proteins were incubated with poly(I:C)-coupled agarose beads. After being washed, the beads were subjected to Western blot analysis using polyclonal antibodies against MDA5 and LGP2. Competition of poly(I:C) binding to MDA5, LGP2a, and LGP2b was conducted by inclusion of 50 μg/ml soluble poly(I:C) in the reaction. (A) Binding affinity of MDA5 with poly(I:C). Lane 1, MDA5 pulled down from poly(I:C)-coupled agarose; lane 2, MDA5 pulled down from coincubation with soluble poly(I:C); lane 3, MDA5 pulled down from poly(C)-coupled agarose. (B) Binding affinity of LGP2a and LGP2b with poly(I:C). Lanes 1 and 3, LGP2a and LGP2b pulled down from poly(I:C)-coupled agarose; lanes 2 and 4, LGP2a and LGP2b pulled down from coincubation with soluble poly(I:C); lanes 5 and 6, LGP2a and LGP2b pulled down from poly(C)-coupled agarose.
DISCUSSION
Discovered in 2004, the RLR family of cytoplasmic viral sensors, to date consisting of RIG-I, MDA5, and LGP2, has become a major focus of research into antiviral innate immunity. In this report, we have identified MDA5 and LGP2 homologues from a teleost species and characterized their functions in antiviral defense against RNA viruses. We have demonstrated that LGP2 is an important sensor for viral PAMPs in addition to MDA5 to activate the host IFN system. Moreover, an LGP2 variant with a deletion of 54 aa residues at the C terminus, likely associated with the nuclear envelope/endoplasmic reticulum, possibly acts as a negative regulator of the LGP2-activated antiviral response by competing for viral PAMPs. Furthermore, our results strongly suggest that MDA5 and LGP2 work independently as cytosolic RNA sensors in fish cells. The present work confirms the presence of a functional RLR system in lower vertebrates and provides an insight into the RLR sensing of viral PAMPs and associated intracellular signaling in teleost fish.
Our previous analysis has shown that the RLR family is well conserved among vertebrates (37). This has been confirmed by recent reports that RIG-I homologues and the associated signaling molecule, MAVS, indeed play an important role in activating fish antiviral responses, in particular the IFN system (2, 14). Like RIG-I homologues in other fish species, the trout MDA5 gene described here encodes a protein which has a structural domain organization similar to that seen in mammalian MDAS homologues, containing two tandem CARDs within the first 200-aa region of the N terminus, a DExD/H box helicase domain and a helicase C-terminal domain in the middle region, and a C-terminal domain of RIG-I near the C terminus. Among the major domains, the two CARDs are least conserved, sharing 34.5% and 33.7% identity with the corresponding human MDA5 CARDs. The second CARD of MDA5 (22.4% aa identity with MAVS CARD) is more closely related to the CARD of trout MAVS than the first CARD (15.3% aa identity with MAVS CARD). However, both CARDs have been shown to interact with MAVS in mammals and fish (2, 12, 27). Overexpression of the CARDs alone in cultured fish cells significantly enhanced type I IFN expression, leading to antiviral protection (2).
The organization of the structural domains in the trout LGP2 is conserved. Like LGP2 molecules in mammals, trout LGP2 has a DExD/H box helicase, a helicase-conserved C-terminal domain, and a CTD but lacks N-terminal CARDs. All key motifs, including the ATP binding motif, the ATPase motif, and RNA unwinding motif, are identical, except for the RNA binding loop, which is 2 aa shorter in trout LGP2. This RNA binding loop was crucial for the binding specificity and affinity with various types of RNA molecules (31). Interestingly, a splicing variant (namely LGP2b), possibly originated from incomplete splicing of an intron in the RNA transcript, was sequenced. The LGP2b mRNA contained an early translation stop and thus translated into a protein with an incomplete CTD, which has a deletion of 54 aa residues at the C terminus. Alignment analysis of the incomplete trout LGP2b CTD with the human LGP2 CTD showed that the trout LGP2b was devoid of the β6, β7, and β8 sheets and the α3 helix (31).
Modulation of RLR expression by viral RNA PAMPs has been well documented. RIG-I, MDA5, and LGP2 were induced by foreign double-stranded RNA and various types of RNA viruses at an early stage of infection (10, 24). In agreement with the studies in mammals, trout MDA5 and LGP2 were upregulated in trout RTG-2 cells by transfection with poly(I:C), with elevated expression of type I IFN, and the antiviral Mx gene. Interestingly, stimulation with extracellular poly(I:C) also enhanced MDA5 and LGP2 expression in both RTG-2 cells and RTS-11 cells, possibly due to an indirect effect of factors, such as IFNs that are induced by poly(I:C). In fact, IFNs, as shown in the present study, are known inducers of RIG-I, MDA5, and LGP2 and are capable of amplifying their responses via a positive feedback mechanism (9). However, it cannot be ruled out that Toll-like receptors (TLRs), such as TLR22, may also play a role in activation of cytosolic RLRs since TLR22 is known to be one of the cell surface receptors that detect extracellular poly(I:C) (17). The fact that the MDA5 and LGP2 transcripts were drastically increased at an early stage of VHSV or SAV infection strongly supports the notion that both MDA5 and LGP2 are likely involved in fish innate antiviral immunity in response to both negative and positive RNA viruses. Furthermore, overexpression of RIG-I molecules in fish cells conferred strong protection against VHS virus (2). These data demonstrate that RIG-I-like RLRs are important sensors in recognizing both single-stranded and double-stranded RNA viruses in fish.
One interesting finding in the present study is that LGP2 appears to act as an activator of the antiviral system in fish, which is in contrast to some reports in mammals where LGP2 was shown to suppress RIG-I- and MDA5-activated IFN responses (13, 24). LGP2 lacks CARDs at the N-terminal region but possesses other domains, including a DExD/H helicase domain, a helicase C-terminal domain, and a C-terminal domain of RIG-I, and is believed to sequester the viral RNA PAMPs from the RIG-I and MDA5 molecules, hence acting as a negative regulator to control excessive production of IFNs (which is detrimental to the host immune system) and keeping the IFN level in check when viral infection diminishes. LGP2 knockout mice showed a higher level of IFNs than the wild-type animals and were more resistant to vesicular stomatitis virus infection (32). However, these LGP-deficient mice exhibited impaired type I IFN production in response to infection with a different virus, encephalomyocarditis virus, and thus were more susceptible to this virus (32). Satoh et al. (26) have confirmed that the positive regulatory roles of LGP2 may be attributed to the ATPase domain within DExD/H. In this study, the trout fibroblast cells stably transfected with the LGP2 construct displayed enhanced resistance against VHSV infection compared with that of untransfected cells, an outcome of the induced innate antiviral factors, such as Mx by LGP2 overexpression in transfected cells.
A trout LGP2 variant (LGP2b) containing an incomplete C-terminal domain of RIG-I was also identified. Trout LGP2b was translated from an RNA transcript containing an intron due to incomplete RNA splicing and was upregulated by poly(I:C) and IFN and during RNA virus infection (Fig. 2). In contrast to MDA5 and LGP2a, overexpression of LGP2b, lacking the 54-aa C-terminal fragment, had little if any effect on Mx expression and cell resistance against VHSV infection in RTG-2 cells (Fig. 6), suggesting that it could play an inhibitory role in LGP2a-elicited antiviral signaling, perhaps as a dominant negative mediator for the LGP2a-elicited IFN response. The similar cellular distributions of LGP2a and LGP2b (Fig. 5) imply that LGP2b may compete for the signaling molecules with LGP2a or directly interact with LGP2a to block downstream signaling. The CTD of LGP2 is essential for RLR's function. In mammals, LGP2 mutants without the RNA binding domain are unable to activate the MDA5-elicited IFN responses. Viruses such as paramyxoviruses also target the LGP2 and MDA5 pathway by interaction between the CTD and the viral V protein to suppress the host antiviral system (20). It is not clear from this study whether the incomplete CTD was capable of binding viral RNA molecules and/or interacting with downstream signaling adaptor proteins. Trout LGP2b containing a shortened CTD with a deletion of 54 aa following the RNA binding motif still had binding affinity with poly(I:C), suggesting that the deleted C-terminal region is not required for RNA ligand binding. The fact that trout LGP2b antagonized the stimulatory effect of LGP2a in induction of Mx expression suggests that the 54-aa C-terminal region is essential for activating the downstream signaling pathway leading to IFN production and that trout LGP2b functions as a negative regulator possibly by competing with LGP2a for viral RNA PAMPs.
The roles of LGP2 in RIG-I- and MDA5-mediated antiviral responses remain patchy and require further investigation. Previous reports indicate that LGP2 may serve as a suppressor to block RIG-I- and MDA5-elicited signaling via MAVS in mammals since it lacks the N-terminal CARDs that are required for interaction with signaling proteins. The LGP2 knockout mice produce a larger amount of type I IFNs than wild-type mice after stimulation with poly(I:C)s and vesicular stomatitis virus infection (24). However, other studies have disputed such findings. Venkataraman et al. found that LGP2 was not the primary negative regulator for IFN production and that LGP2 knockout mice exhibited enhanced resistance to vesicular stomatitis virus infection (32). More recently, LGP2 was suggested to target the signaling pathway upstream of RIG-I and MDA5 (26). It is evident from the present study that both trout MDA5 and LGP2 are able to bind to poly(I:C) to trigger IFN production (Fig. 9). Furthermore, overexpression of MDA5 in transfected cells had no impact on LGP2 protein level and vice versa. These findings, together with that from previous studies, suggest that RIG-I, MDA5, and LGP2 may act in parallel as viral RNA sensors in fish cells (2).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China grant 30830083, the Royal Society of Edinburgh, and the EU FP6 IMAQUANIM project (contract no. 007103).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Besch R., et al. 2009. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 119:2399–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biacchesi S., et al. 2009. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 83:7815–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang M., Nie P., Collet B., Secombes C. J., Zou J. 2009. Identification of an additional two-cysteine containing type I interferon in rainbow trout Oncorhynchus mykiss provides evidence of a major gene duplication event within this gene family in teleosts. Immunogenetics 61:315–325doi:10.1007/s00251-009-0366-y [DOI] [PubMed] [Google Scholar]
- 4. Chang M. X., Chen W. Q., Nie P. 2010. Structure and expression pattern of teleost caspase recruitment domain (CARD) containing proteins that are potentially involved in NF-kappaB signalling. Dev. Comp. Immunol. 34:1–13 [DOI] [PubMed] [Google Scholar]
- 5. Cui S., et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179 [DOI] [PubMed] [Google Scholar]
- 6. Franch R., et al. 2006. Full-length sequence and expression analysis of Toll-like receptor 9 in the gilthead seabream (Sparus aurata L.). Gene 378:42–51 [DOI] [PubMed] [Google Scholar]
- 7. Fringuelli E., et al. 2008. Phylogenetic analyses and molecular epidemiology of European salmonid alphaviruses (SAV) based on partial E2 and nsP3 gene nucleotide sequences. J. Fish Dis. 31:811–823 [DOI] [PubMed] [Google Scholar]
- 8. Gahlawat S. K., Ellis A. E., Collet B. 2009. Expression of interferon and interferon-induced genes in Atlantic salmon Salmo salar cell lines SHK-1 and TO following infection with salmon alphavirus SAV. Fish Shellfish Immunol. 26:672–675 [DOI] [PubMed] [Google Scholar]
- 9. Kang D. C., et al. 2004. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23:1789–1800 [DOI] [PubMed] [Google Scholar]
- 10. Kato H., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 doi: 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawai T., Akira S. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21:317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 doi:10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 13. Komuro A., Horvath C. M. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lauksund S., Svingerud T., Bergan V., Robertsen B. 2009. Atlantic salmon IPS-1 mediates induction of IFNa1 and activation of NF-kappaB and localizes to mitochondria. Dev. Comp. Immunol. 33:1196–1204 [DOI] [PubMed] [Google Scholar]
- 15. Lopez-Doriga M. V., et al. 2001. Isolation of salmon pancreas disease virus (SPDV) in cell culture and its ability to protect against infection by the ‘wild-type’ agent. Fish Shellfish Immunol. 11:505–522 [DOI] [PubMed] [Google Scholar]
- 16. Matsuo A., et al. 2008. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 181:3474–3485 [DOI] [PubMed] [Google Scholar]
- 17. Meijer A. H., et al. 2004. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 40:773–783 [DOI] [PubMed] [Google Scholar]
- 18. Meylan E., Tschopp J., Karin M. 2006. Intracellular pattern recognition receptors in the host response. Nature 442:39–44 [DOI] [PubMed] [Google Scholar]
- 19. Palti Y., et al. 2010. Identification, characterization and genetic mapping of TLR7, TLR8a1 and TLR8a2 genes in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 34:219–233 [DOI] [PubMed] [Google Scholar]
- 20. Parisien J. P., et al. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Phelan P. E., Mellon M. T., Kim C. H. 2005. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio). Mol. Immunol. 42:1057–1071 [DOI] [PubMed] [Google Scholar]
- 22. Pichlmair A., Reis e Sousa C. 2007. Innate recognition of viruses. Immunity 27:370–383 [DOI] [PubMed] [Google Scholar]
- 23. Rodriguez M. F., Wiens G. D., Purcell M. K., Palti Y. 2005. Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics 57:510–519 [DOI] [PubMed] [Google Scholar]
- 24. Rothenfusser S., et al. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268 [DOI] [PubMed] [Google Scholar]
- 25. Saito T., et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Satoh T., et al. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 28. Skjaeveland I., Iliev D. B., Strandskog G., Jorgensen J. B. 2009. Identification and characterization of TLR8 and MyD88 homologs in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 33:1011–1017 [DOI] [PubMed] [Google Scholar]
- 29. Su J., Jang S., Yang C., Wang Y., Zhu Z. 2009. Genomic organization and expression analysis of Toll-like receptor 3 in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 27:433–439 [DOI] [PubMed] [Google Scholar]
- 30. Sumpter R., Jr., et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takahasi K., et al. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J. Biol. Chem. 284:17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Venkataraman T., et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455 [DOI] [PubMed] [Google Scholar]
- 33. Wergeland H. I., Jakobsen R. A. 2001. A salmonid cell line (TO) for production of infectious salmon anaemia virus (ISAV). Dis. Aquat. Organ. 44:183–190 [DOI] [PubMed] [Google Scholar]
- 34. Weston J. H., et al. 2005. Nucleotide sequence variation in salmonid alphaviruses from outbreaks of salmon pancreas disease and sleeping disease. Dis. Aquat. Organ. 66:105–111 [DOI] [PubMed] [Google Scholar]
- 35. Xu L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 36. Yoneyama M., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
- 37. Zou J., Chang M., Nie P., Secombes C. J. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zou J., Tafalla C., Truckle J., Secombes C. J. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 179:3859–3871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.