Abstract
Assembly of herpes simplex virus 1 (HSV-1) occurs in the cytoplasm, where the capsid and tegument bud into host cell membranes. It is at this point that the viral glycoproteins are incorporated into the virion, as they are located at the assembly site. We investigated the role of the Rab GTPases in coordinating the assembly process by overexpressing 37 human Rab GTPase-activating proteins (GAPs) and assessing infectious titers. Rab GTPases are key cellular regulators of membrane trafficking events that, by their membrane association and binding of effector proteins, ensure the appropriate fusion of membranes. We identified that TBC1D20 and RN-tre and their partner Rabs, Rab1a/b and Rab43, respectively, are important for virion assembly. In the absence of Rab1a/b, the viral glycoproteins are unable to traffic from the endoplasmic reticulum to the assembly compartment, and thus unenveloped particles build up in the cytoplasm. The defect resulting from Rab43 depletion is somewhat more complex, but it appears that the fragmentation and dispersal of the trans-Golgi network and associated membranes render these compartments unable to support secondary envelopment.
INTRODUCTION
Herpesviruses are large complex DNA viruses that are composed of four distinct structures, a DNA core, a capsid in which the DNA is enclosed, a proteinaceous tegument, and a host-derived lipid envelope, embedded with viral glycoproteins. The assembly of herpesviruses is a complex process, and the most commonly accepted model is one of envelopment-deenvelopment-reenvelopment. In this model, assembly begins in the nucleus, where the newly synthesized DNA is inserted into preformed capsids. The nucleocapsids then bud at the inner nuclear membrane, into the perinuclear space, followed by fusion with the outer nuclear membrane that releases the nucleocapsids into the cytoplasm (envelopment and deenvelopment). The acquisition of the tegument is thought to occur at two distinct sites, the nucleocapsid and the future envelope. Secondary envelopment (or reenvelopment) occurs when the capsid and envelope protein-associated tegument come together to drive wrapping/budding at trans-Golgi network (TGN)-derived membranes. The resulting virus-containing vesicles will then fuse with the plasma membrane and release the mature virion (reviewed in reference 25).
The location of glycoproteins during the biogenesis of new virus is important for assembly. They must be clustered together in TGN-derived membranes, alongside a subset of tegument proteins, to allow for secondary envelopment. The default trafficking route for all membrane proteins is to the plasma membrane. If this is not the correct fate for the protein, it will contain discrete motifs that interact with proteins involved in vesicle formation and targeting, such as the clathrin adaptor proteins (reviewed in reference 4). Unsurprisingly, several herpesvirus glycoproteins contain characteristic trafficking motifs that ensure they are endocytosed from the plasma membrane and targeted to the TGN. For example, both herpes simplex virus 1 (HSV-1) gB and gE contain a tyrosine motif that binds AP-2 and promotes endocytosis, and gB also contains a dileucine motif that signals for endosome-to-TGN transport (1, 3, 11). Likewise, gM, gK, and pUL20 encode tyrosine motifs and are all localized to the TGN (7, 12). Other glycoproteins, such as gD and gH/L, contain no discernible trafficking motifs and when expressed alone in cells are localized to the plasma membrane (7, 24). However, expression of gM alongside gD or gH/L is sufficient to localize these envelope proteins to the TGN (7).
In addition to specific motifs, numerous cellular proteins control trafficking events. One family of key regulators are the Rab GTPases, which are specific to particular trafficking steps (29). As with all GTPases, these proteins cycle between a GDP-bound “off” state and a GTP-bound “on” state. When bound to GTP the Rab becomes membrane associated, where it binds effectors to ensure correct sorting, motility, tethering, and fusion of appropriate membranes. The cycling between the GDP- and GTP-bound forms is aided by accessory proteins called GDP/GTP exchange factors (GEFs), which will turn the Rab on, and GTPase-activating proteins (GAPs) that hydrolyze GTP, thus turning the Rab off (reviewed in references 2 and 29). Rab GTPases have been shown to be important for the assembly and egress of a number of different viruses, including influenza A virus, hepatitis C virus, and hantavirus (6, 23, 28).
In order to determine which specific Rabs are essential for HSV-1 assembly, we took advantage of the fact that overexpression of a Rab GAP will specifically inactivate the endogenous pool of its target Rab. The GAPs were identified by their conserved catalytic TBC (Tre2/Bub2/Cdc16) domain, which promotes GTP hydrolysis. This unbiased screen has previously been used to identify the Rab proteins required for Golgi complex integrity, Shiga toxin uptake, primary cilia formation, and exosome release (13, 15, 16, 34, 35). We have screened 37 TBC domain-containing human Rab GAPs for the ability to inhibit HSV-1 replication. We identified two Rab GAPs, TBC1D20 and RN-tre, that cause an activity-dependent reduction in infectious HSV-1 and confirmed that the published targets of their GAP activity, Rab1 and Rab43, are indeed important for HSV-1 assembly.
MATERIALS AND METHODS
Cell lines and viruses.
COS7, HeLa, A549, and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. HSV-1 lacking full-length UL36 expression (HSV-1ΔUL36) and a UL36-complementing cell line (HS30), maintained with 200 mg/ml G418, were from P. Desai, John Hopkins University (9). HSV-1 with gE replaced with luciferase (HSV-1ΔgE) has been previously described (5) and was used to prevent nonspecific antibody binding to the Fc receptor formed by the gE and gI complex.
Antibodies.
The antibody reagents used were as follows: actin (AC-40; Sigma), Rab1a (ab27528; Abcam), Rab1b (G-20; Santa Cruz Biotechnology), tubulin (YL1/2; Serotec), pUL48 (LP1; Abcam), green fluorescent protein (GFP; JL-8; Clontech), myc (9E10; Sigma), calnexin (C8.B6; Millipore), TGN46 (Serotec), GM130 (610822; BD Biosciences), gD (LP2 for immunofluorescence, LP14 for Western blotting) (26), and gH (shabba [10]). To generate HSV gB-specific monoclonal antibodies, female BALB/c mice were infected with HSV-1 (strain 17) by ear scarification followed by an intraperitoneal boost 1 month later. Spleens were harvested 3 days later, and B-cell hybridomas were generated as previously described (14). Hybridoma supernatants were screened for reactivity in immunofluorescence assays by using cells transfected with an HSV-1 gB expression plasmid. A cloned hybridoma line secreting antibodies with a strong reactivity to HSV-1 gB was selected and named CB24.
Plasmids.
Plasmids expressing UL36 and VPS4EQ have been previously described (8). The 37 Rab GAPs used in this screen have been previously described also (13).
Complementation assay.
The UL36 complementation was performed as previously described (27). Briefly, COS7 cells were seeded at 2 × 105 cells per well and transfected with 1 μg pcDNA3 or 0.5 μg pcDNA3-UL36 together with 0.5 μg of each of the myc-tagged Rab GAPs by using FuGENE 6 reagent (Roche). After 24 h, cells were infected with HSV-1ΔUL36 at 5 PFU/cell. At 16 h postinfection (hpi), the samples were collected by scraping, followed by sonication and freezing. Infectious virus titers were assessed on HS30 cells.
siRNA.
HeLa or A549 cells were plated at 1.5 × 105 cells per well and were transfected either once (Rab1a/b) or twice (Rab43) with 100 nM total small interfering RNA (siRNA) in Opti-MEM (Invitrogen) using Dharmafect 1 (Thermo Scientific) according to the manufacturer's instructions. The Rab1a and Rab1b SMARTpool siRNA and single oligos, as well as Rab43 single oligo, have been previously described (15). At 48 h after the final transfection, the cells were infected with HSV-1 strain KOS at 5 PFU/cell. Samples were harvested 20 hpi. For plaque assay, cells were scraped into the medium, sonicated to liberate cell-associated virus, and assayed on Vero cells to assess infectious titer. For protein samples, cells were scraped and centrifuged at 5,000 rpm for 5 min and resuspended in lysis buffer (50 mM Tris [pH 7.9], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) supplemented with protease inhibitor (Roche) and incubated on ice for 10 min. Lysates were clarified by centrifugation at 12,000 rpm for 10 min at 4°C. Samples were then analyzed by SDS-PAGE and Western blotting using secondary antibodies linked to infrared dyes (−680 and −800) and imaged using a LI-COR Odyssey infrared imager.
Immunofluorescence.
Cells were seeded on glass coverslips at 2 × 105 cells per well of a 6-well dish. After 24 h they were transfected with either plasmid or siRNA as described above and infected with HSV-1ΔgE to avoid cross-reaction with nonspecific antibodies to the Fc receptor formed by the gE/gI complex. At 20 hpi the cells were fixed in 3% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), permeabilized, and quenched using 0.2% saponin and 50 mM NHCl4. Blocking and antibody dilution were in PBS with 0.2% gelatin, 0.02% saponin, and 0.02% sodium azide. Secondary antibodies used were conjugated to Alexa 488, 568, and 633 (Molecular Probes), and coverslips were mounted in antifade mounting medium (Invitrogen). Images were acquired using a Leica SP2 confocal microscope and processed using ImageJ and Adobe Photoshop. For antibody feeding experiments, at 8 hpi the coverslips were incubated with the appropriate antibody on ice for 60 min. The coverslips were then washed and returned to medium at 37°C for the designated time, at which point they were fixed and processed as described above.
Analysis of released supernatant virus.
A549 cells were seeded at 4.5 × 105 cells in 6-cm dishes. Twenty-four hours later they were transfected either once (Rab1a/b) or twice (Rab43) with 600 nM total siRNA as described above. At 48 h posttransfection, the cells were infected with HSV-1 (KOS) at 5 PFU/ml. At 16 hpi the supernatant was collected and cleared of cellular debris by centrifugation at 5,000 rpm for 5 min. Virus particles were pelleted through a 33% sucrose cushion at 30,00 rpm for 120 min and resuspended in either PBS (Rab1a/b) or sample buffer (Rab43). For Rab1a/b, the sample was subjected to trypsin digestion (200 μg/ml) for 60 min at 37°C, and the reaction was stopped with protease inhibitors (protease inhibitor cocktail; Roche). These samples, along with cell lysate harvested as described above, were separated by SDS-PAGE and analyzed by Western blotting using infrared-conjugated secondary antibodies (LI-COR Biosciences). The LI-COR Odyssey infrared imager was used for scanning, and the Odyssey v3.0 software was used to calculate the integrated intensity for quantification.
Electron microscopy.
A549 cells were seeded on coverslips, transfected with siRNA, and infected as described above. At 18 hpi cells were fixed in 2% PFA plus 1.5% glutaraldehyde (TAAB) in 0.1 M sodium cacodylate buffer, postfixed in 1% osmium tetroxide plus 1.5% potassium ferricyanide, and treated with 1% tannic acid. The cells were subsequently dehydrated in increasing concentrations of ethanol and then embedded in epon (TAAB). Coverslips were removed by plunging the samples into liquid nitrogen. Ultrathin 70-nm sections were stained with lead citrate and observed with a transmission electron microscope (Tecnai G2) operated at 120 kV, and images were taken using an AMT XR60B digital camera running Deben software and processed using Adobe Photoshop.
RESULTS
Screening for effects of Rab GAPs on HSV-1 replication.
The initial screen used an unbiased approach to determine which Rab GTPases act in the assembly of HSV-1. This involved the overexpression of 37 TBC domain-containing putative Rab GAPs in COS7 cells to inactivate their target Rabs, followed by infection with HSV-1 and determination of the progeny virus titer. To ensure that we were only investigating virus production from transfected cells, we used a complementation assay, which involved cotransfecting the cells with a plasmid expressing a Rab GAP and another plasmid expressing the essential virus protein pUL36 (27). The cells were then infected with HSV-1 lacking a functional UL36 gene (HSV-1 ΔUL36), and thus only cells expressing pUL36 from the transfected plasmid were the source of infectious HSV-1. The samples were harvested and virus titers were established on a UL36-complementing cell line to allow plaque formation. The data shown were taken from at least three independent experiments and normalized to the positive control (Fig. 1A).
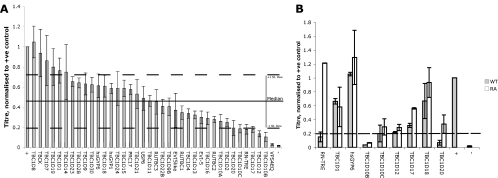
Fig. 1.
Activities of TBC1D20 and RN-Tre are important for production of infectious HSV-1. (A) COS7 cells were cotransfected with a plasmid expressing the essential virus protein pUL36 and plasmids expressing TBC domain proteins as described in Materials and Methods. The positive control was transfected with the UL36 plasmid and pcDNA3, while the negative control was transfected with only pcDNA3. At 24 h posttransfection cells were infected with HSV-1ΔUL36 at 5 PFU/cell, progeny virus was collected 16 hpi, and infectious virus levels were determined by titration on HS30 cells. Titers were normalized to the positive control, and bars represent the means of at least three independent experiments. Error bars are standard errors of the means. The solid line represents the median HSV-1 titer across all samples, and dashed lines represent ±1 standard deviation from the median. (B) Rab GAPs of interest (gray bars) were expressed alongside their inactive controls (white bars) and infected as described above. Titers were normalized to the positive control, and bars represent the means of at least two independent experiments. Error bars are standard errors of the means. The dashed line represents a 5-fold reduction in titer compared to the positive control.
The overexpression of the GAPs showed variable effects on HSV-1 titer compared to the positive control, with the titers being reduced for almost all of the GAPs tested. However, due to the inherent variability in the assay, we decided to consider only those GAPs that resulted in a reduction in infectious progeny of greater than 1 standard deviation from the median. Using this cutoff (which equates to an approximate 5-fold reduction in HSV-1 titer compared to the positive control expressing pUL36 and empty vector), we found that the overexpression of six putative Rab GAPs (RN-tre, TBC1D10B, TBC1D10C, TBC1D12, TBC1D17, and TBC1D20) fell within this range (Fig. 1A). Also included as a control was VPS4EQ, which has been demonstrated to cause an ∼100-fold reduction in infectious titer, due to an assembly defect resulting from the inhibition of the ESCRT pathway (8). To test whether the observed effects were due to overexpression alone, we retested the candidate GAPs from the initial screen, as well as three positive-control GAPs that demonstrated little effect on HSV-1 replication (TBC1D1, hsGYP6, and TBC1D18), alongside catalytically inactive forms, in which a key arginine residue was mutated to an alanine (Fig. 1B). These catalytically inactive forms are unable to stimulate GTP-to-GDP hydrolysis and therefore are unable to bias their target Rabs to the GDP-bound form (35). Thus, any effect Rab GAP R-A mutants may have on titer is not due to their ability to hydrolyze GTP and inactivate their target Rab GTPase.
These data confirmed that the effects of overexpression of TCB1D20 and RN-tre are activity dependent, since only the catalytically active forms of these Rab GAPs caused a 5-fold or greater reduction in infectious titer compared to their inactive forms. However, the reduction in HSV-1 titer upon expression of the mutated forms of TBC1D12, TBC1D17, TBC1D10B, and TBC1D10C indicated that the effects of these Rab GAPs are independent of their ability to hydrolyze GTP. This left us with two candidate regulators of Rab GTPases required for HSV-1 replication. The target Rabs for TBC1D20 and RN-tre have previously been reported as Rab1 and Rab43, respectively (13, 15).
Depletion of Rab1 and Rab43 reduces the yield of infectious HSV-1.
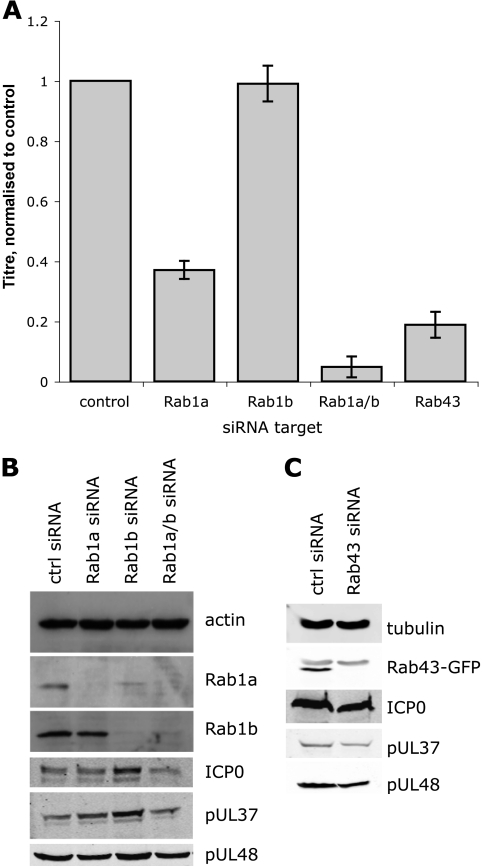
To confirm that Rab1 and Rab43 are indeed essential for the growth of HSV-1, we depleted the cells of these proteins by using previously published siRNA duplexes (13, 15). The efficiency of transfection of siRNA into HeLa cells meant we no longer needed to utilize the complementation assay and instead infected the cells with a wild-type strain of HSV-1, and the progeny virus was collected and titers were determined on Vero cells. The GAP TBC1D20 activates the GTP hydrolysis of the two isoforms of Rab1, Rab1a and Rab1b, so we tested the role for these proteins individually and in concert on the production of HSV-1. While the depletion of Rab1b did not result in a reduction of infectious virus, in cells depleted of Rab1a there was a 60% reduction in infectious titer compared to control cells treated with a nontargeting, control siRNA. Furthermore, when both proteins were depleted together, the resultant titer was less than 5% that of the control cells (Fig. 2A). When the progeny virus was collected, cell lysates were also made to assess depletion. Western blotting showed a good knockdown of both Rab1a and Rab1b (Fig. 2B, upper panels). To rule out effects on viral gene expression, we also considered the levels of a subset of viral proteins, including members of all three temporal classes of gene expression: ICP0 (immediate early), pUL37 (early), and pUL48 (late). The amount of these proteins was similar under all conditions, indicating that there was no significant effect of Rab1a/b depletion on the accumulation of the viral proteins (Fig. 2B, lower panels).
Fig. 2.
Rab1a/b and Rab43 are required for infectious virus production. HeLa cells were transfected once (Rab1a/b) or twice (Rab43) with siRNA duplexes as detailed in Materials and Methods. At 48 h after the final transfection, cells were infected with HSV-1 (MOI, 5), and cell-associated virus was collected 20 hpi. Titers were established on Vero cells, and bars represent the means of three independent experiments, normalized to the control siRNA (A). Parallel samples of infected cells were collected 20 hpi, lysed, separated by SDS-PAGE, and analyzed by Western blotting (B and C). For panel C, cells were additionally transfected with a plasmid expressing Rab43-GFP.
The depletion of Rab43 caused an 80% reduction in the amount of infectious virus (Fig. 2C). We confirmed the siRNA duplex was working as previously published, in the absence of a working antibody, by assessing the depletion of Rab43-GFP in the presence of both the control and Rab43 siRNA (Fig. 2D, upper panel). The presence of similar levels of viral proteins in control cells and those depleted of Rab43 suggested that HSV-1 gene expression was not grossly affected by the absence of Rab43 (Fig. 2D, lower panels). The reduction in virus titer after siRNA depletion of both Rab1a/b and Rab43 was independently confirmed in the human lung epithelial cell line A549 (data not shown). These data suggest that both Rab1 and Rab43 play important roles in the assembly of HSV-1.
The effects of TBC1D20 and Rab1 depletion on HSV-1 glycoproteins.
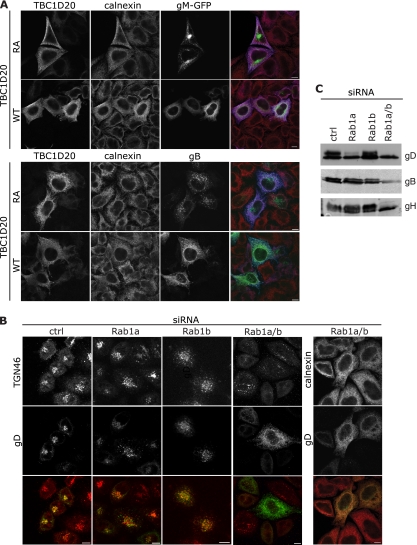
Rab1 is involved in endoplasmic reticulum (ER)-to-Golgi complex transport (31). Therefore, we predicted that in the absence of active Rab1 the viral glycoproteins would be present in the ER rather than colocalizing with TGN markers, as expected from previous data (7, 11, 33). We tested this both by transfection of plasmids expressing individual glycoproteins and by infection. Indeed, in cells expressing inactive TBC1D20 (RA), both gM and gB were localized to perinuclear compartments, reminiscent of the TGN. In the presence of active TBC1D20, both gM and gB localized in a reticular pattern that demonstrated extensive colocalization with the ER marker calnexin (Fig. 3A). Likewise, in infected cells treated with control siRNA, gD colocalized extensively with the TGN marker TGN46. Upon depletion of both Rab1a and Rab1b, gD relocalized to the calnexin-labeled ER and no longer colocalized with TGN46 (Fig. 3B). As predicted from the growth defects, the localization of gD in relation to TGN46 was relatively unchanged in cells depleted of Rab1b, although it was more dispersed, like the TGN itself. Cells depleted of Rab1a showed an intermediate phenotype, with less extensive colocalization of gD with TGN46. We predict this subtle effect of Rab1a depletion on glycoprotein localization may be exacerbated at later time points and cause the small reduction in HSV-1 replication that was observed. For these analyses, infected cells were fixed 8 h postinfection, before HSV-1 induced significant fragmentation of the Golgi complex. These data suggest Rab1 activity, as expected, is required for the transport of viral glycoproteins from the ER to Golgi complex. Since the maturation of glycoproteins occurs within the Golgi stack, in cells depleted of Rab1 we expected to see only the immature form of viral envelope proteins. Analysis of glycoproteins B, D, and H from cells depleted of Rab1a/b indicated that, indeed, only the immature forms were present (Fig. 3C). As with the growth assay, depletion of Rab1a alone showed an intermediate phenotype, with a reduction, but not absence, of the higher-molecular-weight, processed forms of both gD and gH.
Fig. 3.
Rab1a/b is required for transport of glycoproteins from the ER. (A) COS7 cells were cotransfected with a plasmid expressing TBC1D20-myc or TBC1D20RA-myc and HSV-1 gM-GFP (green) or gB (green). Eight hours posttransfection cells were fixed and stained with anti-myc (blue) and anti-calnexin (red). (B and C) HeLa cells were transfected with siRNA duplexes as described in the text. (B) Cells were infected with HSV-1ΔgE (MOI, 3), fixed at 8 hpi, and labeled with TGN46 (red) or calnexin (red) and gD (green). Bars, 10 μm. (C) Cells were infected with HSV-1 (MOI, 5), and 20 hpi cell pellets were lysed, separated by SDS-PAGE, and analyzed by Western blotting with antibodies to gD, gB, and gH.
The incomplete processing of glycoproteins indicated that there could be two alternative explanations for the reduction in HSV-1 titer. This is because glycosylation is essential for infectivity, as shown by experiments in which infected cells were treated with tunicamycin, which blocks N-linked glycosylation (19–21). While a reduction in Rab1 activity would trap all of the envelope proteins in the ER, it is possible that assembly could occur at the ER membrane rather than at post-Golgi complex membranes, but in the absence of glycosylation of envelope proteins, viruses produced in this manner would be noninfectious. However, if the identity of the wrapping membrane that forms the viral envelope is important, then the failure of the glycoproteins to reach the correct compartment would result in an assembly defect.
Analysis of virus assembly in Rab1a/b-depleted cells.
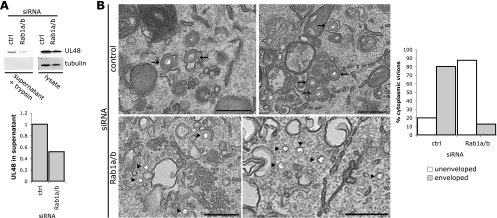
Since plaque assays only provide a readout for infectious virus, we needed to use other approaches to distinguish between an assembly defect and the production of noninfectious virus. First, we collected virus from the supernatant of cells treated with either control or Rab1a/b siRNA. The supernatant was subjected to ultracentrifugation through a sucrose cushion before being resuspended in a small amount of PBS. In preparations from Rab1a/b-depleted cells, we consistently found the presence of cytoplasmic proteins, such as tubulin, but not in those from control cells. While our analyses were conducted before any significant level of cell death had occurred, we found the depletion of Rab1a/b in cells was eventually cytotoxic, presumably due to the essential role of ER-to-Golgi transport in cell viability. In order to remove any signal from nonenveloped or partially enveloped, tegumented capsids due to contamination with cytoplasmic material from lysed cells, but to leave any enveloped virions intact, the samples were treated with the protease trypsin, and then the trypsin was inactivated with protease inhibitors. The lysate and the trypsin-treated samples were analyzed by Western blotting for the presence of the tegument protein pUL48 (Fig. 4A). In the supernatant virus sample, there was a clear reduction in the level of pUL48 upon depletion of Rab1a/b. Quantification of supernatant pUL48 signals and normalization to pUL48 in the cell lysates demonstrated a 50% reduction in the presence of trypsin-resistant pUL48 (i.e., mature enveloped virions) in the presence of Rab1a/b (Fig. 4A). This suggests that the depletion of Rab1a/b does indeed inhibit virion assembly but additionally may reduce the infectivity of any virions that are assembled.
Fig. 4.
Functional Rab1a/b is required for HSV-1 assembly. (A) A549 cells were transfected with Rab1a/b or control siRNA and infected with HSV-1 (MOI, 5). At 18 hpi, supernatant virions were collected as described in Materials and Methods. Virus samples treated with trypsin, as well as parallel cell lysates, were separated by SDS-PAGE and blotted for pUL48 (upper blots) or tubulin (lower blots). Blots were quantified using the LI-COR Odyssey infrared imager, and the Odyssey v3.0 software was used to calculate the integrated intensity for quantification. The graph represents the amount of pUL48 in the supernatant normalized to the pUL48 signal in the cell lysate. (B) A549 cells were grown on coverslips, transfected, and infected (MOI, 3) as described in Materials and Methods. At 16 hpi cells were fixed, processed, and analyzed by transmission EM. Arrows indicate enveloped capsids, and arrowheads indicate unenveloped capsid. Bars, 500 nm.
To further understand the effect of Rab1a/b depletion, siRNA-treated cells were infected with HSV-1 and examined by electron microscopy (EM) to investigate the phenotype at the ultrastructural level (Fig. 4B). The preparation technique used in our EM analysis causes DNA-filled capsids and viruses to appear empty with an unstained core. This makes it easier to identify the unenveloped capsids, which would be otherwise difficult to distinguish from the well-preserved cytoplasm. We observed a clear accumulation of unenveloped capsid in the cytoplasm of cells depleted of Rab1a/b, in contrast to those cells treated with control siRNA, where cytoplasmic virus particles are rare and most capsids observed in the cytoplasm are enveloped. In Rab1a/b-depleted cells, these unenveloped capsids are clustered in the perinuclear area, where membranes are present but the capsids appear unable to engage these membranes for secondary envelopment to occur. Quantification of all cytoplasmic virus particles from at least 12 cells for each condition indicated that in the cells depleted of Rab1a/b ∼90% of the virus in the cytoplasm was unenveloped, compared with ∼20% in cells treated with control siRNA (Fig. 4B). Taken together, these data suggest that Rab1a/b is important for the assembly of HSV-1 particles and may additionally cause a reduction in infectivity of any virions that are assembled, through inhibition of glycoprotein maturation.
Localization of viral glycoproteins in the absence of functional Rab43.
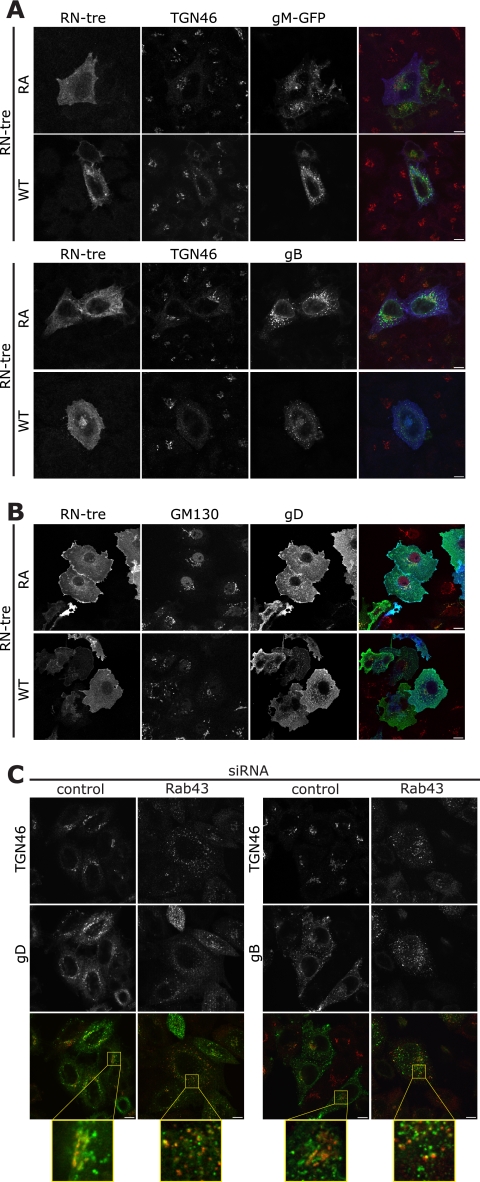
While the cellular role of Rab1 has been well described, the role for Rab43 is less well understood. Rab43 localizes to the TGN/Golgi complex and has been shown to be important in the retrograde trafficking of Shiga toxin from the early endosome to the TGN, as well as in the maintenance of Golgi complex integrity (13, 15). As with Rab1, we initially analyzed cells transfected with plasmids expressing the glycoproteins gM and gB, alongside RN-tre or the inactive mutant of this Rab GAP (Fig. 5A). While there was a striking change in the distribution of both gM and gB, this can be ascribed to the fragmentation of the TGN and Golgi complex in the absence of functional Rab43. The puncta of both glycoproteins still partially colocalized with TGN46, to a similar extent as observed with the catalytically inactive mutant, although the location in the cell had changed. To distinguish between a defect in the delivery of glycoproteins to the plasma membrane or in the endocytic pathway, we took advantage of the fact that gD contains no trafficking motif to mark it for endocytosis; thus, when expressed alone in cells, gD is localized to the plasma membrane. The presence of gD on the plasma membrane in the presence of both the active and inactive forms of RN-tre indicated that there is no defect in the delivery of proteins to the cell surface when Rab43 is inactivated by overexpression of its GAP (Fig. 5B).
Fig. 5.
Rab43 causes Golgi complex fragmentation but is not required for delivery of glycoproteins to the plasma membrane. (A and B) Cos7 cells were transfected with plasmids expressing RN-treRA-myc or RN-tre-myc and gM-GFP (A, upper panels) or gB (A, green, lower panels) or gD (B, all green), fixed 24 h posttransfection, and stained with antibodies to myc (blue) and TGN46 (red) or GM130 (red). (C) HeLa cells were transfected with siRNA and infected (MOI, 3) as detailed in the text. At 8 hpi cells were fixed and labeled with antibodies to gD (green, left panels) or gB (green, right panels) and TGN46 (red). Bars, 10 μm.
In the context of infection of cells depleted of Rab43 by siRNA, the viral glycoproteins were found in puncta throughout the cytoplasm that partially colocalized with TGN46-positive compartments. At early time points of infection (8 h), it was apparent that the distribution of gD and gB was different in the cells depleted of Rab43 compared to the control cells, although partial colocalization with TGN46 was observed in both cases (Fig. 5C). The change in localization is, at least in part, due to the fragmentation of the Golgi complex caused by Rab43 depletion (15). This change in the architecture of the cell makes it difficult to determine if the glycoproteins are now localized in a different compartment.
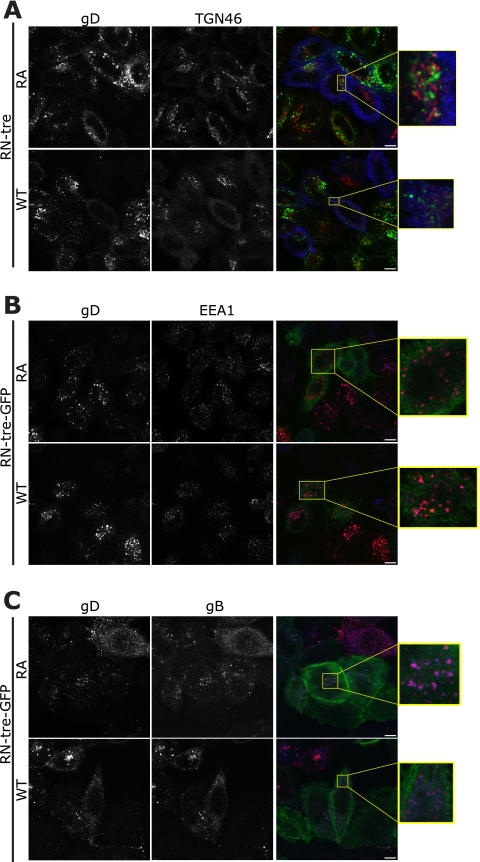
Antibody feeding experiments were used to test for a defect in viral glycoprotein endocytosis. Since previous work considering Shiga toxin pointed toward a defect in trafficking from the early endosomes to the TGN (13), we predicted that the HSV-1 glycoproteins would fail to reach the TGN. Antibody was bound to cells that had been transfected with RN-tre or RN-treRA 4°C for 60 min and infected and chased into the cells at 37°C for up to 60 min. Interestingly, in the case of cells transfected with the inactive mutant of RN-tre, even 60 min after release at 37°C no obvious signal was observed for antibody that had reached the TGN, although they were closely associated (Fig. 6A). As early as 15 min after incubation at 37°C, the glycoproteins gB and gD were present in early endosomes in cells transfected with either the active or inactive form of RN-tre, as shown by EEA1 labeling (Fig. 6B and data not shown), and surprisingly, they remained predominantly associated with EEA1 even 60 min after incubation at 37°C (data not shown). Continuous feeding for 30 min showed that at least gB and gD are localized to the same compartment in the cells independent of the activity of Rab43 (Fig. 6C). These data indicate there is no obvious defect in endocytosis of glycoproteins in the absence of active Rab43, although it was impossible to determine if there was indeed a block in trafficking from the endosomes to the TGN. It is tempting to conclude, therefore, that the reduction in HSV-1 titer when Rab43 is depleted or inactivated is due to the fragmentation and dispersal of the Golgi complex and other endocytic compartments. hsGYP1 and TBC1D22B have also been reported to cause Golgi complex fragmentation (15) but have minimal effects on the production of HSV-1 (Fig. 1A); however, the fragmentation is much less dramatic and the puncta of TGN46 are, in RN-tre-expressing and Rab43-depleted cells, substantially smaller (data not shown). These membrane fragments may not be of a suitable size for envelopment of HSV-1 capsids, or they may be too dispersed in the cell to be able to efficiently engage tegument/capsid.
Fig. 6.
Analysis of glycoprotein localization after internalization from the plasma membrane in infected cells. (A to C) HeLa cells were cotransfected with plasmids expressing RN-tre or RN-treRA-myc (blue) (A) or RN-tre or RN-treRA-GFP (green) (B and C). At 24 h posttransfection cells were infected with HSV-1ΔgE (MOI, 3) for 8 h, whereupon anti-gD antibody was bound at 4°C for 1 h and subsequently incubated at 37°C for 60 min (A), 15 min (B), or 30 min (C). The cells were then fixed and labeled with appropriate secondary antibodies as well as anti-TGN46 (red) or anti-EEA1 (blue). For panel C, anti-gB antibody (blue) was used alongside anti-gD antibody.
Depletion of Rab43 causes an assembly defect.
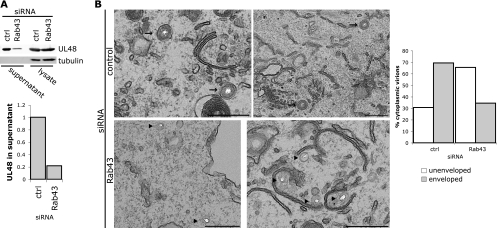
We investigated the role for Rab43 in HSV-1 assembly by analyzing released supernatant virus. Unlike Rab1a/b, depletion of Rab43 from cells did not result in any observed cytotoxicity or the presence of cytoplasmic proteins in preparations of extracellular virions, and thus trypsin treatment of samples was not necessary in these assays. We found that mature extracellular virus, as measured by pUL48 levels, was present at a reduced level in the released virion fraction collected from cells depleted of Rab43. The level of pUL48 in the supernatant was quantified and normalized to pUL48 levels in the lysate, and this showed an 80% reduction in extracellular virus from Rab43-depleted cells, in line with the reduction in the virus titer (Fig. 7A). Upon analysis of infected cells at the ultrastructural level by EM, we also observed an accumulation of unenveloped capsid in the cytoplasm of cells depleted of Rab43, with almost 70% of capsids unenveloped in the cytoplasm, compared to 30% in the control cells (Fig. 7B). However, unlike with Rab1a/b depletion, the capsids were more dispersed throughout the cytoplasm rather than being clustered in the perinuclear area and were not adjacent to clusters of membranes. This correlates with the fragmentation of the Golgi complex and may indicate that a clustered localization of the TGN-derived membranes may be important for forming an effective assembly compartment.
Fig. 7.
Functional Rab43 is required for HSV-1 assembly. (A) A549 cells were transfected with Rab43 or control siRNA and infected with HSV-1 (MOI, 5). At 18 hpi supernatant virions were collected as described in Materials and Methods. The virus samples as well as parallel cell lysates were separated by SDS-PAGE and blotted for pUL48 (upper blot) or tubulin (lower blot). Blots were quantified using the LI-COR Odyssey infrared imager, and the Odyssey v3.0 software was used to calculate the integrated intensity for quantification. The graph represents the amount of pUL48 in the supernatant normalized to the pUL48 signal in the cell lysate. (B) A549 cells were grown on coverslips, transfected, and infected (MOI, 3) as described in Materials and Methods. At 16 hpi cells were fixed, processed, and analyzed by transmission EM. Arrows indicate enveloped capsids, and arrowheads indicate unenveloped capsid. Bars, 500 nm.
DISCUSSION
In this study we demonstrated the importance of two Rab GTPases, Rab1a/b and Rab43, in the production of infectious HSV-1. More specifically, Rab1 activity is important for the trafficking of the viral envelope proteins from the ER to the Golgi complex, where they undergo glycosylation and are eventually incorporated into the forming virus. While it is harder to identify a discrete process that is affected by the depletion of Rab43, we believe it is required to maintain a sufficiently intact Golgi complex and endocytic pathway to allow for secondary envelopment, and it identifies a Rab43-dependent endosome-to-TGN trafficking step as essential for this process. There are three possibilities for a functional role of Rab43 in HSV-1 assembly: (i) a direct role in the delivery of viral glycoproteins to the correct compartment for assembly, with subtle differences in trafficking being masked by the loss of Golgi complex integrity in the cell without active Rab43; (ii) an indirect role of Rab43, with it being required for the delivery of cellular proteins or lipids in order to maintain the integrity of the assembly compartment; (iii) a direct structural role of Rab43 itself in particle assembly, although it was not identified as a virion component in the published mass spectrometry analysis of extracellular virions (22).
The Rab GAP screen has allowed us to successfully identify two Rab GTPases that are required for HSV-1 assembly. Interestingly, these same two Rabs had previously been identified in an independent screen as being essential for the maintenance of Golgi complex integrity (15), highlighting the importance of the Golgi complex to the assembly and infectivity of HSV-1. We are not suggesting that Rab1a/b and Rab43 are the only Rab GTPases involved in HSV-1 assembly, because it is possible that the nature of our screening approach led us to miss the roles of some other Rab GTPases. The Rab GAPs included in the screen have been identified due to their characteristic TBC catalytic domains identified by genome scanning, but discrete functions and target Rabs have not been described for many of these proteins. Indeed, we know that the GAP for Rab6 has, to date, not been identified among the 37 TBC domain proteins that we used (13). Furthermore, due to the variability of the screening data, small effects on HSV-1 titer (2- to 3-fold) may have been hidden by the inherent experimental noise. Nevertheless, our data suggest that Rab1 and Rab43 are among the most crucial Rab proteins for the efficient assembly of infectious HSV-1.
In addition to identifying these Rab GTPases, this screen has highlighted some observations about the potential sites of secondary envelopment. First, in the absence of functional Rab1, viral envelope proteins accumulate in the ER, and virus assembly is less efficient. Our data led us to conclude that the viral envelope must normally be acquired from a post-ER compartment. Second, in the process of investigating the role of Rab43 in HSV-1 assembly, we observed, in the context of infection, that viral glycoproteins are not trafficked back to a TGN46-positive compartment efficiently after passing over the cell surface in HeLa cells. Thus, the major portion of envelope protein we observed in the TGN must be newly synthesized. This could either mean that only newly synthesized envelope proteins are incorporated into virions at the TGN or, interestingly, that secondary envelopment could occur at other post-TGN membranes, in particular, those with markers for the early endosome. Indeed, the exact nature of the compartment where herpesvirus secondary envelopment occurs is still open to speculation, with suggestions of both TGN and endosomal origin (30, 32, 33). It would be interesting to conduct further experiments to distinguish between these possibilities and further characterize the site of secondary envelopment.
While both Rab1 and Rab43 activities are clearly important for HSV-1 assembly, it is likely that other Rab proteins are involved in different aspects of HSV-1 replication in addition to virus assembly. Indeed, it has recently been shown that Rab6 is important for the trafficking of the human cytomegalovirus tegument protein pp150 to the virus assembly compartment (17, 18). Further processes that may involve Rab proteins include endocytosis, in those cells where HSV-1 appears to enter by an endocytic mechanism, as well as Rab proteins involved in the exocytic release of HSV-1 by fusion of the virus-containing compartment with the plasma membrane. These aspects are currently being investigated.
In summary, we have shown for the first time an important role of the cellular trafficking mediators Rab1a/b and Rab43 in the life cycle of HSV-1. Rab1 activity is required for anterograde transport of viral envelope proteins to post-ER compartments for virus assembly, whereas Rab43 appears to be important for the integrity and/or identity of the post-Golgi complex assembly compartments. Further work will shed additional light on the complex interplay of HSV-1 infection and the regulation of the secretory and endocytic pathways in human cells.
ACKNOWLEDGMENTS
We thank Amanda Stuart for A549 cells, Nick Bright for access to EM facilities, Susanne Bell and Birgitte Bruun for technical assistance, and members of the Crump lab for helpful discussions.
This study was funded by the Medical Research Council UK (Research Grant 81404) and the Royal Society (University Research Fellowship to C.M.C.).
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Alconada A., Bauer U., Sodeik B., Hoflack B. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barr F., Lambright D. G. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beitia Ortiz de Zarate I., Kaelin K., Rozenberg F. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 78:1540–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonifacino J. S., Traub L. M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 [DOI] [PubMed] [Google Scholar]
- 5. Browne H., Bell S., Minson T. 2004. Analysis of the requirement for glycoprotein m in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruce E. A., Digard P., Stuart A. D. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crump C. M., et al. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517–3527 [DOI] [PubMed] [Google Scholar]
- 8. Crump C. M., Yates C., Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desai P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desai P. J., Schaffer P. A., Minson A. C. 1988. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J. Gen. Virol. 69:1147–1156 [DOI] [PubMed] [Google Scholar]
- 11. Farnsworth A., Johnson D. C. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foster T. P., Melancon J. M., Olivier T. L., Kousoulas K. G. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262–13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuchs E., et al. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gill M. B., et al. 2006. Murine gammaherpesvirus-68 glycoprotein H-glycoprotein L complex is a major target for neutralizing monoclonal antibodies. J. Gen. Virol. 87:1465–1475 [DOI] [PubMed] [Google Scholar]
- 15. Haas A. K., et al. 2007. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 120:2997–3010 [DOI] [PubMed] [Google Scholar]
- 16. Hsu C., et al. 2010. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 189:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Indran S. V., Ballestas M. E., Britt W. J. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J. Virol. 84:3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Indran S. V., Britt W. J. 2011. A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J. Virol. 85:5213–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katz E., Margalith E., Duksin D. 1980. Antiviral activity of tunicamycin on herpes simplex virus. Antimicrob. Agents Chemother. 17:1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kousoulas K. G., Bzik D. J., DeLuca N., Person S. 1983. The effect of ammonium chloride and tunicamycin on the glycoprotein content and infectivity of herpes simplex virus type 1. Virology 125:468–474 [DOI] [PubMed] [Google Scholar]
- 21. Kuhn J. E., Eing B. R., Brossmer R., Munk K., Braun R. W. 1988. Removal of N-linked carbohydrates decreases the infectivity of herpes simplex virus type 1. J. Gen. Virol. 69:2847–2858 [DOI] [PubMed] [Google Scholar]
- 22. Loret S., Guay G., Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manna D., et al. 2010. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398:21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McMillan T. N., Johnson D. C. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 26. Minson A. C., et al. 1986. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J. Gen. Virol. 67:1001–1013 [DOI] [PubMed] [Google Scholar]
- 27. Pawliczek T., Crump C. M. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowe R. K., Suszko J. W., Pekosz A. 2008. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology 382:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 30. Sugimoto K., et al. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tisdale E. J., Bourne J. R., Khosravi-Far R., Der C. J., Balch W. E. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tooze J., Hollinshead M., Reis B., Radsak K., Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 33. Turcotte S., Letellier J., Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshimura S., Haas A. K., Barr F. A. 2008. Analysis of Rab GTPase and GTPase-activating protein function at primary cilia. Methods Enzymol. 439:353–364 [DOI] [PubMed] [Google Scholar]