Abstract
The viral RNA-dependent RNA polymerase (vRdRp) of paramyxovirus consists of the large (L) protein and the phosphoprotein (P). P is heavily phosphorylated, and it is thought that the phosphorylation of P plays a role in regulating viral RNA synthesis. However, no phosphorylation site within the P protein in paramyxovirus has been identified as playing a positive role in viral RNA synthesis in virus infection. Using mass spectrometry analysis, the threonine residue at position 286 of P of parainfluenza virus 5 (PIV5) was found phosphorylated. Mutation of T286 to alanine (T286A), aspartic acid (T286D), or glutamic acid (T286E) reduced minigenome activity. Recombinant virus containing a mutation at the T286 position (rPIV5-P-T286A) grew slower than wild-type virus; viral mRNA synthesis and protein expression of rPIV5-P-T286A were delayed. Biochemical studies showed that the binding of NP or L protein with the P mutants or tetramer formation by the mutant P proteins was unaltered from that for wild-type P. While we failed to rescue rPIV5-P-T286E virus, several revertant viruses were obtained. All non-wild-type revertants had mutations at T286 and showed defects in both minigenome activity and viral growth. This is the first time that a phosphorylation site within the P protein in paramyxovirus has been found to play a positive role in viral mRNA synthesis and virus growth.
INTRODUCTION
Parainfluenza virus 5 (PIV5) is a prototypical member of the paramyxovirus family, which contains many human and animal pathogens, such as Sendai virus (SeV), mumps virus (MuV), respiratory syncytial virus (RSV), Hendra virus (HeV), and Nipah virus (NiV) (15). The negative-stranded RNA genome of PIV5 contains seven genes yet encodes eight proteins in the order NP-V/P-M-F-SH-HN-L (15). While V mRNA is transcribed faithfully by the V/P gene, P mRNA is produced by the V/P gene through an insertion of two nontemplate guanine residues at a specific sequence during viral RNA transcription (30). The phosphoprotein (P) and large polymerase (L) protein constitute the viral RNA-dependent RNA polymerase complex (vRdRP). The L protein has enzymatic activities capable of initiation, elongation, and termination of RNA synthesis, as well as addition of the 5′ cap structure and 3′ poly(A) sequence (15). The P protein does not have intrinsic enzymatic activity; however, it is an essential cofactor for the polymerase, regulating polymerase activity in viral RNA replication and transcription (15).
The P proteins of all paramyxoviruses are heavily phosphorylated and hence are named phosphoproteins (15). However, the role of P protein phosphorylation in the replication of paramyxoviruses has been an enigma. It was initially suggested that phosphorylation of the P protein was essential for viral RNA synthesis. In paramyxovirus, the P proteins of SeV and RSV have been studied the most extensively. Up to 11 phosphorylation sites of SeV P protein were detected (14, 32), and S249 was identified as the major phosphorylation site (6, 7). However, mutation at S249 showed normal function in the minigenome system. In addition, recombinant SeV containing a mutation at S249 also showed similar growth characteristics in vitro and in vivo, indicating that this phosphorylation site was dispensable for SeV gene expression and pathogenesis (12). Mutating other sites besides S249 resulted in a decrease of over 90% of the phosphorylation level, yet the mutant P protein still had the same activity in the minigenome system (11).
In RSV P protein, two clusters of phosphorylation sites (amino acid residues 116, 117, and 119 and residues 232 and 237) have been identified (5, 9, 21, 24, 34). Phosphorylation of S232 was proposed to regulate RSV transcription; however, that study was based on in vitro assays (33). When all mutations (five) at major phosphorylation sites were incorporated into recombinant RSV using a reverse genetics system, the phosphorylation level of the P protein was reduced by 90%, and yet there was no difference in terms of viral gene expression, indicating that phosphorylation of the P protein was not required for viral gene expression (17). Further studies of the RSV P protein by mass spectrometry identified T108 as a phosphorylation site. Mutation of T108 diminished RSV minigenome activity, possibly through affecting its interaction with M2-1, an important factor for viral RNA synthesis (2). Mutation at S54 of the RSV P protein was shown to affect viral uncoating by using LiCl treatment, which affects phosphorylation of S54 (3, 4). However, the roles of these phosphorylation sites have not been examined in the context of virus infection.
Recently it was shown that S157 of the PIV5 P protein was phosphorylated in infected cells (31). This phosphorylation correlated with decreased viral gene expression, suggesting that phosphorylation at S157 of the P protein plays a role in downregulating viral gene expression (26, 31). Further studies indicate that polo-like kinase 1 (PLK1) associates with the P protein at S157 and phosphorylates the P protein at S308 (26). Phosphorylation of the P protein at S157 and S308 reduces viral gene expression and prevents cytokine induction, benefiting the virus in the long term. This is the first report that phosphorylation of P negatively regulates viral gene expression. However, no phosphorylation site within the P protein has ever been identified as playing a positive role in viral gene expression. Since the P protein is heavily phosphorylated, we hypothesize that phosphorylation at other sites play a positive role in viral gene expression. In this study, we have identified T286 of the PIV5 P protein as a phosphorylation site in infected cells and have found that it played an important, positive role in viral gene expression by upregulating viral mRNA transcription.
MATERIALS AND METHODS
Plasmids, viruses, and cells.
Plasmids expressing the P mutants P-T286A, P-S36A, P-S126A, P-T286S, P-T286D, P-T286E, P-T286K, and P-T286V were generated using standard molecular cloning techniques. Plasmids expressing NP and Flag-L were cloned into the pCAGGS vector as previously described (18, 31). The minigenome plasmid pSMG-RLuc and the replication-defective minigenome plasmid pSMGmut-RLuc have been described (16, 31). P or P mutants with 8 histidines (His) at the N terminus used for protein purification were cloned into the pET15b vector. Plasmids containing the full-length genome for rPIV5-P-T286A and rPIV5-P-T286E viruses were made similarly to rPIV5-CPI+, rPIV5-V/P-S157A, and rPIV5-P-S308A, as previously described (26). The details of plasmid construction and sequences are available on request. To rescue rPIV5-P-T286A and rPIV5-P-T286E viruses from plasmids, 1 μg NP, 0.2 μg P or P mutants, 1.5 μg L, and 3 μg plasmids containing full-length recombinant virus genome were cotransfected into BSR-T7 cells in a 6-well plate. At 4 days posttransfection, the supernatant was collected and used for plaque assay. Single plaques were selected and further amplified in MDBK cells. RNA was extracted from the supernatant using a QIAmp viral RNA minikit, and reverse transcription (RT) was performed with random primers as previously described (26). The reverse transcription product was further amplified by PCR using primer sets as described previously (26). The PCR products were sequenced by the nucleic acid facility at Pennsylvania State University. To recover rPIV5-P-T286E virus, we collected the transfected cells at 6 days posttransfection and cocultured them with Vero cells for 7 days. The supernatant from the cocultured cells was collected for plaque purification. Single plaques were selected and further amplified in MDBK cells. The viral genomes were sequenced as described above. Three individual experiments were performed. All cell lines were incubated at 37°C with 5% CO2. HeLa and MDBK cells were grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. BHK cells were grown in the medium for HeLa or MDBK cells plus 10% tryptose phosphate broth (TPB). Growth medium for BSR-T7 cells was similar to that for BHK cells with the addition of 400 μg/ml G418. The growth medium for infected cells contained 2% FBS (27).
Mass spectrometry analysis.
To determine the phosphorylation sites within the P protein in PIV5-infected cells, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed as previously described (31). HeLa cells in 10-cm plates were infected with PIV5 at a multiplicity of infection (MOI) of 5. At 24 h postinfection, the cells were lysed by whole-cell extraction buffer (WCEB) (50 mM Tris-HCl [pH 8.0], 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, and 10% glycerol), and anti-V5-conjugated agarose (Sigma-Aldrich) was used to precipitate the P protein. After washing, the agarose beads were mixed with one volume of 2× SDS loading buffer (60 mM Tris-HCl [pH 6.8], 40% glycerol, 4% SDS, 200 mM dithiothreitol (DTT), and a few grains of bromophenol blue) and resolved in 10% SDS-PAGE. The gel was stained with Coomassie blue in a solution of 50% methanol and 10% acetic acid. After destaining, the P protein band was excised and digested with trypsin (Promega). The phosphopeptides were enriched using a TiO2 column and analyzed by LC-MS/MS (Waters Q-Tof Ultima mass spectrometer at W.M. Keck Foundation Biotechnology Resource Laboratory, Yale University). The MS/MS spectra were searched against the NCBI database using the automated Mascot algorithm to identify peptides with possible phosphorylation residues of tyrosine (Tyr), threonine (Thr), and serine (Ser).
PIV5 minigenome system and dual luciferase assay.
The PIV5 minigenome system used in this study was described previously (26). Increasing amounts (0.01 to 0.16 μg/well, 24-well plate) of P, P-T286A, P-S36A, P-S126A, P-T286S, P-T286D, P-T286E, P-T286K, or P-T286V were transfected together with other plasmids (0.2 μg pSMG-Rluc, 0.2 μg NP, 0.3 μg L, and 1 ng FF-Luc) into BSR-T7 cells. At 20 to 22 h postinfection (hpi), 1/10 of the lysate from each well was used for the dual luciferase assay (Promega). The ratio of Renilla luciferase (R-Luc) to firefly luciferase (FF-Luc) activity was normalized as relative luciferase activity. Six replicates of each sample were used for statistical analysis. Aliquots of the cell lysate from the minigenome system were used for Western blot analysis. Mouse anti-NP and Pk antibody (mouse anti-P/V) were used together for immunoblotting to detect the expression of NP protein and P or P mutants.
A minigenome system with a defective trailer sequence was established to measure viral mRNA synthesis (16). Because the trailer sequence is essential for viral RNA replication, mutating the sequence resulted in a minigenome system incapable of vRdRp-directed RNA replication. However, since the leader sequence is still intact, the minigenome system is still capable of vRdRp-directed RNA transcription. Thus, this system can measure RNA transcription without interference from RNA replication. The experiments were performed as for the normal minigenome system, except using pSMGmut-RLuc, which has a 6-nucleotide (nt) deletion in the trailer sequence, instead of pSMG-RLuc.
Protein purification and CD.
Circular dichroism (CD) was carried out using His-P, His-P-T286A, His-P-T286D, and His-P-T286E purified from Escherichia coli. Briefly, P and P mutants in the pET15b vector were transformed into BL21(DE3)pLysS competent cells. Single colonies were picked and grown in LB medium with ampicillin (50 ng/ml) and chloramphenicol (34 ng/ml) at 37°C. When the optical density at 600 nm (OD600) reached 0.5 to 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) (final concentration, 1 mM) was added to the medium to induce protein expression for 4 h at 37°C. The P proteins were purified using Ni-charged resin (Novagen) and examined by SDS-PAGE and Coomassie blue staining. The purified proteins were desalted and resuspended in buffer containing 10 mM potassium phosphate (KH2PO4) and 10 mM potassium chloride (KCl) (pH 7.0). Three hundred microliters of 10 μM His-P and His-P mutants was measured on a Jasco-J715 spectropolarimeter using a 0.1-cm-path-length cuvette (10). Three measurements were taken for each sample, and the averages were compared.
Growth curve and plaque assay.
To characterize the rPIV5-P-T286A virus, MDBK cells in 6-well plates were infected with PIV5 or rPIV5-P-T286A at an MOI of 0.01 or 5. Supernatants were collected at different time points and stored at −70°C. BHK cells in 6-well plates were infected using serial dilutions (1:10 to 1:107) of virus. After 1 to 2 h, the infection medium was removed and replaced with 5 ml growth medium (2% FBS, 10% TPB, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 1% low-melting-point agarose in DMEM). The plaques were counted 3 to 4 days postinfection. Three replicates for each time point were collected for statistical analysis.
Western blot.
To compare the viral protein expression levels in PIV5- and rPIV5-P-T286A-infected cells, HeLa cells were infected with PIV5 or rPIV5-P-T286A at an MOI of 5. The medium was replaced, and the cells were lysed at 4, 8, 12, 16, and 20 hpi with WCEB and mixed with 1 volume of 2× SDS loading buffer. Aliquots of the total lysate were resolved in 10% SDS-PAGE. The proteins were transferred to a polyvinylidene difluoride (PVDF) (Millipore) membrane and incubated with mouse anti-P/V antibodies, followed by incubation with horseradish peroxidase (HRP)-labeled anti-mouse secondary antibody. The PVDF membrane was washed and incubated with ECL advance substrate (GE Healthcare) and scanned using a Kodak Image Station 440 system. Rabbit anti-β-actin was used as a protein loading control. Three individual experiments were performed, and one representative result is used for illustration.
Flow cytometry.
Flow cytometry was performed as previously described (26, 28). HeLa cells at 90% confluence were infected with PIV5 or rPIV5-P-T286A at an MOI of 5. The cells were collected at different time points and fixed with 0.5% formaldehyde in phosphate-buffered saline (PBS). After washing, the cells were resuspended in 0.5 ml FBS-DMEM (1:1) and permeabilized with 1.5 ml of 70% ethanol. The cells were incubated with mouse anti-P/V antibody followed by fluorescein isothiocyanate (FITC)-labeled anti-mouse antibody. The fluorescence intensity was measured using a flow cytometer (LSR II; BD). Three replicates were performed for statistical analysis.
Real-time PCR.
To compare levels of viral mRNA and genome in infected cells, real-time PCR was carried out as previously described (31). Briefly, MDBK cells in 6-well plates were infected with PIV5 or rPIV5-P-T286A. Total RNA from the infected cells was extracted using an RNeasy minikit (Qiagen). One-fifth of the total RNA from each sample was used for RT using Superscript III reverse transcriptase (Invitrogen). Oligo(dT)15 was used in RT to detect viral mRNA levels; BH191, which anneals to a region of the genomic RNA in the M gene, was used in RT to measure viral genomes. Two percent of the cDNA was used for real-time PCR on a Step One Plus real-time PCR system using TaqMan universal PCR master mix and custom-made TaqMan gene expression assays with 6-carboxyfluorescein (FAM) dye and nonfluorescent quencher (NFQ) (Applied Biosystems). Relative levels of viral mRNA and viral genome at each time point were determined by calculating ΔCT and normalized with the level of the input genome defined as the genome level at 2 hpi. Three replicates for each sample were used for statistical analysis.
IP-IB.
To detect P-L interaction, BSR-T7 cells were transfected with 3 μg Flag-L together with 1 μg P, P-T286A, P-T286E, or P-T286D. After 24 h, the cells were lysed using WCEB buffer and the lysates were immunoprecipitated with mouse anti-P together with protein G Sepharose beads for 2 to 3 h at 4°C. The immunoprecipitation (IP) products were washed and resolved in SDS-PAGE. The gel was transferred onto a PVDF membrane and immunoblotted (IB) with anti-Flag antibody (Stratagene). Aliquots of the cell lysate were resolved in SDS-PAGE and immunoblotted with antibody against P or Flag to measure the expression levels of P/P mutants and Flag-L.
Immunoprecipitation and DSP cross-linking.
To compare interaction between NP and P/P mutants, BSR-T7 cells in 6-cm plates were transfected with 1 μg NP together with 1 μg P, P-T286A, P-T286E, or P-T286D. After 18 to 20 h, the cells were starved and labeled with [35S]Met/Cys for 3 h. The labeled cells were lysed using WCEB buffer, and half of the lysate was immunoprecipitated with Pk antibody and half with antibody against NP. The IP products were washed and resolved in SDS-PAGE. The gel was dried, and the proteins were visualized using a Typhoon 9700 imager (GE Healthcare). To detect tetramer formation of P, P-T286A, P-T286E, or P-T286D, the transfected BSR-T7 cells were starved and labeled with [35S]Cys/Met for 3 h at 37°C. The labeled cells were incubated with 1 mM disuccinimidyltartarate (DSP) (Pierce, Rockford, IL) in PBS–0.5% NP-40 to cross-link the S-S band as previously described (31). After cross-linking, the cells were lysed using WCEB buffer and the supernatant was immunoprecipitated with Pk antibody. After washing, half of the mixture was added to loading buffer with DTT, and half of the mixture was added to loading buffer without DTT. The mixture was resolved in a 10% SDS gel, and the proteins were visualized with a Typhoon 9700 imager (GE health care).
RESULTS
T286 of the P protein was phosphorylated in PIV5-infected cells.
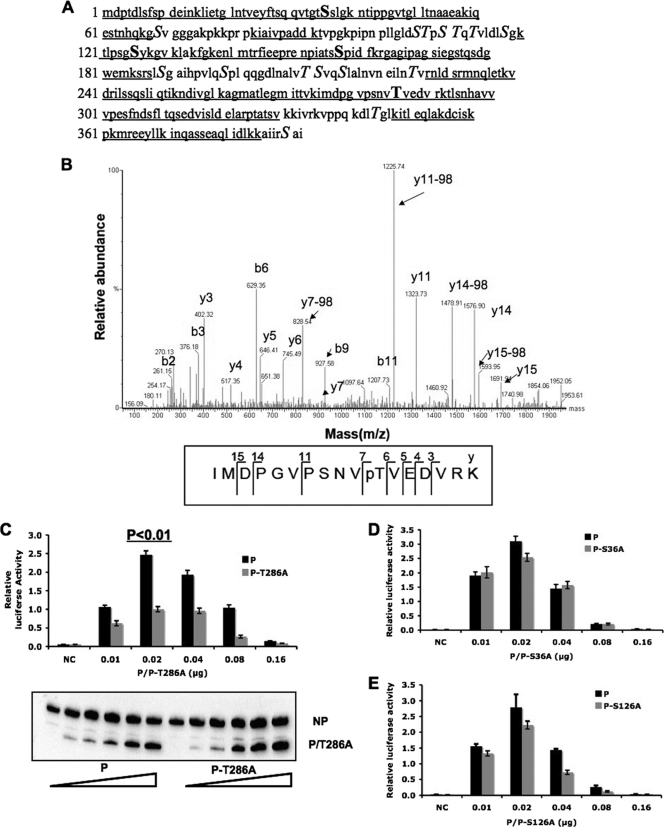
To determine phosphorylation sites within the P protein, the P protein was purified from PIV5-infected cells using affinity purification as described previously (31). The purified protein was then subjected to tryptic digestion and TiO2 enrichment and analyzed by LC-MS/MS. The total coverage of the two mass spectrometry analyses is about 74% (Fig. 1A). The results from two individual experiments are summarized in Table 1. S157 was phosphorylated as described previously (31). Phosphorylation of T286 was detected in both experiments (Fig. 1B). S36 and S126 were identified as phosphorylation sites once. For the peptide from amino acid residue 293 to residue 331, mass spectrometry identified two phosphorylation serines/threonines. However, the exact sites could not be determined.
Fig. 1.
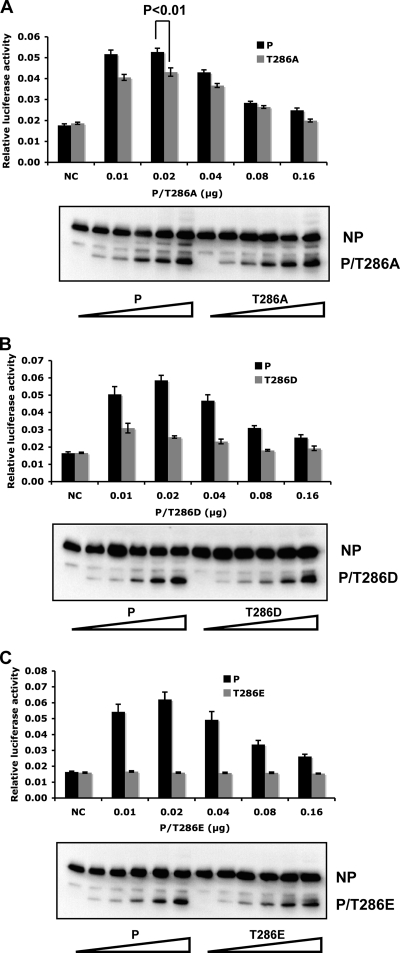
The function of P mutants in a PIV5 minigenome system. (A) Coverage of P by MS. Two MS data sets are combined. Underlined sequences are peptides detected in MS. Bold and underlined residues are potential phosphorylation sites. Capital S and T residues are S and T residues outside the coverage area of MS. (B) Mass spectra showing phosphorylation of T286. The y ions are labeled in the graph. The bottom panel shows the digested and TiO2-enriched fraction, which contains phosphorylated threonine at amino acid 286. The phosphorylation of T286 can be proven by the following: (i) y7-98, a neutral loss of H3PO4 from y7, and (ii) the mass difference between y7 and y6, 182.09, which is equal to the mass of phosphorylated threonine. (C) Minigenome activity of P-T286A. Because the ratio of NP to P is important for minigenome activity, increasing amounts of P or P-T286A were transfected together with other plasmids as described in Materials and Methods. Renilla luciferase is the reporter gene in the minigenome. Firefly luciferase expression was used as a transfection control. The minigenome activity was measured and normalized as the ratio of Renilla luciferase activity to firefly luciferase activity (relative luciferase activity). Error bars represent the standard errors of the means (SEM) of data from 6 replicates. P values were calculated using Student's t test. Western blotting was performed to detect the expression levels of NP and P or P-T286A. NC, negative control which did not have any P. (D and E) The function of P-S36A or P-S126A in the minigenome system. The expression levels of P-S36A or P-S126A were similar to those of wild-type P (data not shown).
Table 1.
LC-MS/MS resultsa
| Phosphorylation site | Peptide | Results |
|---|---|---|
| pS157 | ENPIATSpSPIDEFK | +, + |
| pT286 | IMDPGVPSNVpTVEDVRK | +, + |
| pS126 | TLPSGpSYKGVK | +, − |
| pS36 | LIETGLNTVEYFTSQQVTGTpSSLGK | −, + |
| Unknown two sites (293–331) | TLSNHAVVVPESFNDSFLTQSEDVISLDELARPTATSVK | −, + |
The P protein purified from HeLa cells was digested with trypsin, and the phosphopeptides were enriched by TiO2 and analyzed by LC-MS/MS. Mass spectrometry results of two separate experiments are summarized, and the phosphorylation sites (in bold) or peptide are shown. “+”, positive; “−”, negative.
P-T286A reduced PIV5 minigenome activity.
To study whether phosphorylation of T286 within the P protein affects PIV5 gene expression, T286 was mutated to alanine and the function of P-T286A was compared with that of wild-type P using a minigenome system. Previously, to study replication and transcription of PIV5, we established a minigenome system with a plasmid containing the PIV5 leader, trailer, and a reporter gene (first we used chloramphenicol acetyltransferase [CAT], and then we switched to a luciferase gene) under the control of a T7 RNAP promoter along with plasmids expressing NP, P, and L. Transcription from the T7 promoter results in a negative-strand minigenome. In the presence of NP, P, and L, the reporter gene mRNA is transcribed from the negative-sense RNA genome, resulting in reporter gene activity. The negative-sense RNA also serves as a template for replication (16, 31). Because the ratio of NP to P is critical for the minigenome system, we used a range of concentrations of the P plasmid and performed immunoblotting to monitor the expression levels of P and NP to ensure the optimal concentration for minigenome activity was covered. As shown in Fig. 1C, P-T286A showed lower levels of the minigenome activity, indicating that T286 was important for PIV5 gene expression in the minigenome system. S36 and S126 are two possible phosphorylation sites (Table 1). However, mutating these two sites to alanine had a small effect that was not statistically significant in the minigenome system (Fig. 1D and E), indicating that they are not real phosphorylation sites or their phosphorylation is not important for PIV5 gene expression.
Effect of mutations at T286.
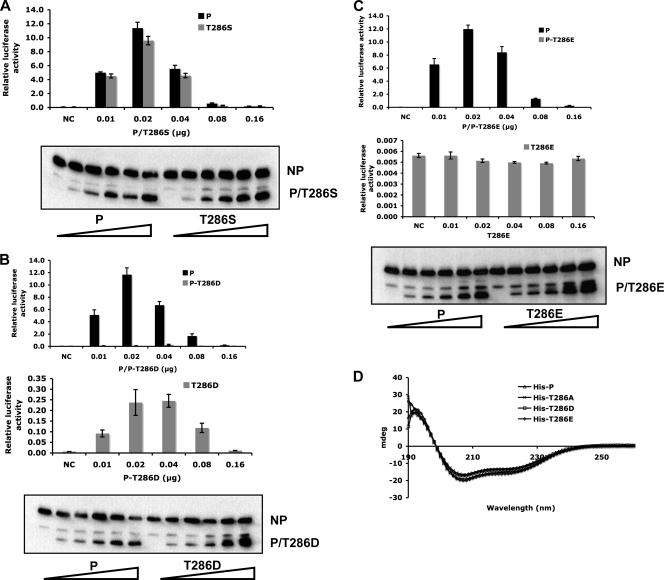
To further confirm the role of T286 phosphorylation in viral gene expression, T286 was mutated to serine (P-T286S), aspartic acid (P-T286D), or glutamic acid (P-T286E), and P-T286S showed function similar to that of P in the minigenome system (Fig. 2A). P-T286D showed very little activity, although there is a significant difference between P-T286D and the negative control (without P) (P < 0.01) (Fig. 2B). The P-T286E mutant showed no activity at all, like the negative control. It is possible that the T-to-E change at position 286 may have caused a gross alteration in structure, resulting in the T286E mutant phenotype. Circular dichroism (CD) was used to examine the secondary structures of the mutants. As shown in Fig. 2D, the P proteins purified from bacteria, His-P, His-T286A, His-T286D, and His-T286E, showed similar CD patterns under a continuous wavelength from 190 to 260 nm, suggesting that the overall protein secondary structure is not affected by the mutations at T286. The caveat of this experiment is that it is possible that the proteins purified from bacteria may not reflect the true structure of a viral protein that is normally expressed in mammalian cells.
Fig. 2.
The effect of mutations at T286. (A) The minigenome activity of P-T286S. (B and C) The minigenome activity of P-T286D or P-T286E was compared with that of wild-type P. The expression of NP and P or P-T286D/P-T286E is shown at the bottom of each figure. To highlight the differences between the T286E and T286D proteins, their respective activities and that of a negative control are compared without wild-type P (middle panel in B and C). NC, negative control which did not have any P. (D) Circular dichroism analysis of P mutants. The P protein fused with a His tag at its N terminus (His-P), His-P-T286A, His-P-T286D, and His-P-T286E were purified from E. coli. Ten micromolar P or P mutants in 10 mM KH2PO4-KCl buffer (pH 7.0) were measured using a Jasco-J715 spectropolarimeter.
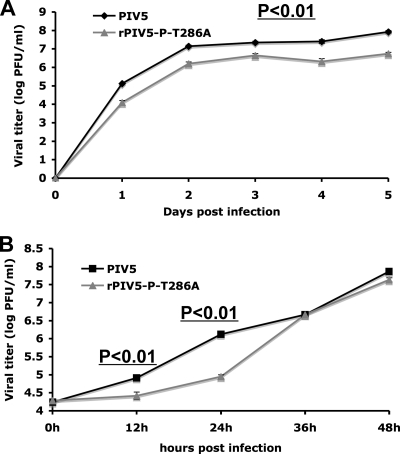
rPIV5-P-T286A grew slower than wild-type virus.
To examine the role of T286 during viral infection, a recombinant virus containing a T-to-A change at position 286 of the P protein (rPIV5-P-T286A) was recovered using a reverse genetics system, and rPIV5-P-T286A was confirmed by sequencing (data not shown). To study the role of T286 phosphorylation of the P protein in infected cells, MDBK cells were infected with PIV5 or rPIV5-P-T286A with a low MOI (MOI = 0.01). The supernatants were collected for plaque assay. As shown in Fig. 3A, the rPIV5-P-T286A titer was lower than that of PIV5, with an approximately 10-fold reduction (P < 0.01) at each time point. With a high MOI (MOI = 5) (Fig. 3B), PIV5 started to release virions at 12 hpi, while there was almost no virion release from rPIV5-P-T286A-infected cells at 12 hpi. The viral titer of rPIV5-P-T286A was lower than that of PIV5 at 12 and 24 hpi (P < 0.01). At 36 hpi and later, rPIV5-P-T286A shows a titer similar to that of PIV5 (Fig. 3B). These data suggest that a lack of phosphorylation at T286 of the P protein resulted in a delay in virus growth.
Fig. 3.
The effect of P-T286A on viral growth. (A) Growth curves of PIV5 and rPIV5-P-T286A in MDBK cells at an MOI of 0.01. Three separate experiments were performed (P < 0.01 for all time points). (B) Growth curves of PIV5 and rPIV5-P-T286A in MDBK cells at an MOI of 5. P values were calculated from three individual experiments.
rPIV5-P-T286A showed delayed protein expression.
To investigate the mechanism of the delayed growth of rPIV5-P-T286A, viral gene expression was examined in HeLa cells infected with PIV5 or rPIV5-P-T286A at an MOI of 5. At 8 and 12 hpi, the viral protein expression levels of rPIV5-P-T286A were lower than those of PIV5, while at the later time points of 16 hpi and 20 hpi, levels were similar (Fig. 4A). These results were further confirmed using flow cytometry by staining both the P and V proteins (Fig. 4B), indicating that rPIV5-P-T286A virus gene expression is delayed but not completely defective. The same phenotype of rPIV5-P-T286A was observed in BSR-T7 cells and MDBK cells (data not shown), indicating that this phenotype is not cell type specific.
Fig. 4.
Viral protein expression in PIV5-P-T286A-infected cells. (A) Western blot. The viral protein expression in PIV5- or rPIV5-P-T286A-infected HeLa cells was compared at different time points at an MOI of 5. “T286A” refers to rPIV5-P-T286A virus. (B) Flow cytometry. PIV5- or rPIV5-P-T286A-infected HeLa cells (MOI of 5) were processed for flow cytometry at different time points. Mean fluorescence intensity (P and V proteins) was used to show viral protein expression level (y axis). Error bars represent the standard errors of the means (SEM).
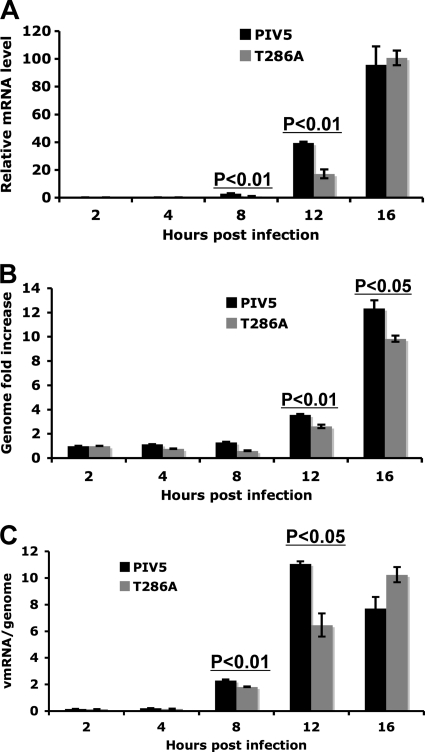
rPIV5-P-T286A affected viral mRNA synthesis.
To further investigate the mechanism of delayed viral protein expression in rPIV5-P-P286A-infected cells, viral mRNA synthesis and genome replication were examined using real-time PCR. At a high MOI (MOI = 5), rPIV5-P-T286A produced approximately half the viral mRNA produced by PIV5 at 8 and 12 hpi. At 16 hpi, the mRNA level of rPIV5-P-T286A is similar to that of PIV5 (Fig. 5A). rPIV5-P-T286A infection also produced lower numbers of viral genomes than PIV5 infection (P < 0.05) (Fig. 5B). There was a decrease in viral mRNA transcribed per viral genome in rPIV5-P-T286A-infected cells as judged by the ratios of viral mRNA over viral genome RNA (Fig. 5C), suggesting that the mutation at T286 reduced viral mRNA transcription. Similar results were obtained for infection at an MOI of 1 (data not shown). These results suggest that rPIV5-P-T286A reduces viral mRNA synthesis, consistent with the differences observed in viral protein expression, as well as the growth curve. To further investigate the effect of mutations of T286 on viral mRNA synthesis, a minigenome system with a defective trailer sequence, in which only viral RNA transcription is allowed, was used. As shown in Fig. 6, mutation at T286 to A, D, or E reduced the reporter gene activity in the transcription-only minigenome system, confirming that the T286 residue played an important role in upregulating viral mRNA transcription. However, these results do not exclude the possibility that the residue may also play a role in upregulating viral RNA genome replication.
Fig. 5.
Viral RNA synthesis in PIV5-P-T286A-infected cells. (A) Viral mRNA levels. MDBK cells were infected with PIV5 or PIV5-P-T286A at an MOI of 5. Total RNA was purified and used for RT. To measure mRNA levels, oligo(dT), primers targeting the HN region (BH193 and BH194), and a FAM dye were used for real-time PCR. Genome levels of PIV5 or rPIV5-P-T286A at 2 hpi were used as the baseline for normalization. (B) Genome replication of PIV5 and PIV5-P-T286A in MDBK cells at an MOI of 5. Oligonucleotide BH191, complementary to a region within the M gene, was used for RT. Genome levels at 2 hpi for each virus were used as a baseline for normalization. (C) Relative viral mRNA levels per genome at each time point are shown in PIV5- or PIV5-P-T286A-infected MDBK cells at an MOI of 1. The experiment was performed similarly to that shown in panel A.
Fig. 6.
Effect of mutations at T286 on a transcription-only minigenome system. A minigenome system identical to the one used for Fig. 1C except with a defective trailer sequence, which resulted in a defect in viral RNA replication but normal viral mRNA transcription, was used. T286A (A), T286D (B), and T286E (C) are analyzed.
P-T286A did not affect formation of the NP-P-L complex.
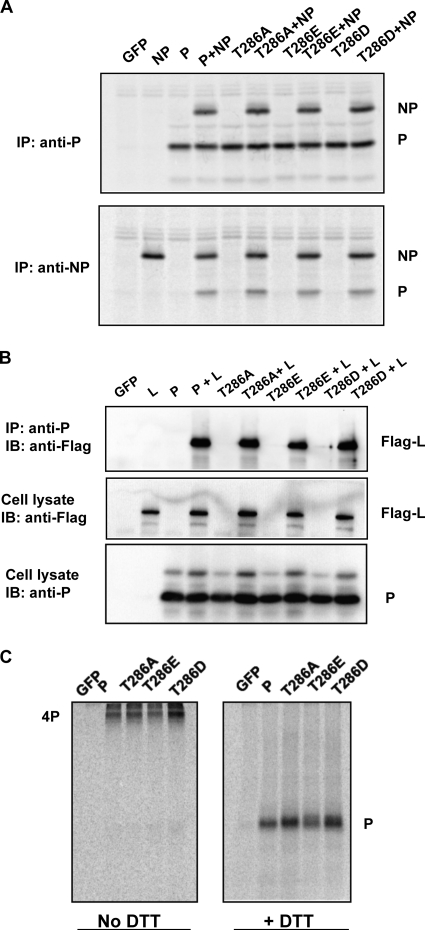
The P protein interacts with NP and L (22, 23). To investigate the mechanism for the defect of P-T286A, P-T286D, and P-T286E, interactions between P, NP, and L were examined. Based on the coimmunoprecipitation and immunoprecipitation followed by immunoblotting (IP-IB) results in transfected cells (Fig. 7A and B), P-T286A, P-T286D, and P-T286E still bound to NP or L protein, similar to the case of the P wild type. P of paramyxovirus is known to form a homotetramer (29). Abilities of the mutants to form tetramers were examined. P-T286A, P-T286D, and P-T286E all formed tetramers (Fig. 7C). These data show that interactions between P and NP, L, or itself were not affected by the mutations at T286.
Fig. 7.
NP-P-L complex formation. (A) Interaction between NP and P/P mutants. BSR-T7 cells were transfected with NP together with P or a P mutant. After labeling with 35S-Cys-Met, the cells were lysed and the supernatants were incubated with antibody against P or NP for coimmunoprecipitation. (B) IP-IB data showing the interaction between Flag-L and P/P-T286A/P-T286E/P-T286D. Pk antibody, which recognizes P, was used for IP, and mouse anti-Flag was used for IB. (C) Tetramer formation of P or P mutants. The transfected BSR-T7 cells were starved and labeled with [35S]Met/Cys. The cells were incubated with DSP to cross-link the S-S band to stabilize the P tetramer. The lysate was incubated with Pk and resolved in SDS-PAGE with or without DTT. The gel was dried, and the proteins were visualized using a Typhoon 9700 system.
Analysis of revertant viruses.
Attempts to rescue rPIV5-P-T286E virus did not succeed. Prolonged coincubation of rPIV5-P-T286E rescue cells with Vero resulted in recombinant viruses. Twenty-six isolates from three separate experiments were obtained. Sequencing results indicated that they all contained revertant mutations at E286, including glutamic acid (E; GAA) to alanine (A; GCA), lysine (K; AAA), valine (V; GTA), aspartic acid (D; GAC), or threonine (T; ACT) (Table 2). The larger number of revertants to A, K, V, and D than to wild-type T is likely a reflection of the number of mutations required to revert back to T; i.e., three mutations are needed to revert back to T, while a single mutation is enough to mutate to A, K, V, or D. This result indicates that T286E is severely defective for the function of the P protein in PIV5 growth, which is consistent with the minigenome data (Fig. 2C). We sequenced the whole genome of one of each T286V, T286D, and T286K revertant virus and did not find other mutations, indicating that T286 of P is a key regulatory site for PIV5 gene expression. All of those revertant viruses grew slower than PIV5 (Fig. 8A). As shown in Fig. 8B and C, both P-T286K and P-T286V showed much less activity than wild-type P in the minigenome system.
Table 2.
rPIV5-P-T286E revertantsa
| Theoretical nucleic acids (amino acid) at position 286 | Nucleic acids found | Amino acid change | Frequency |
|---|---|---|---|
| GAA (E) | AAA | E → K | 9/26 |
| GAA (E) | GCA | E → A | 4/26 |
| GAA (E) | GTA | E → V | 2/26 |
| GAA (E) | GAC | E → D | 4/26 |
| GAA (E) | ACT | E → T | 1/26 |
| GAA (E) | Mixture | Mixture | 6/26 |
Three individual rescues were performed, and 26 plaques in total were examined. The supernatants of the infected cells were used for viral RNA extraction, RT-PCR, and sequencing. The differences in nucleic acid residues and amino acid residues are shown.
Fig. 8.
Effect of revertant mutations. (A) Growth curves of rPIV5-P-T286A/V/D/K compared to those of PIV5 in MDBK cells at an MOI of 0.01. The whole genomes of revertant viruses for T286V, T286D, or T286K were sequenced and used for growth curve analysis. (B) Minigenome activity of P-T286K was compared with that of wild-type P. Western blotting was performed to show the input of NP and P or P-T286K. (C) Minigenome activity of P-T286V was compared with that of wild-type P.
DISCUSSION
In this study, we have identified a phosphorylation site within the P protein of PIV5, T286. Mutating the T residue at 286 to A resulted in a protein that was less efficient than wild-type P (approximately 50%) in a minigenome system and had a delayed and lowered growth rate when incorporated into a recombinant virus, indicating that this phosphorylation plays a critical role in PIV5 growth. Mutating this residue to residues other than serine resulted in a lower level of viral mRNA transcription, suggesting that the phosphorylation of T286 plays a positive role in viral mRNA transcription and viral gene expression. This is the first time that a positive role of a phosphorylation site within the P protein in viral gene expression has been demonstrated for paramyxoviruses. Taken together with findings of other studies showing the role of phosphorylation at S157 and S308 of P of PIV5 in downregulating viral gene expression, these results indicate that phosphorylation does play a role in regulating paramyxovirus gene expression: phosphorylation can upregulate, as well as downregulate, viral gene expression. One limitation of our approach is that mutating a phosphorylation site will result in a potential change in the protein structure due to the change of the residue besides removing the phosphate group. We used CD to compare structures of the P mutants, and we did not detect any significant change in the structures of the mutants, indicating that the phenotype associated with the mutants is likely due to the lack of a phosphate group at the site.
There are likely other phosphorylation sites in addition to S157 and T286. The peptide from amino acid residue 293 to 331 contains two putative phosphorylation sites according to MS results. In addition, the MS analysis covered only approximately 74% of the P polypeptide. Mutating the putative sites S126 and S36 had no impact on minigenome activity, indicating that phosphorylation at these sites may be incidental if they are indeed phosphorylation sites. Further analysis of temporal regulation of the known phosphorylation sites, as well as identification of novel critical phosphorylation sites within the P protein, will provide new knowledge of regulation of viral gene expression.
While the work here indicates that phosphorylation of T286 is important for efficient viral mRNA synthesis, the exact mechanism is not clear. Mutation of T286 to D or E, a phosphomimetic residue, resulted in a mutant with reduced activity in the minigenome system and in a transcription-only minigenome, as with the T286A mutant. This result is reminiscent of mutation at S157 of the P protein; mutating S157 to D or E had the same effect as mutating S157 to A. Phosphorylation of S157 is important for P to bind to the host protein, PLK1 (26). Thus, we speculate that phosphorylation of T286 plays a critical role in association of the P protein with another protein. P is known to interact with other viral proteins, NP and L, and to form a homotetramer (22, 23). Mutating the T286 residue had no detectable effect on its association with NP or L. Mutant P proteins also formed homotetramers as efficiently as wild-type P, suggesting that the partner protein for the function of T286 is not a viral protein. We hypothesize that P likely interacts with a host protein through the T286 site. This is consistent with the data on revertants from the PIV5-T286E rescue. If residue T286 were essential for its interaction with other viral proteins in order to have a role in viral mRNA synthesis, compensatory mutations would have risen in other viral proteins. Instead, the only mutations we have observed were in the P protein at the exact same site, indicating that phosphorylation of T286 is likely important for interacting with a host protein. It is known that P interacts with AKT1, a serine/threonine kinase (27). No difference in interactions between wild-type P or P mutants and AKT1 has been detected (data not shown), indicating that a host protein other than AKT1 may play a role in regulating viral RNA synthesis. We propose that the phosphorylation of T286 enhances association with a host protein, which may result in modification of the P protein itself or the P-associated viral proteins NP and L.
It is not clear which kinase phosphorylates the P protein at T286. Previously, AKT was shown to play an essential role in PIV5 gene expression (27). However, T286 is unlikely to be an AKT phosphorylation site. AKT substrates contain positively charged residues, such as lysine or arginine, at the −3 and −5 positions (1). In addition, the AKT inhibitor IV reduced rPIV5-P-T286A protein expression, as well as phosphorylation of P-T286A (data not shown). A candidate kinase for phosphorylating the P protein at T286 is CK2 (casein kinase 2). T286 (underlined) and adjacent residues (TVED) are consistent with the CK2 consensus phosphorylation motif [pS/pT]XX[E/D] (20). A CK2 inhibitor reduced virus replication in HeLa cells (data not shown), and an in vitro kinase assay showed that CK2 phosphorylated the P protein of PIV5 purified from bacteria (data not shown). However, CK2 phosphorylated P-T286A at a level similar to that for wild-type P. It is possible that there are several potential CK2 sites: mutating one site may not make a significant difference in the overall phosphorylation levels of the P protein. Previous studies suggested that CK2 phosphorylated the P protein from RSV, measles virus, rinderpest virus, and Borna disease virus (8, 13, 19, 25). Due to numerous subunits and isoforms of CK2 and ubiquitous potential CK2 sites, work on the role of CK2 has not been conclusive. Further studies are needed to clarify the role of CK2 and to identify host kinases that phosphorylate the P protein at T286 and other phosphorylation sites.
ACKNOWLEDGMENTS
We thank all the members of Biao He's laboratory for helpful discussions and technical assistance. We are grateful to Kaori Sakamoto for carefully reading the manuscript prior to submission.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases to B.H. (R01AI070847, K02AI65795, and R56AI081816).
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. 1996. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399:333–338 [DOI] [PubMed] [Google Scholar]
- 2. Asenjo A., Calvo E., Villanueva N. 2006. Phosphorylation of human respiratory syncytial virus P protein at threonine 108 controls its interaction with the M2-1 protein in the viral RNA polymerase complex. J. Gen. Virol. 87:3637–3642 [DOI] [PubMed] [Google Scholar]
- 3. Asenjo A., Gonzalez-Armas J. C., Villanueva N. 2008. Phosphorylation of human respiratory syncytial virus P protein at serine 54 regulates viral uncoating. Virology 380:26–33 [DOI] [PubMed] [Google Scholar]
- 4. Asenjo A., Rodriguez L., Villanueva N. 2005. Determination of phosphorylated residues from human respiratory syncytial virus P protein that are dynamically dephosphorylated by cellular phosphatases: a possible role for serine 54. J. Gen. Virol. 86:1109–1120 [DOI] [PubMed] [Google Scholar]
- 5. Barik S., McLean T., Dupuy L. C. 1995. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology 213:405–412 [DOI] [PubMed] [Google Scholar]
- 6. Byrappa S., Hendricks D. D., Pan Y.-B., Seyer J. M., Gupta K. C. 1995. Intracellular phosphorylation of the Sendai virus P protein. Virology 208:408–413 [DOI] [PubMed] [Google Scholar]
- 7. Byrappa S., Pan Y. B., Gupta K. C. 1996. Sendai virus P protein is constitutively phosphorylated at serine249: high phosphorylation potential of the P protein. Virology 216:228–234 [DOI] [PubMed] [Google Scholar]
- 8. Das T., Schuster A., Schneider-Schaulies S., Banerjee A. K. 1995. Involvement of cellular casein kinase II in the phosphorylation of measles virus P protein: identification of phosphorylation sites. Virology 211:218–226 [DOI] [PubMed] [Google Scholar]
- 9. Fuentes S. M., Sun D., Schmitt A. P., He B. 2010. Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression. Future Microbiol. 5:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilmore J. M., et al. 2010. Determinants of affinity and activity of the anti-sigma factor AsiA. Biochemistry 49:6143–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu C., Gupta K. C. 2000. Functional significance of alternate phosphorylation in Sendai virus P protein. Virology 268:517–532 [DOI] [PubMed] [Google Scholar]
- 12. Hu C. J., et al. 1999. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology 263:195–208 [DOI] [PubMed] [Google Scholar]
- 13. Kaushik R., Shaila M. S. 2004. Cellular casein kinase II-mediated phosphorylation of rinderpest virus P protein is a prerequisite for its role in replication/transcription of the genome. J. Gen. Virol. 85:687–691 [DOI] [PubMed] [Google Scholar]
- 14. Lamb R. A., Choppin P. W. 1977. The synthesis of Sendai virus polypeptides in infected cells. III. Phosphorylation of polypeptides. Virology 81:382–397 [DOI] [PubMed] [Google Scholar]
- 15. Lamb R. A., Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication, p. 1177–1204. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 16. Lin Y., Horvath F., Aligo J. A., Wilson R., He B. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338:270–280 [DOI] [PubMed] [Google Scholar]
- 17. Lu B., Ma C. H., Brazas R., Jin H. 2002. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J. Virol. 76:10776–10784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luthra P., Sun D., Silverman R. H., He B. 2011. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazumder B., Adhikary G., Barik S. 1994. Bacterial expression of human respiratory syncytial viral phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology 205:93–103 [DOI] [PubMed] [Google Scholar]
- 20. Meggio F., Pinna L. A. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349–368 [DOI] [PubMed] [Google Scholar]
- 21. Navarro J., Lopez-Otin C., Villanueva N. 1991. Location of phosphorylated residues in human respiratory syncytial virus phosphoprotein. J. Gen. Virol. 72(Pt. 6):1455–1459 [DOI] [PubMed] [Google Scholar]
- 22. Parks G. D. 1994. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J. Virol. 68:4862–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Precious B., et al. 1995. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J. Virol. 69:8001–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanchez-Seco M. P., Navarro J., Martinez R., Villanueva N. 1995. C-terminal phosphorylation of human respiratory syncytial virus P protein occurs mainly at serine residue 232. J. Gen. Virol. 76(Pt. 2):425–430 [DOI] [PubMed] [Google Scholar]
- 25. Schwemmle M., De B., Shi L., Banerjee A., Lipkin W. I. 1997. Borna disease virus P-protein is phosphorylated by protein kinase Cepsilon and casein kinase II. J. Biol. Chem. 272:21818–21823 [DOI] [PubMed] [Google Scholar]
- 26. Sun D., Luthra P., Li Z., He B. 2009. PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS Pathog. 5:e1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun M., et al. 2008. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J. Virol. 82:105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun M., et al. 2004. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 78:5068–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tarbouriech N., Curran J., Ebel C., Ruigrok R. W., Burmeister W. P. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99–109 [DOI] [PubMed] [Google Scholar]
- 30. Thomas S. M., Lamb R. A., Paterson R. G. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Timani K. A., et al. 2008. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J. Virol. 82:9123–9133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vidal S., Curran J., Orvell C., Kolakofsky D. 1988. Mapping of monoclonal antibodies to the Sendai virus P protein and the location of its phosphates. J. Virol. 62:2200–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Villanueva N., Hardy R., Asenjo A., Yu Q., Wertz G. 2000. The bulk of the phosphorylation of human respiratory syncytial virus phosphoprotein is not essential but modulates viral RNA transcription and replication. J. Gen. Virol. 81:129–133 [DOI] [PubMed] [Google Scholar]
- 34. Villanueva N., Navarro J., Mendez E., Garcia-Albert I. 1994. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J. Gen. Virol. 75(Pt. 3):555–565 [DOI] [PubMed] [Google Scholar]