Abstract
The susceptibility of sheep to scrapie is influenced mainly by the prion protein polymorphisms A136V, R154H, and Q171R/H. Here we analyzed the ability of protein misfolding cyclic amplification (PMCA) to model the genetic susceptibility of sheep to scrapie. For this purpose, we studied the efficiency of brain homogenates from sheep with different PrP genotypes to support PrPSc amplification by PMCA using an ARQ/ARQ scrapie inoculum. The results were then compared with those obtained in vivo using the same sheep breed, genotypes, and scrapie inoculum. Genotypes associated with susceptibility (ARQ/ARQ, ARQ/AHQ, and AHQ/ARH) were able to sustain PrPSc amplification in PMCA reactions, while genotypes associated with resistance to scrapie (ARQ/ARR and ARR/ARR) were unable to support the in vitro conversion. The incubation times of the experimental infection were then compared with the in vitro amplification factors. Linear regression analysis showed that the efficiency of in vitro PrPSc amplification of the different genotypes was indeed inversely proportional to their incubation times. Finally, the rare ARQK176/ARQK176 genotype, for which no in vivo data are available, was studied by PMCA. No amplification was obtained, suggesting ARQK176/ARQK176 as an additional genotype associated with resistance, at least to the isolate tested. Our results indicate a direct correlation between the ability of different PrP genotypes to undergo PrPC-to-PrPSc conversion by PMCA and their in vivo susceptibility and point to PMCA as an alternative to transmission studies and a potential tool to test the susceptibility of numerous sheep PrP genotypes to a variety of prion sources.
INTRODUCTION
Scrapie is a fatal neurodegenerative disease of sheep and goats, belonging to transmissible spongiform encephalopathies (TSEs) or prion diseases. TSEs include bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) of cervids, and Creutzfeldt-Jakob disease (CJD) in humans.
TSEs are characterized by the accumulation of the pathological protease-resistant isoform (PrPSc) of the host-encoded cellular prion protein (PrPC) mainly in the central nervous system of affected subjects (36). The abnormal protein may also accumulate in other tissues, such as in the lymphoreticular system and variably in other tissues or body fluids.
Classical scrapie is a contagious disease under natural conditions. It has been observed since the 18th century in Europe and represents the archetype of the TSEs. The susceptibility of sheep to scrapie depends on several factors but is influenced mainly by the strain of the agent and the genetic background of the host, driven primarily by the amino acid sequence of PrP (20, 24). Nor98 or atypical scrapie, first detected in Norway in 1998, is a prion disease that shows distinct phenotypic characteristics compared with classical scrapie, and a spontaneous origin, like that of human sporadic CJD, has been suggested (6).
The ovine PrP gene is highly polymorphic, and several alleles have been demonstrated to influence susceptibility to the disease (4, 5, 8, 19, 31, 45). The most common alleles, ARQ, VRQ, ARR, ARH, and AHQ, are determined by polymorphisms at codons 136, 154, and 171. Knowledge of genotypes associated with resistance in sheep has been obtained in the past by experimental transmission of the disease to sheep and by observational studies (i.e., case-control studies) in the general population (4). The ARR allele has always been found to be associated with protection from scrapie, with very few exceptions in classical scrapie (25) but with most related to Nor98 (6). The VRQ allele is significantly linked to short survival times of experimentally infected sheep and to high susceptibility to classical scrapie in many breeds in field conditions. In breeds in which the VRQ allele is absent or rare (for instance, Suffolk or Sarda breeds), the wild-type ARQ allele is associated with the highest susceptibility to classical scrapie (4, 16). Moreover, using the in vitro cell-free conversion system, the ARR allele showed the lowest conversion efficiency if compared to ARQ or VRQ (39). The homologous Q171R sheep polymorphisms introduced in the mouse PrP gene rendered mouse PrPC inconvertible into PrPSc (37). On the basis of these findings breeding programs have been implemented in Europe and in the United States with the aim of reducing and possibly eradicating scrapie from sheep populations. This will be achieved by reducing the frequency of animals carrying genotypes associated with susceptibility and increasing that of animals with genotypes associated with resistance to scrapie. However, several polymorphisms other than those at codons 136, 154, and 171 have been reported (18, 35, 47, 50) as possibly influencing susceptibility to scrapie. For instance, experimental transmission and case-control studies have shown that the AT137RQ and the ARQK176 sheep alleles are significantly associated with resistance to classical scrapie (47, 50). However, the protective effect of these rare alleles has been documented only with heterozygous sheep, given the extremely low frequency of homozygous sheep in the field. The susceptibility of animals that are homozygous for these alleles still remains to be evaluated before considering them of interest for breeding programs for resistance.
Using a cell-free system that employs purified 35S-labeled PrPC (28), the genetic susceptibility of sheep to classical scrapie has been studied in vitro. A variable propensity of diverse recombinant PrP variants to undergo conversion to the protease-resistant form has been observed (7). However, using the same method, it has also been found that—probably on account of the absence of some host factor—this in vitro cell-free conversion method does not always reflect the biological behavior of prions in vivo (38).
A highly efficient in vitro conversion method, protein misfolding cyclic amplification (PMCA), was developed some years ago (41). This technique speeds up the conversion of PrPC from healthy brain homogenates by the addition of minute amounts of PrPSc, followed by alternating and repeating cycles of sonication and incubation. During PMCA all cell factors that could participate in the conversion event are present in the in vitro reaction. Several findings have shown that PMCA mimics numerous aspects of prion biology. In particular, it has been shown that PMCA is able to amplify infectivity (11). In addition, the biological, biochemical, and infectious properties of distinct prion strains can be faithfully propagated after serial PMCA passages, maintaining their peculiar characteristics (10). Several groups have recently demonstrated the potential of PMCA as a diagnostic tool. Indeed, it was able to detect PrPSc in the blood of hamsters (12), sheep (44), mice (43), and deer (40) infected with prions, in the urine of affected hamsters (21) and deer (23), and in the saliva of scrapie-affected sheep (34) and CWD-affected deer (23).
The aim of this study was to investigate whether the in vitro replication of scrapie prions by PMCA can reproduce the exquisite PrP genetic preferences operating in sheep.
MATERIALS AND METHODS
Source and preparation of substrates.
Ten animals of the Sarda breed were selected from a single flock, with no history of scrapie, on the basis of the PrP genotype, identified by sequencing the complete coding sequence, as previously described (49). Two ARQ/ARQ and two ARR/ARR animals, as well as one animal for each of the following genotypes, ARR/ARQ, ARQ/AHQ, AHQ/ARH ARQ/AT137RQ, ARQ/ARQK176, and ARQK176/ARQK176, were included in the study. Animals were perfused with phosphate-buffered saline (PBS) plus 5 mM EDTA prior to the removal of the brain, and the brain was stored at −80°C.
Ten percent (wt/vol) uninfected cerebral cortex homogenates (substrates) were prepared in ice-cold conversion buffer (PBS containing NaCl 150 mM, 1% Triton X-100, 4 mM EDTA, pH 7.4) and mini-Complete protease inhibitor (Roche). Tissues were homogenized with three passes of 15 s of homogenization in a glass/Teflon 10-ml potter maintained on ice, using a potter's (B. Braun Biotech International) set at 1,200 rpm. Large particulate matter was removed by centrifugation at 1,000 × g for 1 min. Final homogenates were stored at −80°C until use.
The scrapie inoculum used for the in vitro study was the same as that used for the experimental transmission of classical scrapie to Sarda breed sheep (47). It was prepared by pooling brain tissue from eight fully sequenced ARQ/ARQ scrapie-affected Sarda sheep as 10% homogenate. A single prion strain which was indistinguishable from all the other classical scrapie strains from Italy characterized so far was involved in these cases (13).
PMCA.
The inoculum was serially diluted in the substrates, depending on the experiments, from 1:10 (10−2), 1:100 (10−3), 1:1,000 (10−4), 1:10,000(10−5), and 1:100,000 (10−6) (vol/vol). Samples were either frozen at −20°C or immediately submitted to PMCA. Each sample (50 μl) was inserted in a 200-μl PCR tube and placed in the plate holder of a microsonicator (Misonix S4000) programmed to perform cycles, at 37°C, of 30 min of incubation followed by a pulse of 20 s of sonication set at 80% of amplitude, for 48 h (one PMCA round). In all the experiments each dilution of the inoculum was tested as a duplicate. Moreover, two negative controls constituted of 50 μl of substrate were included in each PMCA experiment.
Western immunoblot analysis of PrPSc.
A total of 40 μl of each sample was diluted 1:1 in conversion buffer and digested with proteinase K at a final concentration of 150 μg ml−1 at 38°C for 60 min. An equal volume of a 1:1 butanol-isopropanol mixture was added to the samples, and the samples were centrifuged at 20,000 × g for 5 min.
The supernatant was discarded, and 20 μl of 1× sample-reducing buffer (Invitrogen) was added to the pellets, heated at 90°C for 10 min, and centrifuged at 10,000 × g for 5 min. A total of 15 μl of each sample was loaded into NuPAGE 10% Bis-Tris polyacrylamide gel (Invitrogen). After electrophoresis and Western blotting on polyvinylidene difluoride membranes (Millipore), blots were processed using the SNAP i.d. protein detection system (Millipore). PrP was labeled with the anti-PrP monoclonal antibody 12B2 (2.38 μg ml−1) (30), visualized by the chemiluminescence method (SuperSignal West Femto; Pierce Biotechnology), and detected by the VersaDoc imaging system (Bio-Rad). The amount of each PrPSc band was measured by QuantityOne software (Bio-Rad).
Calculation of amplification factors.
The same serial dilution of PrPSc, equivalent to 0.25, 0.5, 1, and 2 mg per lane of scrapie-infected brain tissue, was loaded in each gel. Quantification of the chemiluminescent signal (QuantityOne software; Bio-Rad) for each dilution provided the standard curve used to estimate the amount of PrPSc in the samples loaded in the same gel, expressed as the milligram equivalent of infected brain.
The amplification factor was calculated for each genotype by quantifying the amount of PrPSc in the last quantifiable post-PMCA dilution (y) and in the 10−2 frozen dilution (x), taking into account the different dilution factors between the last quantifiable post-PMCA dilution and 10−2, using the formula: (y/x) × dilution factor.
The amplification factor was calculated as the mean value (± standard deviation) of three independent experiments for each substrate. The quantitative estimation was obtained by using the same standard curve in each Western blotting (WB) in order to obtain comparable results.
Student's t test (one side) was used to assess whether the mean value of the amplification factor for each genotype was statistically significantly greater than 1 (no amplification of prion protein). The same t test (two tailed) was used to test for the statistical significance of the difference in the mean amplification factor between substrates.
Statistical analysis.
Linear regression analysis was applied to study the relation between the PMCA efficiency expressed as the amplification factor and the in vivo host susceptibility, as estimated by survival times of animals (days postinfection at death), challenged by intracerebral route. For each genotype, the mean value of the PMCA amplification factor and the average survival time were calculated and entered in the regression model as independent and dependent variables, respectively.
Linear regression function was studied for both the bivariate distributions of the PMCA amplification factor and survival times for all the available genotypes after intracerebral inoculation.
Linear regression equation and determination coefficient (R2) were obtained, and the statistical significance of the model was assessed by testing the regression coefficient with the t test (df = n − 2) against the null hypothesis Ho: B = 0.
Data management and analyses were performed using XLSTAT.2009.6.01 (Addinsoft SARL, Paris, France).
RESULTS
PMCA with the ARQ/ARQ substrate.
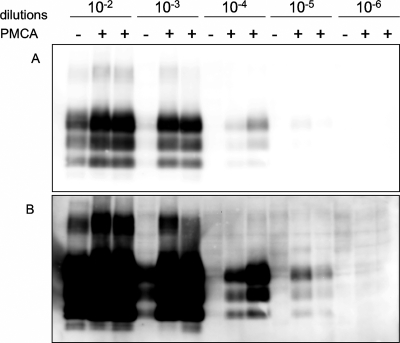
To evaluate the efficiency of sheep brain homogenates as a substrate for PMCA, we first tested the ARQ/ARQ genotype. The inoculum, a scrapie brain pool homogenate (10%, wt/vol) already used in an experimental transmission study of sheep (47), was serially diluted, from 10−2 to 10−6, in the ARQ/ARQ substrate. Samples were subjected to a single round of PMCA in duplicate. When the amount of PrPSc was compared in samples before and after PMCA, a clear increase in PrPSc was observed (Fig. 1A). PK-resistant PrPSc (PrPres) was detected up to the 10−3 dilution in frozen samples, after prolonged exposure of the membranes (Fig. 1B, lane 4). However, using the same blot exposure, it was possible to identify the typical three PrPSc protein bands up to the 10−5 dilutions in post-PMCA samples (Fig. 1B, lanes 11 and 12), evidencing an efficient PrPSc amplification. Post-PMCA samples showed a similar PrPSc molecular profile at all dilutions, which was identical to that observed with the inoculum.
Fig. 1.
ARQ/ARQ sheep brain homogenate used as substrate in PMCA. (A) PK-resistant PrPSc from 10−2 to 10−6 serial dilutions of the inoculum in the substrate, before (−) and after (+) one round of PMCA (in duplicate), as detected by WB. (B) After a longer exposure time for the same blot, it is possible to detect PK-resistant PrPSc up to 10−5 dilutions.
Amplification factors for the various PrP genotypes.
All substrates used in PMCA were first tested for the presence of PrPSc, and none resulted in a positive test. The amount of PrPC in each substrate was measured, by Western blotting, in order to exclude a possible difference in PMCA efficiency due to different PrPC availabilities in substrates. No significant difference in the quantity of PrPC was observed between all substrates.
In order to make a quantitative comparison of differences in the amplification efficiency among different genotypes, the inoculum was serially diluted, from 10−2 to 10−4, in substrates derived from two sheep carrying the ARQ/ARQ genotype and two carrying the ARR/ARR genotype or one carrying the ARQ/AHQ, AHQ/ARH, ARQ/ARR, ARQ/AT137RQ, ARQ/ARQK176, and ARQK176/ARQK176 genotypes. Samples were then submitted to a single round of PMCA.
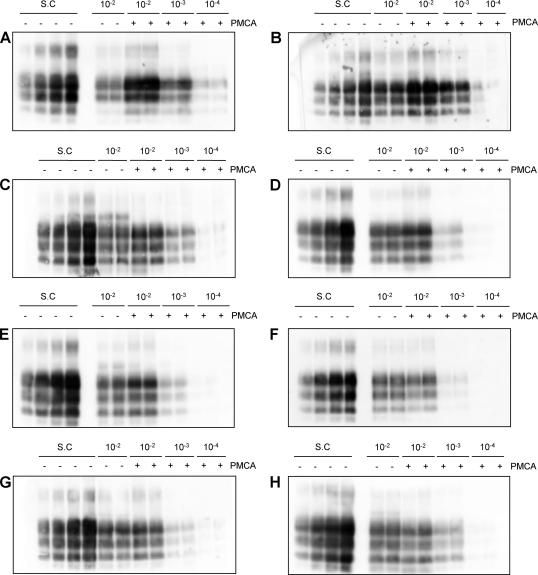
The ARQ/ARQ and ARQ/AHQ substrates showed the highest levels of conversion efficiency. Indeed, there was a clear increase in signal intensity in the post-PMCA samples for both genotypes (Fig. 2A and B). For these substrates the amplification factors were calculated using the 10−3 post-PMCA dilutions, because the intensity of the PrPSc bands of the 10−2 post-PMCA dilutions fell outside the range of the standard curve (Fig. 2A and B). The mean amplification factors obtained for the two ARQ/ARQ substrates were 9.9 ± 2.8 (mean ± SD) and 10.4 ± 1.2, respectively. The ARQ/AHQ substrate gave an amplification factor of 8 ± 2.9.
Fig. 2.
Comparison of PMCA conversion efficiencies of different substrates. Representative WBs used to calculate the amplification factor for each genotype are shown. (A) ARQ/ARQ; (B) ARQ/AHQ; (C) AHQ/ARH; (D) ARQ/ARR; (E) ARR/ARR; (F) ARQ/AT137RQ; (G) ARQ/ARQK176; (H) ARQK176/ARQK176. In each blot the standard curve (S.C) and duplicates of serial dilutions of the inoculum in each substrate are loaded before (−) and after (+) one round of PMCA.
A slight increase in PrPSc intensity was also detected after the PMCA using the AHQ/ARH substrate. Using this substrate, the PrPSc intensity of the 10−2 post-PMCA samples was different from those for the previous genotypes included within the standard curve range (Fig. 2C). The 10−2 dilutions were then used for amplification efficiency determination. The mean amplification factor obtained for the AHQ/ARH substrate was 3.6 ± 2.0. This amplification factor was significantly lower than that of ARQ/ARQ (P = 0.002) and marginally lower than that of ARQ/AHQ (P = 0.090).
In contrast, no significant amplification was observed using the ARR/ARR, ARQ/ARR, ARQ/AT137RQ, ARQ/ARQK176, and ARQK176/ARQK176 substrates (Fig. 2D, E, F, G, and H). Indeed, all of them showed a PrPSc signal intensity for the 10−2 frozen samples similar to that of the 10−2 samples after one round of PMCA. The mean amplification factors obtained for the two ARR/ARR and for the single ARQ/ARR, ARQ/AT137RQ, ARQ/ARQK176, and ARQK176/ARQK176 substrates were 1.3 ± 0.3, 1.6 ± 0.6, 1.3 ± 0.4, 1.9 ± 1.3, 1.3 ± 0.2, and 2.0 ± 1.6, respectively. These amplification factors were not significantly different from 1 (P > 0.05; one tailed), indicating a failure of PrPC from these brain homogenates to undergo seeded conversion into PrPSc. Importantly, we found no significant differences between the two ARQ/ARQ or the two ARR/ARR substrates, indicating an insignificant individual effect on the level of amplification by PMCA. All negative controls used in each PMCA experiment remained negative for PrPSc (data not shown).
Comparison between in vitro and in vivo data.
In a previous study (47), we analyzed the effect of the PrP genotype on the susceptibility to classical scrapie of sheep experimentally challenged by the intracerebral or oral route. We observed different survival times in the ARQ/ARQ, ARQ/AHQ, AHQ/AHQ, and ARQ/ARH genotypes, indicating a variable degree of susceptibility. In these experiments, all animals carrying at least one ARR or the AT137RQ and the ARQK176 alleles failed to develop the disease, clearly indicating a protection from scrapie.
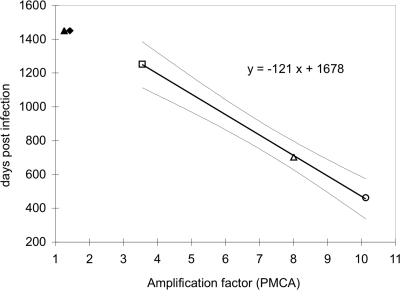
Given that the same sheep breed, PrP genotypes, and scrapie inoculum were used for the in vivo and in vitro experiments, we tested the correlation between the in vivo genetic susceptibility and the efficiency of in vitro conversion obtained by PMCA. A linear regression model was used to analyze the relationship between the amplification efficiency obtained by PMCA and the survival time observed following scrapie infection by intracerebral route (Table 1). The resulting best-fit linear regression function, indicating the PMCA amplification factor and days postinfection with x and y, respectively, was y = −121x + 1,678 (Fig. 3). A negative linear relationship between amplification efficiency and survival time was observed. The coefficient of determination (R2) for the association between PMCA amplification factor and survival time after experimental infection was R2 = 0.999 for the intracerebral challenge, with a P value of >0.05. These results show that the regression coefficients differ significantly from 0, indicating that the regression models fit the data much better than the null hypothesis (no relationship between the variables) at least within the range of the observed data.
Table 1.
Mean values of the amplification factor by PMCA and survival times (days postinfection) after oral and intracerebral scrapie infection, for each genotype
| Genotype | Amplification factor (PMCA) | dpia | |
|---|---|---|---|
| Oral | i.c. | ||
| ARQ/ARQ | 10.1 | 833 | 462 |
| ARQ/AHQ | 8 | 1,115 | 703 |
| AHQ/AHQ | 790 | ||
| ARQ/ARH | >1,550 | 1,083 | |
| AHQ/ARH | 3.6 | 1,252 | |
| ARQK176/ARQK176 | 2 | ||
| ARQ/AT137RQ | 1.9 | >1,550 | |
| ARR/ARR | 1.4 | >1,550 | >1,450 |
| ARQ/ARR | 1.3 | >1,550 | >1,450 |
| ARQ/ARQK176 | 1.2 | >1,550 | |
dpi, days postinfection; i.c., intracerebral.
Fig. 3.
Linear regression models describing the relationship between amplification factor by PMCA and survival times of sheep infected by intracerebral route: ARQ/ARR (▴), ARR/ARR (♦), AHQ/ARH (□), ARQ/AHQ (▵), and ARQ/ARQ (○). Light gray continuous lines indicate confidence bands in the model. Open symbols refer to animals that died of scrapie, filled symbols to animals that were still alive at the time of follow-up.
The absence of incubation times for resistant animals did not permit the use of additional genotypes for the computation of the linear regression. However, for these genotypes an amplification factor not statistically different from 1 was observed, indicating a clear correlation of resistance with inability to sustain replication by PMCA in our experimental setup.
DISCUSSION
Several studies have demonstrated the ability of PMCA to sustain in vitro conversion of PrPC into PrPSc, using different mammalian species as the source of substrates and inocula (17). The PMCA setup in our laboratory was very sensitive, being able to detect a dilution of 10−5 of the scrapie inoculum after a single PMCA round. This allowed us to have a wide range of measurement, permitting us to detect minor differences in the amplification efficiencies of different substrates. The high sensitivity was obtained by coupling a single round of PMCA with a highly sensitive PrPSc detection by Western blot analysis.
The main aim of our study was to establish whether PMCA could reproduce the genetic susceptibility of sheep to scrapie. In a previous study (47), we analyzed the effect of the PrP genotype on the susceptibility of sheep to experimental challenge with scrapie. Overall, our data show that the susceptibility of the genotypes observed in vivo correlates with the conversion efficiency obtained in vitro by PMCA. Indeed, the ARQ/ARQ genotype, which was associated with the highest susceptibility to classical scrapie, was also the most efficiently converted by PMCA. The ARQ/AHQ genotype showed a slightly less efficient conversion than ARQ/ARQ, thus reflecting the observation that sheep with the ARQ/AHQ genotype show, in vivo, longer survival times than ARQ/ARQ sheep. Moreover, a lower risk has been found to be associated in field conditions with the ARQ/AHQ than with the ARQ/ARQ genotype (3). In our previous intracerebral challenge with scrapie (47), sheep carrying the ARQ/ARH genotype showed much longer survival times than sheep with the ARQ/ARQ, ARQ/AHQ, and AHQ/AHQ genotypes. Moreover, when the ARH and AHQ alleles were combined, the incubation time was longer than that of AHQ/AHQ animals. In PMCA experiments, the amplification factor obtained for the AHQ/ARH genotype was significantly lower than that of ARQ/ARQ and marginally lower than that of ARQ/AHQ. These results clearly suggest that the rank order of susceptibility obtained from the in vivo study, ARQ>AHQ>ARH, was reproduced also by in vitro experiments. Linear regression analysis highlighted the strong correlation between the PMCA amplification factor and the in vivo susceptibility of different PrP genotypes. The statistical significance obtained and the good fit of the linear regression support the ability of PMCA to replicate in vitro the genotype susceptibility phenomenon observed in vivo. This result clearly indicates the potential of PMCA as a tool to be used to predict the genetic susceptibility of sheep and as an alternative to experimental challenge.
The resistance to classical scrapie conferred by the ARR allele in an autosomally dominant manner is widely accepted. In fact, only three classical scrapie cases carrying the ARR/ARR genotype have been reported to date (22, 25). Accordingly, PMCA detected no evident amplification using ARQ/ARR and ARR/ARR genotypes. Recent studies indicate that some rare PrP variant may also confer protection from classical scrapie (47, 50), and this is of particular interest for scrapie control strategies, taking into consideration the existence of sheep breeds in which the ARR allele is rare or absent (1, 46) and the potential risk deriving from the selection in favor of a single allele. The control of scrapie in goats, which do not have the ARR allele, also represents a challenge for health policies, given that two BSE cases have been detected so far in goats (15, 26). In our study we included the ARQ/AT137RQ and ARQ/ARQK176 genotypes, which have recently been shown to be significantly associated with classical scrapie resistance in Italy. Indeed, sheep carrying the ARQK176 and AT137RQ alleles were resistant to experimental challenge (47), and no sheep carrying these variants were found to be positive in outbreaks with a high prevalence of scrapie (50). In our PMCA experiments, the ARQ/AT137RQ and ARQ/ARQK176 genotypes were unable to produce newly formed PrPSc, thus reproducing what was observed in vivo. Given the low frequency of these alleles and the consequent rarity of homozygous sheep, their protective effect has been studied only with heterozygous animals. To determine definitively the susceptibility level of rare alleles and their potential usefulness for scrapie eradication plans, the susceptibility of homozygous animals needs to be evaluated. As a matter of fact, it has been reported that in mice inoculated with some scrapie strains, PrP heterozygous individuals may be less susceptible than homozygous individuals (9), in accordance with a phenomenon known as overdominance of heterozygosis (14). Given the obvious advantages offered by PMCA over expensive and time-consuming in vivo experiments, we tested the ability of a brain substrate with the homozygous ARQK176/ARQK176 genotype to support PrPSc amplification by PMCA. An amplification factor not significantly different from 1 was observed, thus suggesting that the ARQK176 confers resistance to the classical scrapie isolate used in this study in both heterozygosis and homozygosis.
The result of our experiment could have been biased by the use of a single animal as a donor of substrate for several genotypes tested. However, the results obtained with ARQ/ARQ and ARR/ARR genotypes, for which two substrates were used, clearly indicate that individual variability has a negligible effect, if compared with genotype, on the PMCA amplification factor.
Among genotypes for which a significant amplification factor was observed, the AHQ/ARH genotype is the one with the lowest value, thus suggesting an association with a low susceptibility rather than resistance to classical scrapie. However, in previous experiments we showed that sheep carrying the ARH allele were susceptible after intracerebral scrapie challenge but were resistant to oral challenge with the same inoculum (47). This partial discrepancy between PMCA and in vivo experiments highlights how PMCA reflects the intracerebral better than the oral challenge. On this point it could be speculated that PMCA should be able to reflect scrapie replication in the brain, but not the complex pathogenetic pathway occurring in vivo, which also involves prion replication in peripheral tissues. Scrapie replication in tissues other than the brain may indeed have features which were not modeled by our PMCA conditions, which used brain-derived substrates for prion conversion.
Based on these considerations, it can be assumed that animals with genotypes having PMCA amplification factors statistically higher than 1 are more likely to be susceptible to classical scrapie after experimental challenge by the intracerebral route, but not necessarily by the oral route. On the other hand, the oral route is more similar to natural infection, so animals with a genotype showing an amplification factor not significantly different from 1, indicating a low susceptibility even to intracerebral challenge, should be considered particularly resistant to classical scrapie in field conditions.
In conclusion, PMCA could be a valuable alternative to the experimental transmission of classical scrapie to sheep, offering a rapid means to test the susceptibility of different PrP genotypes to several scrapie sources. In a similar manner the use of PMCA could be of incomparable value in studying the association with the resistance or susceptibility of candidate genotypes in goats, for which a genetic selection plan for scrapie resistance is still not available (48).
Given the high sensitivity of PrPSc detection by PMCA, this technique has been envisaged as a powerful in vitro diagnostic tool (12, 27, 29, 44). The sensitivity obtained in our lab is very high, and it is therefore predictable that with additional rounds this technique should be extremely sensitive and able to detect PrPSc expected in samples such as skeletal muscles, kidneys, salivary glands, or mammary glands from scrapie-infected sheep (2, 32, 33, 42, 51).
ACKNOWLEDGMENTS
We thank J. Langeveld (CVI, Lelystad, Netherlands) for supplying 12B2 antibody.
This research was supported by the Italian Ministry of Health, Department of Veterinary Public Health, Nutrition and Food Safety, and the European network of excellence NeuroPrion (FOOD-CT-2004-506579).
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Acutis P. L., et al. 2004. Low frequency of the scrapie resistance-associated allele and presence of lysine-171 allele of the prion protein gene in Italian Biellese ovine breed. J. Gen. Virol. 85:3165–3172 [DOI] [PubMed] [Google Scholar]
- 2. Andréoletti O., et al. 2004. PrPSc accumulation in myocytes from sheep incubating natural scrapie. Nat. Med. 10:591–593 [DOI] [PubMed] [Google Scholar]
- 3. Baylis M., et al. 2004. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 85:2735–2740 [DOI] [PubMed] [Google Scholar]
- 4. Baylis M., Goldmann W. 2004. The genetics of scrapie in sheep and goats. Curr. Mol. Med. 4:385–396 [DOI] [PubMed] [Google Scholar]
- 5. Belt P. B., et al. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 76(3):509–517 [DOI] [PubMed] [Google Scholar]
- 6. Benestad S. L., Arsac J. N., Goldmann W., Noremark M. 2008. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet. Res. 39:19. [DOI] [PubMed] [Google Scholar]
- 7. Bossers A., de Vries R., Smits M. A. 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 74:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bossers A., Schreuder B. E., Muileman I. H., Belt P. B., Smits M. A. 1996. PrP genotype contributes to determining survival times of sheep with natural scrapie. J. Gen. Virol. 77(10):2669–2673 [DOI] [PubMed] [Google Scholar]
- 9. Bruce M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49:822–838 [DOI] [PubMed] [Google Scholar]
- 10. Castilla J., et al. 2008. Cell-free propagation of prion strains. EMBO J. 27:2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castilla J., Saa P., Hetz C., Soto C. 2005. In vitro generation of infectious scrapie prions. Cell 121:195–206 [DOI] [PubMed] [Google Scholar]
- 12. Castilla J., Saa P., Soto C. 2005. Detection of prions in blood. Nat. Med. 11:982–985 [DOI] [PubMed] [Google Scholar]
- 13. Di Bari M. A., et al. 2008. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J. Gen. Virol. 89:2975–2985 [DOI] [PubMed] [Google Scholar]
- 14. Dickinson A. G., Fraser H. 1979. An assessment of the genetics of scrapie in sheep and mice, p. 367–385. In S. B. Prusiner and W. J. Hadlow (ed.), Slow transmissible diseases of the nervous system, vol. 1 Academic Press, New York, NY [Google Scholar]
- 15. Eloit M., et al. 2005. BSE agent signatures in a goat. Vet. Rec. 156:523–524 [DOI] [PubMed] [Google Scholar]
- 16. Elsen J. M., et al. 1999. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch. Virol. 144:431–445 [DOI] [PubMed] [Google Scholar]
- 17. Fernández-Borges N., de Castro J., Castilla J. 2009. In vitro studies of the transmission barrier. Prion 3:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldmann W., et al. 2006. Ovine prion protein variant A136R154L168Q171 increases resistance to experimental challenge with bovine spongiform encephalopathy agent. J. Gen. Virol. 87:3741–3745 [DOI] [PubMed] [Google Scholar]
- 19. Goldmann W., et al. 1990. Two alleles of a neural protein gene linked to scrapie in sheep. Proc. Natl. Acad. Sci. U. S. A. 87:2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldmann W., Hunter N., Smith G., Foster J., Hope J. 1994. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 75(5):989–995 [DOI] [PubMed] [Google Scholar]
- 21. Gonzalez-Romero D., Barria M. A., Leon P., Morales R., Soto C. 2008. Detection of infectious prions in urine. FEBS Lett. 582:3161–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groschup M. H., et al. 2007. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerg. Infect. Dis. 13:1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haley N. J., Seelig D. M., Zabel M. D., Telling G. C., Hoover E. A. 2009. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One 4:e4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hunter N. 1997. PrP genetics in sheep and the applications for scrapie and BSE. Trends Microbiol. 5:331–334 [DOI] [PubMed] [Google Scholar]
- 25. Ikeda T., et al. 1995. Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J. Gen. Virol. 76(10):2577–2581 [DOI] [PubMed] [Google Scholar]
- 26. Jeffrey M., et al. 2006. Immunohistochemical features of PrP(d) accumulation in natural and experimental goat transmissible spongiform encephalopathies. J. Comp. Pathol. 134:171–181 [DOI] [PubMed] [Google Scholar]
- 27. Jones M., et al. 2007. In vitro amplification and detection of variant Creutzfeldt-Jakob disease PrPSc. J. Pathol. 213:21–26 [DOI] [PubMed] [Google Scholar]
- 28. Kocisko D. A., et al. 1994. Cell-free formation of protease-resistant prion protein. Nature 370:471–474 [DOI] [PubMed] [Google Scholar]
- 29. Kurt T. D., et al. 2007. Efficient in vitro amplification of chronic wasting disease PrPRES. J. Virol. 81:9605–9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langeveld J. P., et al. 2006. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet. Res. 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laplanche J. L., et al. 1993. PrP polymorphisms associated with natural scrapie discovered by denaturing gradient gel electrophoresis. Genomics 15:30–37 [DOI] [PubMed] [Google Scholar]
- 32. Ligios C., et al. 2007. Intraepithelial and interstitial deposition of pathological prion protein in kidneys of scrapie-affected sheep. PLoS One 2:e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ligios C., et al. 2005. PrPSc in mammary glands of sheep affected by scrapie and mastitis. Nat. Med. 11:1137–1138 [DOI] [PubMed] [Google Scholar]
- 34. Maddison B. C., et al. 2010. Prions are secreted into the oral cavity in sheep with preclinical scrapie. J. Infect. Dis. 201:1672–1676 [DOI] [PubMed] [Google Scholar]
- 35. Moum T., et al. 2005. Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. J. Gen. Virol. 86:231–235 [DOI] [PubMed] [Google Scholar]
- 36. Oesch B., et al. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell 40:735–746 [DOI] [PubMed] [Google Scholar]
- 37. Perrier V., et al. 2002. Dominant-negative inhibition of prion replication in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 99:13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piening N., et al. 2006. Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo. J. Biol. Chem. 281:9373–9384 [DOI] [PubMed] [Google Scholar]
- 39. Raymond G. J., et al. 1997. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature 388:285–288 [DOI] [PubMed] [Google Scholar]
- 40. Rubenstein R., et al. 2010. A novel method for preclinical detection of PrPSc in blood. J. Gen. Virol. 91:1883–1892 [DOI] [PubMed] [Google Scholar]
- 41. Saborio G. P., Permanne B., Soto C. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810–813 [DOI] [PubMed] [Google Scholar]
- 42. Siso S., et al. 2006. Prion protein in kidneys of scrapie-infected sheep. Vet. Rec. 159:327–328 [DOI] [PubMed] [Google Scholar]
- 43. Tattum M. H., Jones S., Pal S., Collinge J., Jackson G. S. 2010. Discrimination between prion-infected and normal blood samples by protein misfolding cyclic amplification. Transfusion 50:996–1002 [DOI] [PubMed] [Google Scholar]
- 44. Thorne L., Terry L. A. 2008. In vitro amplification of PrPSc derived from the brain and blood of sheep infected with scrapie. J. Gen. Virol. 89:3177–3184 [DOI] [PubMed] [Google Scholar]
- 45. Tranulis M. A., Osland A., Bratberg B., Ulvund M. J. 1999. Prion protein gene polymorphisms in sheep with natural scrapie and healthy controls in Norway. J. Gen. Virol. 80(4):1073–1077 [DOI] [PubMed] [Google Scholar]
- 46. Tsunoda K., et al. 2010. Prion protein polymorphisms and estimation of risk of scrapie in East Asian sheep. Biochem. Genet. 48:13–25 [DOI] [PubMed] [Google Scholar]
- 47. Vaccari G., et al. 2007. Prion protein alleles showing a protective effect on the susceptibility of sheep to scrapie and bovine spongiform encephalopathy. J. Virol. 81:7306–7309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaccari G., et al. 2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet. Res. 40:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vaccari G., et al. 2001. PrP genotype in Sarda breed sheep and its relevance to scrapie. Brief report. Arch. Virol. 146:2029–2037 [DOI] [PubMed] [Google Scholar]
- 50. Vaccari G., et al. 2009. Protective effect of the AT137RQ and ARQK176 PrP allele against classical scrapie in Sarda breed sheep. Vet. Res. 40:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vascellari M., et al. 2007. PrPSc in salivary glands of scrapie-affected sheep. J. Virol. 81:4872–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]