Abstract
On the prereceptor-engaged HIV-1 envelope glycoprotein (Env) spike, epitope access by the membrane-proximal external region (MPER)-directed broadly neutralizing antibodies 2F5 and 4E10 remains unresolved. Data on binding to cell surface Env and entry data using primary isolates suggest inaccessibility of the 2F5 and 4E10 epitopes on the viral spike prior to receptor engagement, but trimer gel shift analysis and slow kinetics of shedding induced by 2F5 and 4E10 indicate otherwise. Therefore, it remains unclear if the epitopes themselves are formed in their antibody-bound state (or at least sampled) prior to receptor/coreceptor engagement or if receptor interactions both expose and form the MPER epitopes, presumably in the putative prefusion transitional intermediate. Here, we performed antibody-virus “washout experiments” using both lab-adapted and a panel of clade B primary isolates to analyze MPER accessibility. The neutralization activity of 2F5 and 4E10 against lab-adapted viruses and sensitive and moderately resistant viruses was largely unaffected by relatively rapid antibody-virus washing, suggesting direct interaction with the “static” spike. However, for more neutralization-resistant viruses, the 2F5 and 4E10 antibodies could neutralize only under the “no antibody-virus wash” conditions, implying that the MPER epitopes were not accessible prior to receptor engagement. Accessibility in the washout conditions could be precisely predicted by the relative resistance to neutralization in a standard neutralization format. These data are consistent with a model in which the local MPER antibody epitope conformations may be sampled on the native spike but are occluded to antibody by local steric or distal quaternary constraints adopted by highly resistant HIV-1 isolates.
INTRODUCTION
The HIV-1 gp160 envelope glycoprotein (Env) precursor is cleaved by cellular furins to generate the noncovalently associated gp120 and gp41 trimeric Env complex. The gp120 subunit binds to the primary receptor, CD4, and, following conformational changes, also interacts with the CCR5 coreceptor. The gp41 trans-membrane Env then mediates virus-to-cell membrane fusion, resulting in the entry of viral genomic information into the target cell (8, 11, 14, 24, 64–66). The genetic variability of Env, coupled with the inaccessibility of conserved epitopes, makes the generation of antibodies capable of neutralizing a broad array of primary isolates (i.e., broadly neutralizing) a substantial challenge. Viral entry into cells can be blocked in vitro by relatively rare broadly neutralizing antibodies that are elicited during the course of natural infection. Since viral half-life in vivo is relatively brief (t1/2, 30 to 60 min) (40, 57), HIV-1 neutralizing antibodies need to act rapidly to inactivate infectious virus. Hence, most in vitro neutralization assays, which assess the ability of antibody to interfere with HIV entry, are traditionally performed with approximately 1-h incubation times of antibody-virus (41, 42, 70).
Neutralizing antibodies can either efficiently and directly recognize the prereceptor-engaged native spike on the virus surface or require receptor engagement to better expose specific neutralizing determinants. Broadly neutralizing antibodies capable of directly accessing the static spike often map to the Env gp120 subunit (5, 26, 36, 62, 63, 67). The gp41-directed broadly neutralizing antibodies 2F5 and 4E10 are known to recognize contiguous and continuous epitopes within the gp41 region of Env. In addition, in relatively rare broadly neutralizing patient sera, the specificity of the broad neutralizing activity can be mapped to the gp41 membrane-proximal external region (MPER) (21, 37, 52). In these sera, the neutralizing activity appears similar to the specificity displayed by 4E10 (22, 35, 52), and in one report, the activity could be mapped to the 2F5 epitope region (60). The gp41-directed neutralizing MPER-specific antibodies may bind directly to free virus, or they may neutralize virus during the process of receptor-triggered entry. Current models suggest that MPER access is achieved after receptor engagement and during formation of the putative transitional fusion intermediate and that the transitional intermediate may be required to fully form the MPER neutralizing epitopes into the structurally defined “antibody-bound” conformations (10, 16–18, 20). When the 2F5 and 4E10 antibodies can access their epitopes during the HIV entry process has been incompletely explored previously in the literature, but the precise timing of accessibility remains unresolved (1, 4, 10, 19, 55). In addition, several studies have demonstrated differences between the Envs of lab-adapted viruses and primary isolates, suggesting that there could be distinct rules of accessibility for each class of virus (13, 29, 33, 43, 44, 46, 56, 58, 59, 69). A very recent study reports the ability of many antibodies to induce shedding of the HIV-1 Env, including the MPER antibodies described here, but often with slow kinetics approaching 18 h (50).
Using fluorescence-activated cell sorting (FACS)-based cell surface staining employing gp120-directed neutralizing and nonneutralizing antibodies, we demonstrated previously that there is a direct correlation between efficient recognition of the cleaved functional spike and the neutralization capacity of a given antibody (45). In a second study, we demonstrated that 2F5 and 4E10 do not efficiently recognize cleaved, tail-truncated JR-FL oligomers expressed on the surface of transfected cells, but when the same Env is rendered cleavage defective, efficient binding is observed. The nonneutralizing cluster I and cluster II gp41 antibodies do not bind to these highly cleaved spikes, similar to previous reports for the nonneutralizing gp120-directed antibodies (9, 45). Incubation of cleaved spikes with soluble CD4 marginally increased spike recognition by the 2F5 and 4E10 neutralizing antibodies, consistent with previous studies (15, 53).
In the present study, we sought to explore epitope accessibility of the gp41 neutralizing antibodies, 2F5 and 4E10, either on the functional spike or during receptor-mediated entry. We sought to determine if these antibodies bind to the static spike on the surface of the HIV-1 or require target cell/receptor engagement to gain access to their MPER binding sites. We first confirmed that the binding of 2F5 and 4E10 to full-length, cleaved JR-FL spikes was inefficient, as we have previously reported for tail-truncated JR-FL spikes (9). To investigate the kinetics of neutralization mediated by the MPER-directed neutralizing antibodies, we performed a modified version of an antibody-virus washout assay using viruses containing envelope glycoproteins derived from both lab-adapted viruses and primary isolates. Following specificity and validation of the antibody washout assay in the context of viral entry, we confirmed that neutralizing but not nonneutralizing antibodies directed to gp120 could directly access their epitopes in the context of primary isolates. We found that viruses generated with the Env derived from either lab-adapted viruses or particular primary isolates displayed direct accessibility of their contiguous 2F5 and 4E10 epitopes, whereas more resistant viruses required receptor engagement on target cells to provide access to the 2F5 and 4E10 epitopes. We demonstrate that, for the resistant viruses JRCSF and JR-FL, we were able to render direct access on the static spike by generating selected point mutations either in Env variable regions (V1/V2 or V3) or in the gp41 region of the viral Env. We confirmed that the mutated viruses were CD4 dependent and the Env spikes were not in the receptor-triggered state. Taking these results together, we conclude that the inefficient binding of 2F5 and 4E10 to most primary isolates is due to the inaccessibility of their cognate epitopes. Based upon direct accessibility in the more sensitive but CD4-dependent isolates, we propose that inaccessibility is not likely due to the formation of epitope after receptor engagement but is likely due to steric occlusion resulting from quaternary Env packing. These data have important implications for the structure or exposure of discrete epitopes in the context of the prereceptor-engaged HIV-1 spike, the generation of global resistance to HIV neutralizing antibodies, and the design of HIV-1 vaccine candidates.
MATERIALS AND METHODS
Plasmids.
pSG3ΔEnv containing the luciferase reporter gene and the plasmids expressing envelopes (molecular clones) of HIV-1 primary isolates JR-FL, CAAN5342.A2, TRO.11, PVO.4, BAL, ADA, SF162.LS, JRCSF, mutant JRCSF, mutant JR-FL, and the T-cell-line-adapted HIVMN and HIVHXBc2 were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP) except as noted below. The 2F5 epitope containing recombinant chimeric HIV-2 viral envelope (7312-A-C3, referred to here as HIV2-C3) was kindly provided by George Shaw (23).
Cell lines and antibodies.
TZM-bl cells (CD4+ CXCR4+ CCR5+) were obtained from the NIH ARRRP, and 293T cells were purchased from ATCC. Both cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal calf serum (HIFCS), 20 mM l-glutamine, 100 U 110/ml penicillin, and 100 μg/ml streptomycin. Anti-HIV-1 monoclonal antibodies (MAbs) were obtained from the following sources: 2G12 (61), 4E10, and 2F5 (MPER) IgGs were gifts from H. Katinger (Polymun); b12 (CD4 binding site; CD4bs) (6), 7B2 (cluster I), and 22B (cluster II) IgG were gifts from J. Robinson, Tulane University. The anti-RSV MAb was obtained from Medimmune Inc. (MD).
Synthetic peptides.
Wild-type (WT) and mutant 2F5 peptide were purchased from New England Peptide (NEP; Gardner, MA), and two-domain soluble CD4 (sCD4, contributed by Progenics Pharmaceuticals) was obtained from the NIH ARRRP. T20 peptide was obtained from New England Peptide (Gardner, MA).
Transient transfection of envelope glycoprotein expression plasmids.
293T cells were transfected following the protocol described before (9, 45). Briefly, 1 day prior to transfection, 10 × 106 293T cells in DMEM and containing 10% heat-inactivated fetal calf serum (HIFBS) and 1% penicillin-streptomycin were seeded in a 150-mm tissue culture dish. The cells were transfected with the pSVIII expressor plasmids encoding JR-FL cleavage-competent WT Env, along with cotransfection of the tat expression plasmid, pctat, using Fugene6 (Roche) at a DNA/Fugene6 ratio of 1:3 and 5 μg total DNA per 1 × 106 cells.
FACS staining of cell surface HIV-1 Env.
Fluorescence-activated cell sorting (FACS) staining was performed as previously described (9, 32, 45). Forty-eight hours following transfection, the cells were harvested and washed in FACS buffer (phosphate-buffered saline [PBS], 5% HIFBS, 0.02% azide) and stained with a panel of monoclonal antibodies that were also used in viral neutralization assays. The transfected cells were suspended in FACS buffer and were incubated with the antibodies for 1 h at room temperature (RT). The monoclonal antibody-cell mixture was washed extensively in FACS buffer, and phycoerythrin (PE)-conjugated goat anti-human secondary antibody (Sigma) was added for 1 h at a 1:200 dilution, followed by extensive washing to remove unbound secondary antibody. The antibody-PE-stained cells were analyzed by FACS on a BD SLRII instrument.
Virus production.
Pseudotyped viruses (PSVs) were produced by transient cotransfection of 293T cells using the HIV-1 env-deleted backbone plasmid, pSG3ΔEnv, and the Env-complementation plasmid, pCAGGS-JR-FL (45), at a ratio of 3:1. A 3:1 ratio of the transfection reagent, Fugene (Roche, Indianapolis, IN), to DNA was used for transfection. Cell culture supernatants containing viruses were collected 2 days posttransfection. All other envelope plasmids are in pcDNA3.1. The I675V mutant of JR-FL was made by site-directed QuikChange mutagenesis (Stratagene, Cedar Creek, TX).
Antibody-virus washout experiments.
From a starting concentration of 2 mg/ml, 12.5 μl of 5-fold serially diluted antibodies in PBS were added to 487.5 μl of DMEM containing 10% HIFCS and 15 μl of either JR-FL or HXBc2 pseudovirus such that the final concentrations of antibodies were 50 μg/ml to 0.08 μg/ml in a total volume of 500 μl. In the “no inhibitor” control, the same volume of PBS was added instead of antibody. The reaction mixture was incubated for 30 min at 37°C. The 250-μl reaction mixture was diluted to 10 ml by complete DMEM and centrifuged at 25,000 rpm in an SW41 swinging rotor for 2 h at 4°C. The virus pellet was then washed two additional times with 10 ml of PBS. During the washing steps, the virus-antibody complex was centrifuged at 40,000 rpm in an SW41 swinging rotor for 20 min at 4°C. After a final wash, 250 μl of DMEM was added to the washed virus pellet and suspended by gentle shaking at 4°C for 30 min. Next, 100 μl of the suspended virus was used to infect 100 μl of TZM-b cells (0.2 × 106/ml) in duplicate. From the remaining 250 μl of reaction mixture, an equal volume of the antibody virus mixture was used as a “no washout” control. Plates were incubated at 37°C in a CO2 incubator for 2 days. After 2 days, the luciferase assay was done as described previously (41). The data were then plotted to determine the neutralization mediated by the antibodies under “wash” or “no wash” conditions. In the case of peptide inhibitors, the concentrations of the peptide were 50 μg/ml and 100 μg/ml and served as controls to confirm that complete washing of neutralizing ligands had occurred.
RESULTS
2F5 and 4E10 bind inefficiently to cleaved, full-length JR-FL Env spikes.
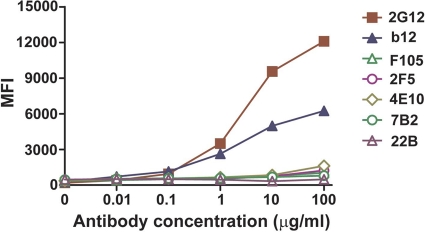
Previously we reported inefficient binding of 2F5/4E10 to cleaved but tail-truncated JR-FL cell surface Env spikes. Tail truncation was performed to increase the levels of Env on the cell surface, which allows a higher signal-to-noise ratio to better perform the binding analysis. JR-FL Env was studied previously, and now, because of its highly efficient cleavage by cellular furins (27, 45). In our previous study (9), however, we did observe some increased sensitivity of tail-truncated JR-FL viruses to several neutralizing ligands, in part consistent with previous reports that Env tail truncation can impact cell surface Env antigenicity. Here, we examined 2F5 and 4E10 antibody access to full-length JR-FL Env spikes and again observed very inefficient recognition of cleaved JR-FL Env spikes by 2F5 and 4E10. These data suggest that the MPER is not directly accessible to these neutralizing antibodies on cell surface, fully cleaved spikes (Fig. 1).
Fig. 1.
Antibody binding to cell surface JR-FL Env monitored by FACS. Mean fluorescence intensity (MFI) values of both neutralizing (b12, 2G12, 2F5, and 4E10) and nonneutralizing (F105, 7B2, 22B) antibodies are shown for cleavage-competent JR-FL Env expressed on 293T cells. The data were derived from representative experiments performed in duplicate. The standard errors between duplicates were minimal (less than 0.01) and are not shown since the error bars are obscured by the symbols.
Neutralization by MPER-directed antibodies under “no wash” but not “with wash” conditions indicate limited direct spike access.
Previously, we demonstrated that only gp120-directed broadly neutralizing antibodies efficiently recognize cleaved JR-FL functional spikes on the cell surface. In contrast, the gp41-directed neutralizing antibodies, 2F5 and 4E10, do not efficiently recognize cleaved JR-FL functional spikes on the cell surface, implying that they bind to their epitopes after receptor/coreceptor activation of the spike (9). Therefore, we sought to assess if the 2F5 and 4E10 antibodies interact directly with virus spikes to neutralize a range of viruses displaying differential neutralization sensitivity. When antibody recognition of the Env spike is examined in the context of inhibiting viral entry, one then detects interaction of the antibody only with cleaved, entry-mediating functional spikes. Thus, unlike directed binding analysis to the functional spike, which requires near-complete precursor cleavage for straightforward interpretation of the data, the use of entry as an indirect readout of antibody-functional spike interaction greatly expands the repertoire of viral Envs that can be examined (assuming that there is sufficient precursor cleavage to permit viral entry).
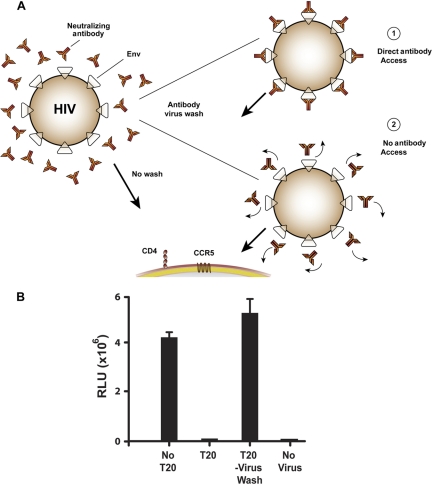
To assess at which juncture of the entry process 2F5 and 4E10 access their respective MPER epitopes, we performed neutralization assays, either using the traditional manner of preincubating antibody with virus for 30 to 60 min and then adding the mixture to target cells (“no antibody-virus wash”) (Fig. 2 A) or, following incubation of antibody with virus, performing an “antibody-virus wash” by ultracentrifugation of the virus in the presence of a large excess of buffer to separate unbound antibody from the virus pellet (see Materials and Methods). To validate the antibody-virus washout assay, we first incubated the known neutralizing peptide ligand, T-20, with both lab-adapted virus, HXBc2, and the primary isolate, JR-FL (71). Because this peptide potently inhibits virus entry by binding to the receptor activated transitional intermediate that exposes the HR1 coiled-coil region of gp41, but only after receptor engagement (12, 25, 30, 31, 64), we used this compound to confirm complete washout from the system of a neutralizing ligand that cannot access its determinant in the absence of receptor-induced activation. Accordingly, the virus was incubated either with buffer alone or with the T-20 peptide at a concentration 200-fold in excess of its 50% inhibitory concentration (IC50). If the T-20-virus mixture was subjected to washing, the T-20 peptide was not able to neutralize either the HXBc2 or JR-FL viruses, whereas it did so efficiently under no virus-antibody wash conditions. These results indicated that we could perform the assay under conditions where there was complete removal of the inhibitor from the viral entry assay system for both antibody-sensitive and relatively antibody-resistant viruses with similar results (Fig. 2B). Note also that the wash method used here did not greatly affect overall viral entry for either virus. Furthermore, we did not observe any noticeable change of viral titer when we left the half of the virus-antibody mixture (no wash) at either RT or 4 or 37°C for the approximately 4-h time period that it takes to perform the washout assay.
Fig. 2.
Virus entry into TZM-bl cells with and without inhibitory antibodies and with and without ligand-virus washing. (A) Schematics of “no antibody-virus” wash and “antibody-virus wash” of the antibody-virus mixture in the assay used in this study. (B) Positive- and negative-controls for the JR-FL virus, with and without washing, are shown as bar graphs. Relative luciferase units (RLU) generated from target cell lysates are shown on the vertical axis. Left to right: virus with no T-20 added to target cells; virus incubated with T-20 at 100 μg/ml prior to target cell interaction; virus incubated with T-20 followed by washing prior to target cell interaction; no virus added to target cell (negative control).
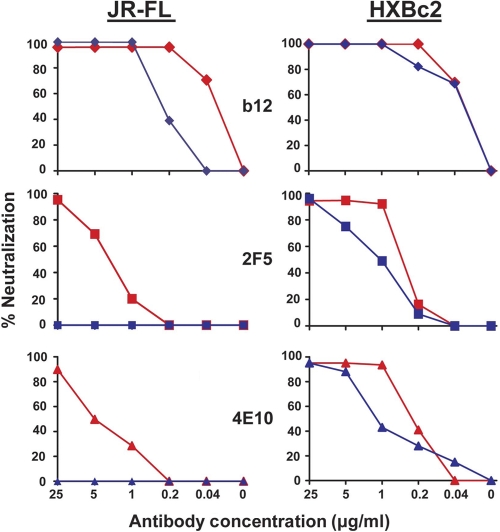
We next performed the antibody-virus wash assay using the broadly neutralizing, gp120-directed, CD4 binding site antibody b12. As expected, the antibody could neutralize HXBc2 either without (normal neutralization format) or with washing of the antibody-virus mixture. It was shown previously that b12 interferes with CD4 engagement, so logically it must be able to access its epitope prior to primary receptor interaction as previously observed (3, 49, 51). We next assessed the gp41-directed neutralizing antibodies. Interestingly, both 2F5 and 4E10 retained the ability to neutralize HXBc2 following washing of the antibody-virus mixture (Fig. 3 A). These data indicate that the 2F5 and 4E10 epitopes are readily accessible on the prereceptor-activated spike of the lab-adapted virus and imply that the conformation of the epitopes recognized by 2F5 and 4E10 are likely sampled on the lab-adapted Env spike prior to receptor engagement, allowing avid antibody interaction and efficient neutralization. We confirmed the specificity of binding of 2F5 to free HXBc2 virus under our antibody-virus wash condition by using both the WT and a point-mutated 2F5 epitope peptide that cannot be efficiently recognized by 2F5. In the antibody-virus wash assay, only the WT 2F5 peptide could block neutralization (and antibody binding to virus); the mutant peptide could not do so under either no antibody-virus wash (standard entry inhibitor format) or antibody-virus wash conditions (see Fig. S1 in the supplemental material). These data suggest that the binding of 2F5 antibody to HXBc2 in the washout assay is epitope specific.
Fig. 3.
Neutralization of JR-FL and HXBc2 virus by the antibodies b12, 2F5, and 4E10 with and without washing. Left panels (b12, 2F5, and 4E10), preincubation with JR-FL virus with or without washing prior to incubation with target cells. Right panels (b12, 2F5, and 4E10), preincubation with HXBc2 virus with or without washing prior to incubation with target cells. The red curves and the blue curves indicate “no antibody-virus” wash and “antibody-virus” wash, respectively.
Next, we sought to assess access of 2F5 and 4E10 to their epitopes on an HIV-1 primary isolate. We began the analysis with well-described JR-FL isolate and analyzed MPER epitope accessibility under our established assay conditions. As expected, JR-FL was neutralized by b12, a gp120-specific CD4 binding site antibody, either without or with any antibody-virus wash step over a broad range of concentrations (Fig. 3B). The potent neutralization exerted by b12 even after the antibody-virus washing confirmed high-affinity binding and indicated that b12 could directly access its epitope on the prereceptor-engaged viral Env spike. There was a slight loss of b12 neutralization potency of virus following the wash steps, but this likely reflects some detectable but limited dissociation of the antibody from the functional spike during the hours of the washing process itself, a factor to consider in further interpretations of the data presented here. As an additional control, the nonneutralizing CD4bs antibody, F105, did not neutralize JR-FL under any conditions tested (data not shown).
Having further validated the system with the well-described gp120-directed neutralizing antibodies, we next examined 2F5 and 4E10 neutralization kinetics of JR-FL. We confirmed that both MPER-directed antibodies could neutralize JR-FL under the standard “no antibody-virus wash” conditions, but, following washing of the antibody-virus mixture, the ability of either antibody to neutralize JR-FL was completely lost (Fig. 3). These data, although indirect, indicate either that the 2F5 and 4E10 antibodies do not bind to the prereceptor-engaged functional spike of the JR-FL virus or that binding was not rapid or of high-enough affinity to neutralize under the conditions of the assay.
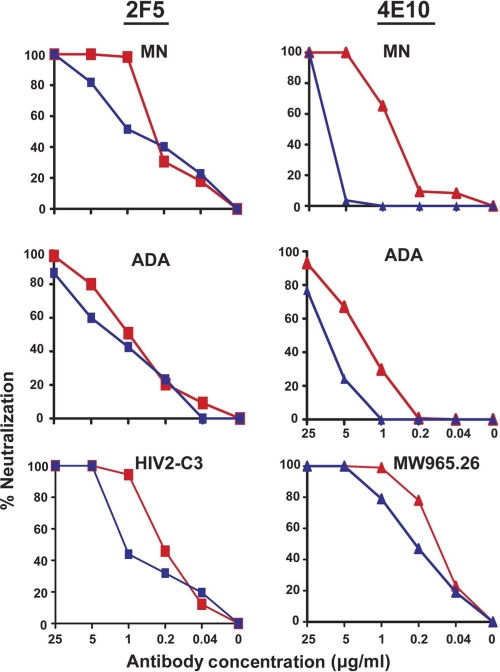
To further test the model, we performed similar analyses with other neutralization-sensitive viruses. We began analysis with the lab-adapted virus, MN, a primary isolate, ADA, an HIV-2 isolate engineered to contain the WT HIV-1 2F5 epitope sequence, HIV-2 C3 (23), and a clade C primary isolate, MW 965.26, which is relatively more sensitive to 4E10. As before, washing of the antibody-virus mixtures had little impact on the ability of 2F5 to neutralize the MN virus and ADA isolate (Fig. 4). Similarly, antibody-virus washing had little effect on the ability of 2F5 to neutralize the engineered HIV-2 C3 virus (Fig. 4). The antibody 4E10 could not be washed out from the MW965.26 virus-antibody complex. It is to be noted that this primary clade C virus is resistant to 2F5 antibody in a standard neutralization assay.
Fig. 4.
Resistance to washing of antibodies 2F5 and 4E10 from the viruses, MN, ADA, MW965.26, and HIV2-C3 after “antibody-virus” complex formation. Antibodies 2F5 and 4E10 preincubated with MN with or without washing prior to incubation with target cells are shown in the top row. The same antibody-virus wash experimental data are shown in the middle panels for the primary isolate ADA. The bottom panels show HIV2-C3 virus with 2F5 and MW965.26 with 4E10 only.
Antibody accessibility to the MPER on a set of HIV-1 isolates is differential.
We next examined a set of primary HIV-1 isolates that displayed a range of neutralization resistance to both MPER-directed antibodies and to antibodies that target other regions of Env. Besides JR-CSF and JR-FL, we examined the neutralization-sensitive isolate, SF162, the moderately resistant isolates, BaL and ADA, and the more neutralization-resistant isolates, CAAN, TRO, and PVO.4. Also as part of this survey, we included another well-described neutralization-resistant virus, JR-CSF, and three JR-CSF variants that contained point mutations in either the V1/V2 or V3 regions. As shown in Table 1, the V-loop point mutations rendered JR-CSF generally more sensitive to a number of antibody specificities, suggesting that these mutations likely induce a “global, spike opening” phenotype. One model that derives from the effects of many point mutations in V1/V2 or V3 is that the variable loops are interacting with each other in the trimeric Env to occlude a variety of neutralizing epitopes. Therefore, we made selected single point mutant changes in either the V3 or V1/V2 regions of the JR-CSF viral Env. Such mutations should render the virus globally sensitive, consistent with the global sensitivity observed when the complete V1/V2 region is deleted in full-length virus (7, 39, 47). Our previous data were also consistent with the global increase in sensitivity to serum neutralization by selected V1, V2, and V3 single residue alterations (4, 63). We tested the effect of point mutations at I309A and I307A in the V3 region and Y177A and L179A in the V1/V2 region of JRCSF envelope. WT JR-CSF and the mutant viruses were neutralized by b12 either without or with an antibody-virus wash step over a broad range of concentrations (data not shown). The potent neutralization exerted by b12 even after the antibody-virus washing confirmed that it could directly access its epitope on the prereceptor-engaged viral Env, likely indicating near irreversible, high-affinity b12 binding.
Table 1.
Comparison of the neutralization sensitivities of wild-type and mutant JRCSF virus
| gp120 domain | Mutation | Neutralization potency relative to WT JRCSF (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CD4 | b12 | VRC01 | VRC03 | b6 | F425 | X5 | 2F5 | ||
| V2 | Y177A | >10,000 | 2,416 | 89 | 61 | >21,008 | 41,100 | >100,000 | 683 |
| L179A | 1,526 | 19 | 13 | 25 | >10 | 5 | 2 | 3 | |
| V3 | 1307A | >10,000 | 7,399 | 119 | <1 | >120 | >2,000 | >200 | 17 |
| 1309A | >10,000 | 3,493 | 82 | 89 | >27,933 | 6,850 | >156,250 | 1,984 | |
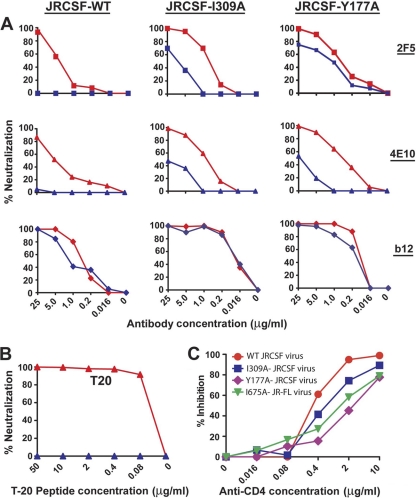
We next examined 2F5 and 4E10 neutralization kinetics of the primary isolate, JR-CSF. We confirmed that both antibodies could neutralize JR-CSF under the standard “no antibody-virus wash” conditions, but following washing of the antibody-virus mixture, the ability of either ligand to neutralize JR-CSF entry into target cells was completely lost (Fig. 5). These data, although indirect, again indicate that the 2F5 and 4E10 antibodies either do not bind to the static functional spike of JR-CSF or that binding was not rapid or durable enough (that is, not of high-enough affinity) for neutralization under the conditions of the assay. Interestingly, the Env I309A and Y177A point mutations rendered JR-CSF partially “antibody-virus wash” resistant in regard to the 2F5 and 4E10 antibodies, implying some direct access to these variant spikes (Fig. 5). A similar effect was observed for two other mutant (I307A and L179A) JRCSF viruses (data not shown). The increased accessibility of the 2F5 and 4E10 epitopes was paralleled by increased sensitivity by the CD4bs ligands b12, b6, and sCD4 as well as the V3-directed antibody F425. These data are consistent with the concept that the variable loop mutations allow a global opening of the primary isolate spike, affecting loop-dependent, quaternary packing, which likely evolved by neutralizing antibody selection pressure.
Fig. 5.
Accessibility of WT JRCSF, V3, and V1/V2 mutant JRCSF for 2F5, 4E10, and T-20 under “no antibody-virus” and “antibody-virus” wash conditions. (A) Results for the antibodies 2F5, 4E10, and b12 preincubated with either JRCSF or mutant JRCSF virus with or without washing prior to incubation with the target cells are shown. (A) Left, data for WT JRCSF; middle, V3 mutant JRCSF virus or JRCSF-I309A data; right, V1/V2 mutant JRCSF virus or JRCSF-Y177A data. The red curves depict “no antibody-virus wash” data sets, and the blue curves depict “antibody-virus wash” data sets. (B) Percent inhibition mediated by T-20 peptide preincubated with JRCSF-I309A virus with or without washing prior to incubation with target cells (TZM-bl). The red curves and the blue curves represent “no antibody-virus” and “antibody-virus” wash, respectively. (C) Percent inhibition of the entry of WT and mutant virus to the target cells in the presence of differing concentrations of the anti-CD4 antibody.
Another alternative explanation for these data is that, besides subtle exposure of neutralizing determinants, the mutations allowed or caused more substantial shifts to CD4 independence, as is seen by V1/V2 deletion in the ADA virus context (34). We confirmed that the mutant viruses did not acquire CD4 independence by performing the entry assay using TZM-bl cells in the presence of various concentrations of entry-inhibiting anti-CD4 antibodies starting at 10 μg/ml (OKT4a; eBioscience, Inc., San Diego, CA). In the presence of anti-CD4 antibodies, entry of the mutant viruses into TZM-bl cells was inhibited to an extent similar to that of WT virus (Fig. 5), indicating that the mutated viruses were CD4 dependent and they were not in a CD4-like triggered state. As an additional control to confirm the specificity of the CD4 dependence assay, a CD4-independent virus, HX8, a kind gift of Robert Doms (28), was not inhibited in its ability to enter TZM-bl cells in the presence of the same concentration of the anti-CD4 antibody (not shown). Additionally, to rule out the possibility that a shift toward the receptor-triggered state of Env was imbued by virtue of the introduced mutations, we performed the antibody-virus wash experiments in the presence of the T-20 peptide as described above. The T-20 peptide, starting at a concentration of 100 μg/ml, was completely washed out after virus incubation and centrifugation phase. However, the T-20 peptide was found to potently inhibit viral entry in the no wash, standard inhibitory conditions in the presence of target cells. This observation suggested that the envelopes were not in a receptor-triggered state, i.e., not in a prehairpin conformation (Fig. 5). As an additional control, we also tested the binding ability of both cluster 1 and cluster 2 antibodies to the I309A mutant JRCSF. These antibodies could not inhibit the mutant virus in either the standard “no antibody-virus wash” or the experimental antibody-virus wash conditions, consistent with our observation that the mutant was not in a CD4-bound or cleavage-defective state (data not shown).
We made similar V2 and V3 mutations in JR-FL, but interestingly they did not render JR-FL globally or 2F5/4E10 neutralization sensitive or “wash resistant” as they did in the JR-CSF context. However, the I675V substitution in the gp41 region of JR-FL Env, as previously described by Shen et al., (54), did render the mutant virus resistant to antibody-virus wash of both the 2F5 and 4E10 neutralizing antibodies (see Fig. S2 in the supplemental material). This mutated JR-FL also required CD4 for efficient entry, as it was inhibited in the presence of anti-CD4 antibodies (Fig. 5).
MPER differential resistance is precisely quantifiable.
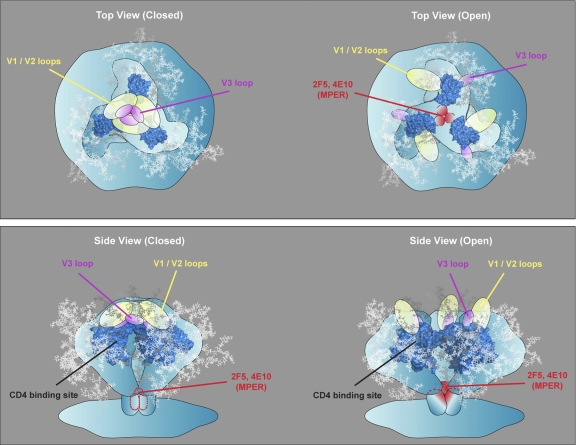
We observed a correlation between the neutralization sensitivity of viruses and the resistance to washing out MPER-directed neutralizing antibodies. There was a clear IC50 cutoff value for which these antibodies were washed out from free virus. We found that the more sensitive the virus, the more difficult it was to wash out (Table 2). These data reinforce a model that regions of MPER epitopes in the static spikes of most primary isolates are inaccessible and occluded by steric barriers not present in the presumed more-open conformation of lab-adapted viral envelopes (Fig. 6).
Table 2.
Correlation between the sensitivity to neutralization and the resistance to washing of MPER-directed neutralizing antibodiesa
| Antibody | Virus | IC50 (μg/ml) for no wash condition | Resistant under wash condition (IC50 [μg/ml]) |
|---|---|---|---|
| 2F5 | |||
| MN | 0.246 | Yes (4.45) | |
| HIV2-C3 | 0.246 | Yes (1.28) | |
| HXBc2 | 0.38 | Yes (1.15) | |
| JR-FL-mut41 (I675V) | 0.44 | Yes (6.35) | |
| JRCSF-mut V3 (I309A) | 0.55 | Yes (4.95) | |
| JR-FL-deICT(+) | 0.736 | Yes (ND) | |
| JRCSF-mut V1/V2 (Y177A) | 0.8 | Yes (0.8) | |
| ADA | 1.18 | Yes (2.27) | |
| SF162 | 1.29 | Yes (20.64) | |
| JR-FL | 3.21 | No (>25) | |
| REJ04541.67 | 3.59 | No (>25) | |
| SC422661.8 | 4.115 | No (>25) | |
| BaL | 6.1 | No (>25) | |
| CAAN | 6.47 | No (>25) | |
| JRCSF | 8.8 | No (>25) | |
| TRO | >25 | No (>25) | |
| PVO | >25 | No (>25) | |
| 4E10 | |||
| MW965.26 | 0.1092 | Yes (0.96) | |
| HXBc2 | 0.24 | Yes (1.75) | |
| MN | 0.32 | Yes (4.5) | |
| JRCSF-mut V1/V2 | 0.49 | Yes (ND) | |
| JRCSF-mutV3 | 0.75 | Yes (4.45) | |
| JR-FL-mut41 | 1.53 | Yes (ND) | |
| ADA | 2.39 | Yes (5.5) | |
| CAAN | 2.66 | No (>25) | |
| SF162 | 4.16 | No (>25) | |
| JRCSF | 4.53 | No (>25) | |
| TRO | 4.7 | No (>25) | |
| JR-FL | 7.475 | No (>25) | |
| JR-FL deICT(+) | 8.7 | No (>25) | |
| BaL | ND | No (>25) | |
| PVO | >25 | No (>25) |
Shading indicates viruses that are not resistant to washing, indicating no direct 2F5 or 4E10 antibody access.
Fig. 6.
Model depicting MPER neutralizing antibody access to primary isolate and lab-adapted virus functional spikes before and after CD4 engagement. Upper panels: left, the more tightly packed primary isolate spike does not permit access of neutralizing ligands before CD4 engagement (top view); right, the more open architecture of the lab-adapted virus allows direct binding to the functional spike by neutralizing antibodies, implying that the epitopes are formed prior to CD4 engagement (top view). Lower panels: left, the more tightly packed primary isolate spike does not allow MPER-directed antibodies to bind before receptor engagement (side view); right, the more open architecture of the primary isolates after receptor engagement and repositioning of primary isolate Env elements (perhaps variable regions) allow accessibility of neutralizing ligands to already preformed MPER determinants (side view).
In support of this interpretation, we performed a washout analysis with the fully wash-resistant JR-FL in the presence of a subneutralizing concentration of soluble CD4 (sCD4) in an attempt to partially activate the functional spike, potentially presenting a more open conformation where we might observe an effect on 2F5 accessibility to its epitope on the virus prior to target cell engagement. The pretreatment of virus with subneutralizing levels of sCD4 permitted some access of 2F5 to its epitope on JR-FL prior to interaction with target cells, as there was an increased resistance to washing out of 2F5 from virus in the presence of sCD4 (see Fig. S3 in the supplemental material).
DISCUSSION
In this study, we demonstrated that the gp41 MPER 2F5 and 4E10 neutralizing epitopes, as well as the b12 neutralizing determinant, were exposed in both the neutralization-sensitive lab-adapted viruses and selected primary HIV-1 isolates by antibody-virus washout analysis. In contrast, on highly resistant HIV-1 isolates, the gp41 neutralizing determinants remained largely occluded until receptor-coreceptor engagement on CD4+/CCR5+ target cells, even though the b12 neutralizing determinant was directly accessible. Furthermore, in two viruses where the 2F5 and 4E10 antibodies could be completely washed from the virus prior to interaction to target cells, selected point mutants in either the gp120 variable regions or gp41 rendered these viruses wash resistant relative to 2F5 and 4E10. In addition, these point mutant viruses demonstrated global sensitivity to antibodies that cannot neutralize the WT parental isolates. We also determined that on laboratory-adapted viruses where 2F5/4E10 appear to directly access their respective epitopes prior to receptor engagement, such access is not because the viruses are in a CD4-triggered state. Entry by these viruses is fully dependent on an accessible CD4 CDR2 region on the cell surface of target cells. It has been suggested that MPER-directed antibodies might bind directly to lab-adapted viral spikes (2), but to our knowledge this is the first report that shows direct binding of MPER-directed antibodies to bona fide virus prior to interaction with target cells. We confirmed a similar dependence upon CD4 for entry on JR-CSF and JR-FL variant isolates rendered directly accessible to 2F5 and 4E10 by the selected V loop mutations or previously reported sensitivity-enhancing mutations in gp41.
One model has been proposed that the HIV-1 MPER epitopes are not readily accessible to neutralization on primary isolates and that the MPER antibody epitope conformations may be fully formed only during the putative fusion transitional intermediate state (10, 19, 48). In this model, the binding of 2F5 and 4E10 to Env occurs during receptor-induced formation of the putative fusion intermediate. Simultaneous exposure and formation of the MPER neutralizing epitopes may occur in this scenario. Indirect analysis in a previous study indicated that 2F5/4E10 cannot access their epitopes on the prereceptor-engaged virus spike (19). However, JR-FL trimer gel shift assays, which tightly correlate with neutralizing antibody epitope accessibility on the virus, indicated that 4E10, and especially 2F5, binds to static HIV-1 primary isolate spikes (2). Here, we present data that strongly suggest that MPER exposure to neutralizing antibody requires receptor engagement on relatively resistant tier 2 and tier 3 primary isolates. However, on the relatively sensitive tier 1a/1b isolates, the 2F5/4E10 epitopes are directly accessible to antibody. In fact, the ability of 2F5 or 4E10 to bind directly to virus, and to be resistant to washing from free virus, can be quantitated and predicted by neutralization potency in the absence of the antibody-virus wash step. Additionally, we demonstrate that if a tier 2 isolate is rendered globally sensitive, then conventionally nonneutralizing antibodies can access their epitopes and neutralize virus.
The data suggest that at least parts of the primary Env spike, namely, the MPER and the coreceptor binding site, are preformed and that engagement of CD4 predominantly repositions Env elements, creating a more open structure akin to that of a lab-adapted isolate. This may also occur in tier 1a/1b viruses. Such a model would not require a transitional intermediate, CD4-induced state to form the MPER or the coreceptor binding site but would suggest that at least these elements of Env are sampled and CD4 engagement permits access indirectly, most likely by repositioning variable gp120 Env elements that normally occlude access. Because V3 is involved in chemokine receptor interaction, and V1/V2 and V3 are thought to be proximal in the context of the functional spike (38), it is plausible that they may occlude the gp120 chemokine receptor binding region. Such a model would be consistent with effects of CD4, which was suggested to reposition V1/V2 in the context of the monomer (68). If repositioning of these variable regions simultaneously impacts exposure of the MPER, then access of 2F5 and 4E10 to their cognate epitopes may occur indirectly as a consequence of disrupting gp120 loop-dependent protomer-protomer interaction. Once protomer-protomer interactions are disrupted, 2F5 and 4E10 may gain access to the MPER through CD4-induced gaps on either the side or perhaps even the top of the spike (Fig. 6). Resistance to antibody washout, or conversely, MPER accessibility, can be quantified in which the IC50 derived by a modified neutralization assay predicts precisely whether the antibody can be washed out from the free virus. There are other possibilities to explain these data, such as that the MPER antibodies actually do access their epitopes on the more resistant viruses but do not irreversibly alter the Env prereceptor-engaged spike and therefore are removed by washing (Joseph Sodroski, personal communication). In addition, our analysis presented here was performed with relatively short incubation times of antibody with virus. Traditionally, in vitro neutralization assays are performed with short incubation times (30 to 60 min) in an attempt to mirror in vivo physiologic conditions where virus is inactivated or removed from an infectious state relatively rapidly (40). Recently, it has been shown that MPER-directed neutralizing antibodies 2F5 and 4E10 can induce shedding of gp120 from the virus with slow kinetics, approaching 18 h. One can envision that under some circumstances, such as the initial portal of entry, HIV may persist for longer intervals and a process that impacts long-lived virus may be an additional and important mechanism (50). Such slow inactivation of HIV might be related to protection when viral inoculum levels are low and a slow and indolent infection process occurs in mucosal regions prior to the establishment of the high levels of viremia established in the gut or lymphoid tissues during acute or even chronic HIV-1 infection.
An intriguing possibility exists regarding selection pressure for viral resistance to 2F5 and 4E10. Based upon the relatively straightforward principle that antibody selection pressure will necessitate viral escape (i.e., anti-V3 antibodies and the resistance of most circulating isolates to such antibodies) one might assume that the occlusion of the MPER in the more resistant HIV isolates is driven by direct selection pressure exerted by 2F5- or 4E10-like antibodies. Although MPER peptide binding antibodies appear relatively abundant in the patient sera of some cohorts, others report a frequency of MPER binding antibodies at a representation of approximately 30% (20). Sera in which the broad and potent neutralizing activity can be mapped to the MPER are even rarer; this type of activity has been reported in only a few cases. It appears possible that steric occlusion of the MPER may result from rearrangements in quaternary packing to provide escape from antibody pressure on the variable loops or the CD4 binding site.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Corbaci, Nadege Pelletier, and Christian Poulsen for technical assistance with the figures.
We are grateful to IAVI, the Bill and Melinda Gates Foundation, and the NIH intramural research program for the funding for this work.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Alam S. M., et al. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106:20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binley J. M., et al. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binley J. M., et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blish C. A., Nguyen M. A., Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burton D. R., et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 [DOI] [PubMed] [Google Scholar]
- 6. Burton D. R., et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 7. Cao J., et al. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Center R. J., et al. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakrabarti B. K., et al. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retroviruses. doi:10.1089/aid.2010.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan D. C., Kim P. S. 1998. HIV entry and its inhibition. Cell 93:681–684 [DOI] [PubMed] [Google Scholar]
- 11. Choe H., et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 12. Cladera J., Martin I., Ruysschaert J. M., O'Shea P. 1999. Characterization of the sequence of interactions of the fusion domain of the simian immunodeficiency virus with membranes. Role of the membrane dipole potential. J. Biol. Chem. 274:29951–29959 [DOI] [PubMed] [Google Scholar]
- 13. Daar E. S., Li X. L., Moudgil T., Ho D. D. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. U. S. A. 87:6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dalgleish A. G., et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 15. de Rosny E., Vassell R., Jiang S., Kunert R., Weiss C. D. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 78:2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dimitrov A. S., et al. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46:1398–1401 [DOI] [PubMed] [Google Scholar]
- 17. Eckert D. M., Kim P. S. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810 [DOI] [PubMed] [Google Scholar]
- 18. Follis K. E., Larson S. J., Lu M., Nunberg J. H. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frey G., et al. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 105:3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gallo S. A., et al. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36–50 [DOI] [PubMed] [Google Scholar]
- 21. Gray E. S., et al. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 83:11265–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gray E. S., et al. 2008. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J. Virol. 82:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray E. S., et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hallenberger S., et al. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358–361 [DOI] [PubMed] [Google Scholar]
- 25. Harrison S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haynes B. F., Montefiori D. C. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:579–595 [DOI] [PubMed] [Google Scholar]
- 27. Herrera C., Klasse P. J., Michael E., Kake S., Barnes K., Kibler C. W., Campbell-Gardener L., Si Z., Sodroski J., Moore J. P., Beddows S. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338:154–172 [DOI] [PubMed] [Google Scholar]
- 28. Hoffman T. L., et al. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. U. S. A. 96:6359–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hwang S. S., Boyle T. J., Lyerly H. K., Cullen B. R. 1992. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science 257:535–537 [DOI] [PubMed] [Google Scholar]
- 30. Jacobs A., et al. 2008. HIV-1 envelope glycoprotein-mediated fusion and pathogenesis: implications for therapy and vaccine development. Vaccine 26:3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones P. L., Korte T., Blumenthal R. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404–409 [DOI] [PubMed] [Google Scholar]
- 32. Koch M., et al. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387–400 [DOI] [PubMed] [Google Scholar]
- 33. Koito A., Harrowe G., Levy J. A., Cheng-Mayer C. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kolchinsky P., Kiprilov E., Bartley P., Rubinstein R., Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M., et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ly A., Stamatatos L. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Markowitz M., et al. 2003. A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J. Virol. 77:5037–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mascola J. R., et al. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 76:4810–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Montefiori D. C., Robinson W. E., Jr., Schuffman S. S., Mitchell W. M. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Brien W. A., Chen I. S., Ho D. D., Daar E. S. 1992. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J. Virol. 66:3125–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orloff S. L., Kennedy M. S., Belperron A. A., Maddon P. J., McDougal J. S. 1993. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J. Virol. 67:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pancera M., Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156 [DOI] [PubMed] [Google Scholar]
- 46. Parren P. W., et al. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raja A., Venturi M., Kwong P., Sodroski J. 2003. CD4 binding site antibodies inhibit human immunodeficiency virus gp120 envelope glycoprotein interaction with CCR5. J. Virol. 77:713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rits-Volloch S., Frey G., Harrison S. C., Chen B. 2006. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 25:5026–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roben P., et al. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruprecht C. R., et al. 2011. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 208:439–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saphire E. O., et al. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155–1159 [DOI] [PubMed] [Google Scholar]
- 52. Sather D. N., et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sattentau Q. J., Zolla-Pazner S., Poignard P. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713–717 [DOI] [PubMed] [Google Scholar]
- 54. Shen X., et al. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc. Natl. Acad. Sci. U. S. A. 107:5972–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen X., et al. 2009. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J. Virol. 83:3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Si Z., Phan N., Kiprilov E., Sodroski J. 2003. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res. Hum. Retroviruses 19:217–226 [DOI] [PubMed] [Google Scholar]
- 57. Simon V., Ho D. D. 2003. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 1:181–190 [DOI] [PubMed] [Google Scholar]
- 58. Sullivan N., Sun Y., Li J., Hofmann W., Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sullivan N., et al. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomaras G. D., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Trkola A., et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. VanCott T. C., et al. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 65. Willey R. L., Bonifacino J. S., Potts B. J., Martin M. A., Klausner R. D. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U. S. A. 85:9580–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu L., et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183 [DOI] [PubMed] [Google Scholar]
- 67. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wyatt R., et al. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y. J., Fredriksson R., McKeating J. A., Fenyo E. M. 1997. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology 238:254–264 [DOI] [PubMed] [Google Scholar]
- 70. Zhou J. Y., Montefiori D. C. 1997. Antibody-mediated neutralization of primary isolates of human immunodeficiency virus type 1 in peripheral blood mononuclear cells is not affected by the initial activation state of the cells. J. Virol. 71:2512–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zwick M. B., et al. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79:1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.