Abstract
Cellular proteins play many important roles during the life cycle of all viruses. Specifically, host cell nucleic acid-binding proteins interact with viral components of positive-stranded RNA viruses and regulate viral translation, as well as RNA replication. Here, we report that nucleolin, a ubiquitous multifunctional nucleolar shuttling phosphoprotein, interacts with the Norwalk virus and feline calicivirus (FCV) genomic 3′ untranslated regions (UTRs). Nucleolin can also form a complex in vitro with recombinant Norwalk virus NS6 and -7 (NS6/7) and can be copurified with the analogous protein from feline calicivirus (p76 or NS6/7) from infected feline kidney cells. Nucleolin RNA levels or protein were not modified during FCV infection; however, as a consequence of the infection, nucleolin was seen to relocalize from the nucleoli to the nucleoplasm, as well as to the perinuclear area where it colocalizes with the feline calicivirus NS6/7 protein. In addition, antibodies to nucleolin were able to precipitate viral RNA from feline calicivirus-infected cells, indicating a direct or indirect association of nucleolin with the viral RNA during virus replication. Small interfering RNA (siRNA)-mediated knockdown of nucleolin resulted in a reduction of the cytopathic effect and virus yield in CrFK cells. Taken together, these results demonstrate that nucleolin is a nucleolar component that interacts with viral RNA and NS6/7 and is required for feline calicivirus replication.
INTRODUCTION
The Caliciviridae family of small positive-stranded RNA viruses includes viruses that infect both animals and humans, causing a wide range of diseases. Human caliciviruses (HuCVs), which encompass the genera Norovirus and Sapovirus, have received increased attention because of their established role as etiologic agents of acute gastroenteritis (19). In particular, noroviruses (NoVs) are the most commonly detected pathogens both in sporadic cases and outbreaks of gastroenteritis (19, 53). Since HuCVs are not conveniently propagated in cell culture, other animal caliciviruses, such as the rabbit hemorrhagic disease virus, the murine norovirus (MNV), and feline calicivirus (FCV), have been used as surrogates to study various aspects of the calicivirus life cycle in cell culture (59).
It has been widely established that the replicative cycle of positive-strand RNA viruses requires interactions between viral components and various host cell factors (2, 35, 36). In particular, the interaction of host cell nucleic acid-binding proteins with structured RNA elements, usually present at the 5′ and 3′ ends of the viral genome, has been extensively implicated in viral translation and replication (2). These interactions often contribute to the host specificity, tissue tropism, and pathology of infections.
One of the best-studied mammalian RNA viruses, for which the role of host-virus RNA structure interactions has been extensively characterized, is poliovirus (PV). Poliovirus translation and negative-strand RNA synthesis are coordinated by the interaction of a complex of host proteins: poly(C) binding protein (PCBP) and poly(A) binding protein (PABP), viral RNA, and polymerase precursor 3CD (4, 23, 33). In addition, the immunodepletion of nucleolin, a host RNA binding protein that interacts with the PV 3′ untranslated region (UTR), from a PV cell-free replication system inhibits early yields of PV virions (59). Nucleolin has also been implicated in poliovirus translation via interaction with the viral internal ribosome entry site (IRES) (37).
Numerous other host cell factors have been implicated in the life cycle of several positive-strand RNA viruses. The translation and replication of hepatitis C virus (HCV) RNA depends on several host cell proteins, such as La and polypyrimidine tract-binding protein (PTB) (15) and Rck/p54, LSm1, and PatL1, which regulate the fate of cellular mRNAs from translation to degradation in the 5′-to-3′ deadenylation-dependent mRNA decay pathway. The requirement of these proteins for efficient HCV RNA translation is linked to the interaction with cis-acting RNA elements located in the 5′ and 3′ UTRs of the viral genome (51). These proteins are also implicated in the replication of brome mosaic virus and the bacteriophage Qβ (22, 46). Other host factors, such as TIAR and the closely related protein TIA-1, components of stress granules (SG), bind to the 3′ stem-loop minus-strand RNA of the West Nile virus, facilitating replication (44). Finally, the cellular proteins La, PTB, Y box-binding protein 1, PABP, and the translation elongation factor eEF-1α bind to the dengue virus 3′ UTR (14). In particular, PTB translocates from the nucleus to the endoplasmic reticulum (ER) during dengue virus infection and plays a role in modulating dengue virus replication (1).
For caliciviruses, in vitro interaction between several host cell nucleic acid-binding proteins and the 5′ and 3′ ends of Norwalk virus (NV) (30, 31) and FCV genomic RNA (42) have been reported. PCBP, La, hnRNP-L, poly(A) binding protein, and PTB were identified among the proteins that bound to the NV 3′ UTR. However, other proteins, with molecular masses from 120 to 33 kDa, that also bound to the same region were not identified (31). Recently, it was established that PTB is required for efficient FCV replication in a temperature-dependent manner (42). Moreover, it was observed that as the levels of viral proteins rise during the course of virus infection, the nuclear-cytoplasmic shuttling of PTB is altered, causing an increase in the cytoplasmic levels of this protein and an inhibition of viral translation initiation, contributing to the stimulation of viral RNA replication (42). In the present study, we report the identification of a host cell protein with a molecular mass of 105 kDa that interacts with the 3′ UTR of the NV and FCV genomes as nucleolin. FCV infection had no apparent effect on the steady-state levels of either nucleolin RNA or protein; however, FCV infection resulted in nucleolin relocalization from the nucleoli to nucleoplasm and the perinuclear area, where it colocalizes with the FCV NS6/7 proteins. Finally, using small interfering RNA (siRNA) against nucleolin, we showed a marked inhibitory effect on FCV replication in CrFK cells, confirming a functional role for nucleolin in the calicivirus life cycle.
MATERIALS AND METHODS
Cells and virus infection.
HeLa cells were grown in Dulbecco's minimal essential medium supplemented with 10% newborn calf serum, 5,000 U/ml of penicillin, and 5 μg/ml of streptomycin. The culture medium was changed every other day until the cells reached confluence. CrFK cells obtained from the American Type Culture Collection (ATCC) (Rockville, MD) were grown in Eagle's minimal essential medium with Earle's balanced salt solution (BSS) and 2 mM l-glutamine (EMEM) that was modified by the ATCC to contain 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, 1.5 g/liter sodium bicarbonate. The medium was supplemented with 10% horse serum, 5,000 U of penicillin and 5 μg/ml of streptomycin. Both cell lines were grown in a 5% CO2 incubator at 37°C. CrFK infection with the FCV F9 strain (obtained from the American Type Culture Collection) was performed as previously described (50).
UV treatment of FCV was conducted as previously described, with minor modifications (55). Briefly, virus stocks (1 ml at 8 × 106 PFU/ml) were placed on ice and irradiated with UV light (254 nm, Ultralum UV lamp) for 15, 30, 45, 60, and 90 min at a distance of 5 cm. UV-treated viruses were analyzed for infectivity on CrFK cells to confirm inactivation. Virus irradiated for 45 min, which resulted in a complete loss of infectivity, was used in immunofluorescence assays to control for any nonspecific effects of host cell proteins which may be present in the virus preparations.
In vitro transcription.
Two RNA molecular species that correspond to the complete 3′ UTRs from NV (nucleotides 7588 to 7654) and FCV (nucleotides 7707 to 7699) were produced by in vitro transcription, using T7 RNA polymerase, from two PCR-amplified cDNAs containing the respective regions. The NV cDNA 3′ UTR was obtained by PCR as described previously (31). The FCV 3′ UTR cDNA was obtained by one-step reverse transcription (RT)-PCR from infected CrFK cells, using a sense primer that contains the bacteriophage T7 promoter sequence (5′ TAATACGACTCACTATAGGGTCATATATCCCTTTGGG 3′) and an antisense primer (5′ CCCTGGGGTTAGGCGCAGG 3′). The RT-PCR was performed using a murine leukemia virus (MLV) reverse transcriptase kit (Invitrogen) at 40°C for 30 min, followed by 35 cycles of 94°C for 1 min, 55°C for 30 s, and 68°C for 30 s using a Perkin-Elmer Cetus DNA thermocycler. The resulting amplicon was purified by using a QIAquick gel extraction G-50 kit (Qiagen) before being used as the template for RNA synthesis. After in vitro transcription (Epicentre Biotechnologies), the reaction mixture was treated with DNase RQ1 (Promega) at 37°C for 30 min in the presence of RNase inhibitors (Promega) to remove the DNA template. Unincorporated nucleotides were removed by precipitation. For the synthesis of radiolabeled transcripts, [α32P]UTP or [α32P]ATP was included in the transcription reaction mixture.
Quantitative RT-PCR detection of nucleolin.
The levels of expression of the nucleolin gene in FCV-infected CrFK cells were determined by using relative quantification of gene transcripts and the expression of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as a housekeeping gene control. Briefly, 6-well cell culture plates were infected with FCV at a multiplicity of infection (MOI) of 10. After 1 h of adsorption at 37°C, the wells were rinsed with phosphate-buffered saline (PBS), and the cells in each well were collected at 0, 2, 4, and 8 h postinfection (hpi). The RNA was extracted with Trizol reagent (Invitrogen), following the manufacturer's instructions, and treated with DNase I (New England Biolabs). The resulting RNA quantity and quality were assayed using a NanoDrop spectrophotometer (Thermo Fisher Scientific). One microgram of RNA was reverse transcribed using Improm II reverse transcriptase (Promega) according to the manufacturer's instructions, with oligo(dT) as a primer. The quantitative PCR step was developed with recombinant DNA polymerase (Invitrogen) and Evagreen (Biotium), using the following primers: for nucleolin, Fw (5′ TGCAAGAAGCCAGCCGTCCAA 3′) and Rv (5′ CCGAACAGAGCCGTCGAACGATT 3′), and for GAPDH, Fw (5′ GGAAGGTGAAGGTCGGAGTC 3′) and Rv (5′ GAAGATGGTGATGGGATTTCC 3′). The data were analyzed with the comparative threshold cycle (ΔΔCT) method. Nontemplate and non-reverse transcriptase controls were included for each time point and for each gene transcript to be detected. The reaction was performed three times in duplicate using a 7300 real-time PCR system (Applied Biosystems).
Preparation of cell extracts.
For NP-40 extracts, cell monolayers were washed twice with cold PBS and once with detachment buffer (40 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl). After the final wash, cells were resuspended in RSB–NP-40 buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 1% NP-40), incubated for 20 min at 4°C, and centrifuged at 13,000 × g for 5 min in a Sorvall GSA rotor. Lysates were adjusted to 1 mM CaCl2, treated with micrococcal nuclease (15 μg per ml), and incubated at 20°C for 15 min. The nuclease was inactivated with 2 mM EGTA, and the lysate centrifuged in a Sorvall MC 12 microcentrifuge at 13,000 × g for 15 min at 4°C. The supernatants were aliquoted, the concentrations of proteins in each extract were determined with the Bradford assay (10), and the samples stored at −70°C until use.
Isolation of replicative complexes from cells.
The membrane fractions corresponding to FCV replicative complexes were obtained as described previously (28). The presence of nucleolin, protein disulfide isomerase (PDI; Santa Cruz Biotechnology, Santa Cruz, CA), and NS6/7 (41) was determined by Western blotting.
Electrophoretic mobility shift assay (EMSA).
Thirty micrograms of cell extracts from HeLa and CrFK cells were preincubated for 15 min at 4°C with the same amount of tRNA in a buffer containing 10 mM HEPES (pH 7.4), 0.1 mM EDTA, 0.2 mM dithiothreitol (DTT), 8 mM MgCl2, 4 mM spermidine, 3 mM ATP, 2 mM GTP, and 10% (vol/vol) glycerol in a final volume of 10 μl. Where indicated, monoclonal antinucleolin (C23-MS-3) and anti-GAPDH antibodies (0.6 μg) (Santa Cruz Biotechnology, Santa Cruz, CA) were added separately to the corresponding reaction mixture and incubated for 30 min on ice. Amounts of 4 × 105 cpm of 32P-labeled NV or FCV RNAs were added to each reaction mixture and incubated for 15 min at 4°C. Before loading the gels, a final incubation with 20 units of RNase A and 20 μg of RNase T1 was performed for 15 min at room temperature. The RNA-protein supershifted complexes were analyzed by electrophoresis through a 6% polyacrylamide (acrylamide-bisacrylamide, 80:1) gel in 0.5× TBE buffer (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA [pH 8.3]) and run at 20 mA for 4 h. The gels were dried and autoradiographed.
Immunoprecipitation assay.
A UV cross-linking of HeLa cell extracts with the 32P-labeled NV 3′ UTR was performed as described previously (31), and reaction mixtures were precleared by incubation with 10 μl of protein G-agarose resin (Roche) for 2 h at 4°C followed by centrifugation at 13,000 × g for 5 min. The resultant supernatants were incubated overnight at 4°C with monoclonal antinucleolin or anti-GAPDH antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). The immune complexes were immobilized on protein G-agarose beads saturated with 2% bovine serum albumin for a further 2 h at 4°C. Unbound material was removed by five washes with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 0.05% Nonidet P-40). Bound proteins were analyzed by SDS-PAGE followed by autoradiography.
Pulldown assay.
Two hundred micrograms of HeLa cell protein extract preabsorbed with Ni-nitrilotriacetic acid (NTA) agarose resin (Qiagen) was brought to a final volume of 500 μl with interaction buffer (20 mM NaH2PO4, 40 mM NaCl [pH 8], 0.5% Triton X-100), mixed with 50 μg of Norwalk virus recombinant NS6/7 protein coupled to Ni-NTA agarose resin or Ni-NTA alone as a control, and mixed overnight at 4°C. The beads were then collected by a 2-min centrifugation at 380 × g at 4°C and then washed three times with 500 μl of washing buffer (20 mM NaH2PO4, 200 mM NaCl [pH 8], 1% Triton X-100, 10 mM imidazole) for 10 min at room temperature. The complexes were eluted from the beads by the addition of loading buffer, boiled for 10 min, and electrophoresed on 10% SDS–polyacrylamide gels. Proteins were transferred to nitrocellulose for Western immunoblotting. Briefly, the membranes were blocked for 1 h with 5% skim milk and incubated overnight at room temperature with the mouse monoclonal antinucleolin antibody diluted 1:5,000 in 0.05% Tween 20–PBS solution. Then, the membranes were incubated for 2 h at room temperature with goat anti-mouse IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:10,000. The proteins were detected with an enhanced chemiluminescence kit (SuperSignal West Femto; Pierce). The same membranes were washed with Western blot stripping buffer (Pierce) and incubated with anti-NS6/7 antibody (41).
Coimmunoprecipitation assay.
Fifty micrograms of infected CrFK NP-40 protein extracts were precleared by incubation with 10 μl of protein G-agarose resin for 2 h at 4°C followed by centrifugation at 13,000 × g for 5 min. The clarified supernatant was then incubated with either a polyclonal antinucleolin (Santa Cruz Biotechnology, Santa Cruz, CA) or antiactin antibody (kindly provided by M. Hernández, CINVESTAV) overnight at 4°C on a rotator (Cole-Parmer). The immunocomplexes were immobilized on 10 μl of protein G-agarose resin saturated with 2% bovine serum albumin for an additional 2 h at 4°C. After five washes with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 0.05% Nonidet P-40), bound proteins were analyzed by SDS-PAGE and subjected to Western blotting as described above.
Coimmunoprecipitation of viral RNA during infection.
The interaction of nucleolin with FCV viral RNA during infection was studied by using RNA coimmunoprecipitation. Briefly, CrFK cells were infected with FCV at an MOI of 3 50% tissue culture infective doses (TCID50)/cell. Cell lysates were prepared 5 hpi using polysomal lysis buffer (10 mM HEPES, pH 7.0, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT, and protease inhibitors) and used in subsequent immunoprecipitations. Immunoprecipitations were carried out using a mouse monoclonal antibody to nucleolin (Invitrogen) and rabbit polyclonal antibody to the viral RNA polymerase NS6/7 or control purified rabbit IgG using protein A/G-Sepharose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Viral RNA coimmunoprecipitated in the immune complexes was isolated using the GenElute RNA purification system (Sigma) and subsequently RT-PCR amplified. Reverse transcription was performed using SuperScript III (Invitrogen) and viral-RNA-specific primer IGRDG-64 (TTATCAAACTTCGAACACATCACAGTG) according to the manufacturer's protocol. cDNA was then subjected to PCR using the primers IGRDG-58 (TGTACCTGTCACTTGGAATCTC) and IGRDG-64 to produce a 583-bp product encompassing nucleotides 4732 to 5314 of the FCV genome.
Immunofluorescence assays.
CrFK cells (1.5 × 105) were seeded in a 6-well plate containing coverslips pretreated with poly-l-lysine (0.1%) and grown overnight. At the times indicated, infected CrFK cells were washed once with C buffer (10 mM MES [morpholineethanesulfonic acid] hydrate, 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, and 5 mM glucose) and treated for 5 min with C buffer containing 4% paraformaldehyde and 0.5% Triton X-100 at room temperature. After another round of three washes with PBS, the cells were incubated with 2 ng/μl of antinucleolin antibody at 4°C overnight and washed again as described previously. The cells were incubated for 60 min at room temperature with anti-mouse antibody coupled to Alexa Fluor 488 (Invitrogen). The cells were washed three times with PBS, incubated with the anti-NS6/7 antibody diluted 1:100 at 4°C overnight, washed again, and incubated with anti-rabbit antibody labeled with Alexa Flour 594 (Invitrogen) for 1 h at room temperature. The cells were washed three times with PBS and incubated with 1 μg/μl of 4′,6′-diamidino-2-phenylindole (DAPI) for 2 min. The cells were washed six times with PBS and three times with distilled water. Finally, the coverslips were removed and treated with Vectashield (Vector Laboratories). The samples were examined by confocal microscopy.
Confocal microscopy.
For confocal microscopy, a Leica TCS-SP5 multiphotonic confocal laser scanning device fitted to a DMI 6000 fluorescence microscope (see Fig. 4A) or a Leica TCS-SPE fitted to a DMI 6000 fluorescence microscope (see Fig. 6A) was used. Images were taken with a 63× lambda blue 1.4 or 63× lambda blue 1.3 oil immersion lens, respectively. Images were acquired as 8-bit TIFF files and imported into Power Point. The individual images were pseudocolored in their respective RGB (red-green-blue) channels.
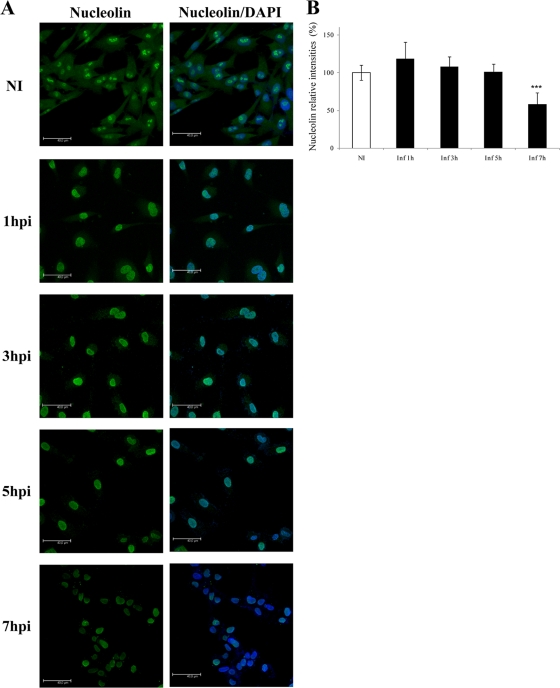
Fig. 4.
Subcellular localization of nucleolin during FCV infection. (A) Monolayers of CrFK cells were infected with FCV at an MOI of 10 or mock infected (NI). At 1, 3, 5, and 7 hpi, cells were fixed and stained for nucleolin (polyclonal antinucleolin, green). The cells were examined using confocal microscopy. DAPI was used for nuclear (blue) staining. Omission of the primary antibody resulted in no nucleolin signal (data not shown). Scale bars, 40 μm. Images correspond to a z-stack of 15 slices. (B) Fluorescence intensities in the nucleoli and nucleoplasm at 0, 1, 3, 5, and 7 hpi were measured with LAS AF Lite software and are expressed as percentages of the relative intensities of the uninfected cells. Error bars show standard deviations. ***, P < 0.001.
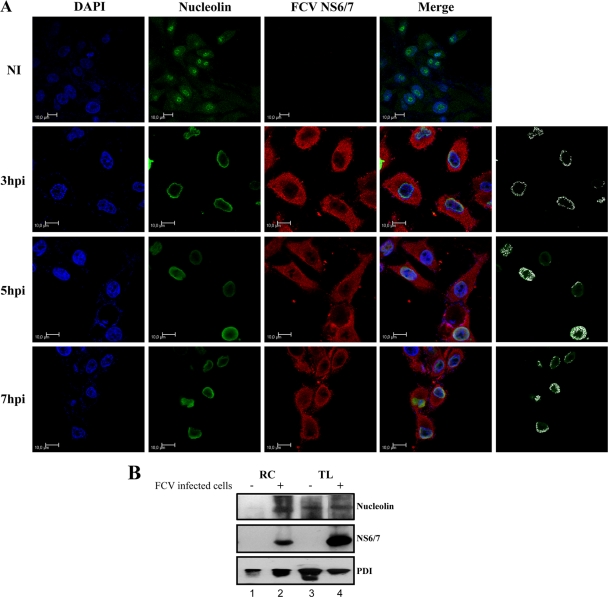
Fig. 6.
Nucleolin and NS6/7 colocalization during FCV infection. (A) Monolayers of CrFK cells were left uninfected (NI) or infected with FCV for 3, 5, and 7 h at an MOI of 10, fixed, and stained for NS6/7 (red) and nucleolin (polyclonal antinucleolin, green). DAPI was used for nuclear (blue) staining. Confocal microscopy was used to observe 0.5-μm-thick optical sections of uninfected and infected cells. Merged images of nucleolin and FCV NS6/7 and colocalization of FCV NS6/7 and nucleolin at 3, 5, and 7 hpi are highlighted in white. Colocalization analysis was performed using LAS AF confocal software. Scale bars, 10 μm. Images depict single confocal slices taken from z-stacks. (B) Proteins associated with total extracts (TL) or RC pellet (RC) were obtained from mock-infected (lanes 1 and 3) and FCV-infected cells (lanes 2 and 4) as described in Materials and Methods. The presence of nucleolin, FCV NS6/7, and PDI as an ER marker was evaluated by Western blotting.
Design and synthesis of nucleolin siRNAs.
The carboxyl-terminal nucleotide sequence of feline nucleolin was obtained from total RNA from CrFK cells by RT-PCR using a pair of oligonucleotides designed to amplify the carboxyl-terminal region of the murine nucleolin (Fwd 5′-GCGGCCGCATGGTGAAGCTCGCAAAG-3′ and rev 5′-GCGCCGCATGTCAGAACCAACTACACC-3′). This amplicon was sequenced and used as a template for the design of siRNAs targeting nucleolin mRNAs (17). Two siRNA duplexes, siRNA #1 (5′-GCUUUAAAUUCCUGUAAUAAA-3′) and siRNA #2 (5′-CACUUUUGGCUAAAAUCUGGC-3′), were obtained from Applied Biosystems (México).
siRNA-mediated knockdown of nucleolin.
For siRNA-mediated knockdown of nucleolin expression, transfections were carried out according to the protocol recommended by the manufacturer (siPORT amine transfection agent; Applied Biosystems, Mexico). Briefly, CrFK cells were plated in a 6-well plate to reach 60% confluence. After 24 h, 5 μl of siPort and 60, 80, and 100 nM siRNAs for nucleolin were mixed separately with 100 μl Opti-MEM, respectively, for 10 min at room temperature. The two mixtures were combined, allowed to incubate at room temperature for 10 min, and then diluted to 1 ml with 800 μl Opti-MEM. A 200-μl aliquot of the mixture was added directly to the cells, and transfection with the siRNAs was carried out at 37°C for 8 h, followed by the addition of 1 ml growth medium and additional incubation for 24, 48, and 72 h. CrFK cells were treated in the same way with a nontargeting 20- to 25-nucleotide siRNA as a control (Santa Cruz Biotechnology, Santa Cruz, CA). Mock-transfected cells were treated with transfection reagent only. After transfection, cells were infected with FCV at an MOI of 10 and then lysed 5 h postinfection. The levels of the nucleolin, hnRNP A1, viral protease-polymerase NS6/7, and NS3 (p39) proteins were determined by Western blotting. The viability of untreated and nucleolin siRNA-treated cells was determined at 48 h using a CellTiter 96 assay (Promega), following the manufacturer's instructions.
One-step growth curve.
CrFK cells previously transfected with specific siRNAs for nucleolin or a nontarget siRNA used as a control were infected with FCV at an MOI of 10. At various times postinfection, cell supernatants were collected and virus yield determined by plaque assay. In addition, in a parallel experiment, the cytopathic effect was evaluated by phase-contrast microscopy.
Computation of RNA structure.
Computer analysis of the RNA secondary structures was performed using Mfold2 software (64) with default settings through the Web interface at http://mfold.rna.albany.edu/?q=mfold/RNA=Folding=Form2.3. The VARNA Web applet at http://varna.lri.fr/ was used to draw RNA secondary structures.
RESULTS
Nucleolin interacts with the 3′ UTR of NV and FCV.
We have previously established that the polyadenylated NV 3′ UTR was able to specifically interact with several cellular proteins present in HeLa cell extracts (31). Three of these proteins were identified as La, PTB, and PABP. Among the unidentified proteins, one of the most abundant was a 105-kDa protein. The molecular mass of this protein was compatible with the molecular mass of nucleolin, a cellular multifunctional nucleolar protein that is implicated in cell proliferation and growth (25, 47, 56), as well as the life cycle of many RNA viruses (34, 39, 61).
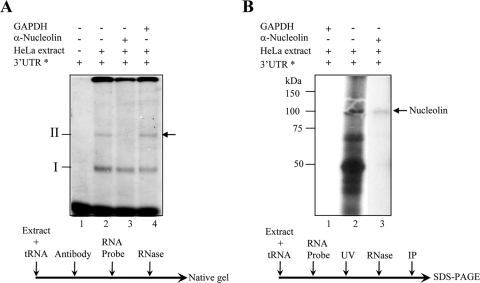
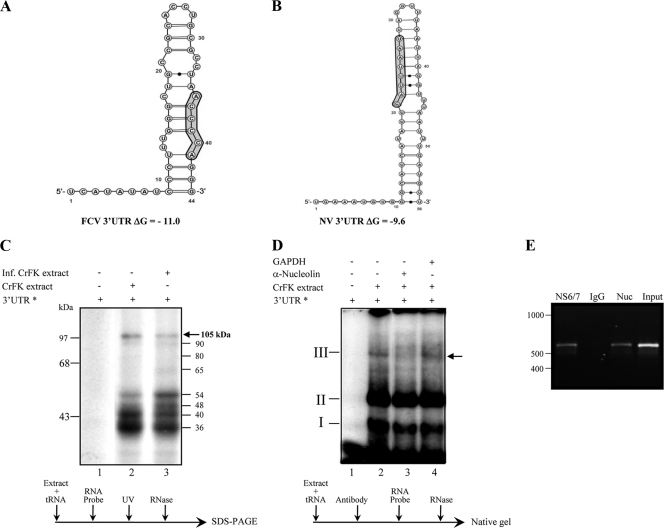
In order to elucidate whether nucleolin was present in the ribonucleoprotein (RNP) complex formed between the polyadenylated NV 3′ UTR and HeLa cell proteins, an EMSA was performed in the presence of an antinucleolin antibody. As described previously (31), two well defined complexes, referred to as I and II, were observed when the NV 3′ UTR was incubated with HeLa protein extracts (Fig. 1 A). When a monoclonal antinucleolin antibody was incubated with HeLa protein extracts prior to the addition of the RNA, the formation of complex II was abolished (Fig. 1A), while the same amount of control anti-GAPDH antibody did not alter the formation of either complex (Fig. 1A). The inhibitory effect of antinucleolin antibody on complex II formation indicated that nucleolin was present in this complex and that the antibody binding ablated the nucleolin-3′ UTR interaction. To date, the precise components of complex I are not known, but it may contain one or more of the factors we previously identified as also interacting with the NV 3′ UTR, namely, PCBP, La, hnRNP-L, PABP, or PTB. The interaction between the NV 3′ UTR and nucleolin was further confirmed by immunoprecipitation of a 105-kDa protein following UV cross-linking to the [α-32P]UTP-labeled NV polyadenylated 3′ UTR with the antinucleolin antibody (Fig. 1B). This result and the inability of the anti-GAPDH antibody to immunoprecipitate any labeled protein (Fig. 1B) confirm that nucleolin binds to the polyadenylated NV 3′ UTR. The identities of some of the lower-molecular-mass proteins shown in Fig. 1B, such as PTB and La protein, have been previously determined (31). Since nucleolin was able to interact with the NV 3′ UTR in vitro, it is possible that this interaction would also take place during virus replication, playing some role in the NV life cycle. However, because HuCVs cannot be cultured in cells, we used FCV as a model system to examine the role of nucleolin in the calicivirus life cycle. Computational analysis indicated that although the primary sequence is considerably different, the predicted secondary structures of the 3′ UTRs from both NV and FCV are very similar (Fig. 2). The prediction for FCV and NV 3′ UTRs showed stem-loop structures with ΔG values of −11.0 and −9.6, respectively (Fig. 2A and B).
Fig. 1.
Nucleolin binds to the Norwalk virus 3′ UTRs. (A) Super-mobility shift assay of the [α-32P]UTP-labeled NV 3′ UTR RNA incubated with HeLa cell extracts in the absence (lanes 1 and 2) or presence of human antinucleolin (lane 3) or anti-GAPDH (lane 4) antibody. Lane 1, free RNA. The positions of complexes I and II are indicated. The arrow indicates the position of the complex competed with antinucleolin antibodies. (B) [α-32P]UTP-labeled NV 3′ UTR RNA was cross-linked with HeLa cell extracts (lane 2) and immunoprecipitated with human antinucleolin antibody (lane 3) or anti-GAPDH antibody (lane 1). The arrow indicates the position of the immunoprecipitated labeled nucleolin. The asterisks indicate [α-32P]UTP-labeled NV 3′ UTR RNA. The order of the addition of various reagents is indicated below each experiment.
Fig. 2.
Predicted secondary structure of the feline calicivirus 3′ UTR and its binding to nucleolin. (A and B) Secondary structure of the complete FCV 3′ UTR (A) and the complete NV 3′ UTRs (B) predicted using Mfold 2 software (http://mfold.rna.albany.edu/?q=mfold/RNA=Folding=Form2.3). Potential nucleolin binding sites, as discussed in the text, are shaded. (C) [α-32P]-UTP-labeled FCV 3′ UTR (lanes 1 and 2) RNA was UV cross-linked without (lane 1) or with noninfected (lane 2) or infected (lane 3) CrFK cell extracts. (D) Super-mobility shift assay of the [α-32P]UTP-labeled FCV 3′ UTR RNA incubated with CrFK cell extracts (lanes 2 to 4) in the absence (lanes 1 and 2) or presence of human antinucleolin (lane 3) or anti-GAPDH (lane 4) antibody. The positions of complexes I, II, and III are indicated. The arrow indicates the position of complex III. (E) Antiserum to the FCV NS6/7 protein or nucleolin precipitated viral RNA from FCV-infected cells. Viral RNA was coimmunoprecipitated from FCV-infected CrFK cell extracts using an antibody directed against viral protease-polymerase (NS6/7) or an antinucleolin antibody (Nuc), respectively. Purified IgG was used as a negative control. RNA was extracted from the immunoprecipitated complex or from an aliquot of the input lysate and subjected to RT-PCR using FCV-specific primers as detailed in Materials and Methods. RT-PCR products were visualized on a 1% agarose gel.
To analyze whether nucleolin binds to the FCV 3′ UTR, a UV cross-linking assay was performed using an [α-32P]UTP-labeled FCV 3′ UTR and extracts enriched with cytoplasmic membrane-associated proteins from uninfected and infected CrFK cells (Fig. 2C). Proteins with molecular masses of 30 to 105 kDa from uninfected and infected cells were detected bound to this region (Fig. 2C). In addition, proteins with molecular masses of 48 and 65 kDa were observed when extracts from infected cells were used. To determine whether the 105-kDa protein was nucleolin, a supershift assay was performed in the presence of an antinucleolin antibody. The interaction of the FCV 3′ UTR with proteins present in CrFK cell extracts resulted in the formation of two major complexes and one minor complex, referred to hereinafter as I, II, and III, respectively (Fig. 2D), although additional minor complexes were also formed. As observed for the NV 3′ UTR-nucleolin complex, a monoclonal antinucleolin antibody inhibited the formation of complex III, while anti-GAPDH antibody had no effect (Fig. 2D), confirming that nucleolin was present in complex III.
To validate that nucleolin associates with FCV RNA during virus replication, an RNA-coimmunoprecipitation assay was performed (Fig. 2E). Nucleolin was immunoprecipitated from FCV-infected CrFK cell extracts, and the copurified RNA was extracted from the immunoprecipitated complex and subjected to RT-PCR using FCV-specific primers. As expected, FCV RNA was copurified with the viral RNA polymerase (NS6/7) but also with nucleolin. No viral RNA was amplified when an irrelevant purified rabbit IgG antibody was used. Taken together, these results confirm that nucleolin is associated with FCV viral RNA during viral replication.
Nucleolin RNA and protein levels remain unmodified during FCV infection.
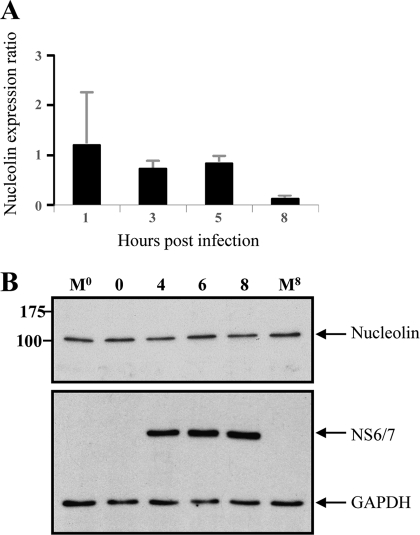
The expression and subcellular localization of cellular proteins implicated in viral replication are often modified during the course of infection. Thus, to determine whether nucleolin expression is modified during FCV infection, the amount and integrity of nucleolin RNA and protein were analyzed up to 8 hpi (Fig. 3). The nucleolin RNA (Fig. 3A) and protein levels detected by Western blotting (Fig. 3B) and by quantitative Western blotting (data not shown) remained constant throughout the course of viral infection and were detected in amounts similar to the amounts observed in noninfected cells, indicating that the gross levels of nucleolin RNA and protein were not altered during FCV infection.
Fig. 3.
Nucleolin RNA and protein expression during FCV infection. (A) Total RNA extracted from FCV-infected CrFK cells at 1, 3, 5, and 8 h was subjected to quantitative RT-PCR using specific primers. Levels of expression of the nucleolin gene were obtained by relative quantification of the gene transcript, and expression of the GAPDH gene was used as a housekeeping gene control. Error bars show standard deviations. (B) Total cellular extracts from uninfected cells prepared at either time zero or 8 h (M0 and M8) and from FCV cells infected at an MOI of 3, obtained at 2, 4, 6, and 8 hpi, were analyzed by Western blotting using human antinucleolin (upper panel) or anti-FCV NS6/7 (lower panel) antibody. GAPDH was used as the loading control. Molecular sizes, in kDa, are shown on the left.
Nucleolin is relocalized from the nucleolus to the nucleoplasm after FCV infection.
Given that nucleolin is a shuttling protein present in the membrane, cytoplasm, nucleus, and most prominently, the nucleolus of a cell, it was important to determine the subcellular localization of the nucleolin during FCV infection. This aspect was analyzed by indirect immunofluorescence using confocal microscopy. As expected, in uninfected cells, staining of nucleolin with Alexa Fluor 488 was observed to be concentrated in nucleolus foci, with less intense staining elsewhere in the nucleoplasm and cytoplasm (Fig. 4 A, green). At 1 hpi, an evident movement of nucleolin to the nucleoplasm is observed, although a weak staining is still present in nucleolus foci and cytoplasm. At 3 and 5 hpi, nucleolin was found almost exclusively in the nucleoplasm. Finally, at 7 hpi, nucleolin distribution was largely heterogeneous through the nucleoplasm in comparison to the distribution in mock-infected controls (Fig. 4A). In agreement with the levels of nucleolin observed by Western blot analysis (Fig. 3B), the overall fluorescence intensities of nucleolin in the nuclei of infected cells showed no significant differences from those observed in uninfected cells. However, a reduction in the fluorescence intensities was observed at 7 hpi (Fig. 4B), which correlates with the evident cytopathic effect reported at these times of infection. No relocalization of nucleolin was observed in cells infected with UV-inactivated virus (data not shown), confirming that the observed redistribution of nucleolin occurs as a consequence of active viral replication.
Nucleolin associates with recombinant NV NS6/7 and FCV NS6/7 proteins during FCV replication.
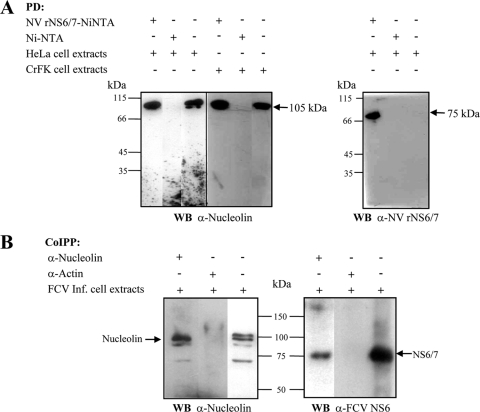
Since the 3′ UTR is an important element for RNP complex formation and for the initiation of the negative-strand RNA synthesis, we hypothesized that nucleolin could also be part of the replication complex (RC) formed with RNA-dependent RNA polymerase. To test this possibility, we first analyzed the ability of recombinant NV NS6/7 to interact with nucleolin. For this purpose, a nickel agarose pulldown assay using a polyhistidine-tagged NV NS6/7 fusion protein was performed. Purified recombinant NV NS6/7 was bound to Ni-NTA resin (Ni-NTA-NV NS6/7) and incubated with HeLa or CrFK cell extracts, and the interacting proteins analyzed by Western blotting. Ni-NTA-His-NV NS6/7 but not Ni-NTA resin alone was able to precipitate nucleolin present in cell extracts (Fig. 5 A, left). The extracts used in this assay were largely devoid of nucleic acid as the result of micrococcal nuclease digestion; therefore, we can conclude that the observed NS6/7-nucleolin interaction is likely to be protein mediated and not to occur as a result of binding an RNA intermediate.
Fig. 5.
Recombinant NV NS6/7 protein and FCV NS6/7 protein coprecipitate with nucleolin. (A) Recombinant Norwalk virus protein (NV rNS6/7) coupled to Ni-NTA resin or Ni-NTA resin alone was incubated with HeLa or CrFK cell extracts as indicated. After several washes, the precipitated proteins were analyzed by SDS-PAGE, transferred to a nitrocellulose membrane, and analyzed by Western blotting using human antinucleolin (right) or anti-NV rNS6/7 (left) antibody. Molecular masses of nucleolin and NV NS6/7 are indicated to the right of the panels. PD, pulldown. (B) Antinucleolin or antiactin antibody was incubated with total extracts from FCV-infected cells, and immunocomplexes were immobilized in protein G-agarose resin, precipitated, and analyzed by SDS-PAGE. The transferred proteins were analyzed by Western blotting using human antinucleolin (left) or anti-FCV NS6 (right) antibody. Co-IPP, coimmunoprecipitation. Arrows indicate migration of nucleolin and NV rNS6/7. Input cell extracts are shown in the third lane of each panel.
Given that the recombinant NV NS6/7 protein was associated with nucleolin, the ability of the FCV NS6/7 protein to interact with nucleolin during virus infection was analyzed. The NS6/7 protein coimmunoprecipitated with a 105-kDa band, which corresponds to full-length nucleolin in CrFK cells infected with FCV (Fig. 5B, right). Additional bands of 98 and 70 kDa were also detected. The molecular masses of these products might correspond to specific degradation peptides generated by nucleolin autocatalysis, as previously described, most likely as a result of the particular conditions of the assay (11, 13). No detection of FCV NS6/7 was observed when the immunoprecipitation assay was performed with antiactin antibody, suggesting that the immunoprecipitation of NS6/7 by the antinucleolin antibody was specific (Fig. 5B, right). As observed with NV, the interaction between FCV NS6/7 and nucleolin is likely to be a protein-protein interaction and not an RNA-mediated interaction, as all extracts used in these assays were treated with micrococcal nuclease. Taken together, these results indicate that nucleolin and the FCV NS6/7 protein interact during the course of FCV infection.
Nucleolin colocalizes with FCV NS6/7 in the perinuclear region.
Since nucleolin relocalizes to the nucleoplasm after FCV infection and because we found that nucleolin coprecipitates with FCV NS6/7 in infected cells, the subcellular localization of both nucleolin and FCV NS6/7 was analyzed by confocal microscopy. The FCV NS6/7 protein was detected by indirect immune staining with Alexa Fluor 594 (Fig. 6 A, red). From 3 hpi, the FCV NS6/7 was observed mostly throughout the cytoplasm of the infected cells; however, at 7 hpi, the majority of infected cells display a spherical shape. Nucleolin was labeled by indirect immune staining with Alexa Fluor 488 (Fig. 6A, green) and observed at 3, 5, and 7 hpi throughout the nucleus. Analysis of the merged images in optical sections indicated that both NS7 and nucleolin were present in the perinuclear area (Fig. 6A). The colocalization of the two proteins is highlighted in white.
To further determine if nucleolin was present within FCV RCs, as would be predicted based on our RNA-coimmunoprecipitation data, RCs from both mock-infected and FCV-infected cells were prepared as previously described (28). Western blot analysis was used to examine the presence of nucleolin and FCV NS6/7, as well as protein disulfide isomerase (PDI) as a marker for the ER. Nucleolin, FCV NS6/7, and PDI were detected in the RCs obtained from FCV-infected cells, while nucleolin was not observed in the corresponding fractions from mock-infected cells (Fig. 6B, lanes 4 and 3, respectively). In contrast, nucleolin was present at equal levels in total cellular lysates prepared from mock-infected and FCV-infected cells. These results indicate that nucleolin, FCV NS6/7, and PDI are present in RCs from FCV-infected cells.
Inhibition of the expression of nucleolin results in the reduction of viral protein synthesis and FCV replication.
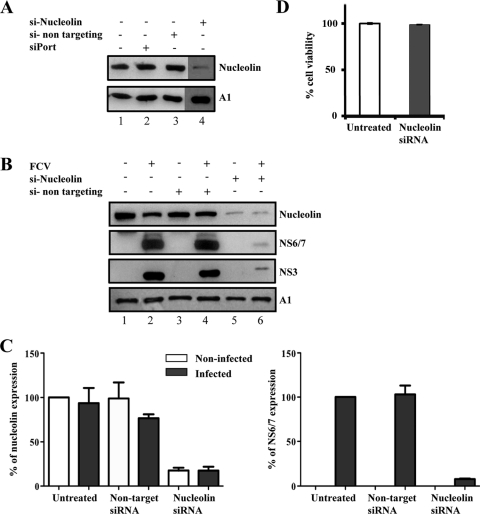
The results presented above show that in FCV-infected cells, nucleolin undergoes relocalization to the nucleoplasm as well as the perinuclear area, where it colocalizes with the FCV NS6/7 protein, suggesting that this association could have a role during viral replication. To begin to determine the role of nucleolin in the FCV life cycle, CrFK cells were transfected with two specific siRNAs directed toward nucleolin and the levels of nucleolin were detected by Western blot analysis. Transfection of nucleolin-specific siRNAs for 48 h resulted in an 82% decrease in nucleolin levels compared to the levels in cells transfected with the nontargeting siRNA, as estimated by densitometry (Fig. 7 A). Nucleolin gene silencing was observed for at least 72 h (data not shown).
Fig. 7.
Effect of nucleolin silencing on the expression of FCV NS6/7. (A) CrFK cells were either left untreated (lane 1), treated with siPort reagent alone (lane 2), or transfected with a nontargeting siRNA (lane 3) or with two siRNAs specific for feline nucleolin (lane 4). The levels of nucleolin expression were evaluated by Western blotting. (B) CrFK cells were either not treated (lanes 1 and 2) or transfected with a nontargeting siRNA (lanes 3 and 4) or with two siRNAs specific for feline nucleolin (lanes 5 and 6) for 48 h. Cells were then either mock infected or infected with FCV (MOI 10) for 5 h, and the expression levels of nucleolin, FCV NS6/7, NS3 or p39, and hnRNP A1 (A1), used as the loading control, were evaluated by Western blotting in FCV-infected (lanes 2, 4, and 6) and uninfected (lanes 1, 3, and 5) cells. (C) Nucleolin and NS6/7 band intensities were quantified by densitometric analysis using Quantity-One software (Bio-Rad) and are expressed as the percentage in relation to the hnRNP A1 expression level. Error bars represent the standard deviation from three independent experiments. (D) Forty-eight hours after siRNA transfection, cell viability of untreated and nucleolin siRNA-treated cells was measured as described in Materials and Methods. Error bars show standard deviations.
To analyze the effect of nucleolin knockdown on the production of viral proteins, transfected CrFK cells were infected with FCV, and after 5 hpi, the levels of NS6/7 were monitored by Western blotting. Nucleolin knockdown resulted in a 92% reduction in the NS6/7 levels compared to the levels in cells transfected with the nontargeting siRNA (Fig. 7B and C). The reduction in the expression of NS6/7 as a result of the siRNA-mediated knockdown of nucleolin levels further suggests that nucleolin is important for FCV replication. As expected, a similar level of reduction was observed for the NS3 protein, confirming that nucleolin knockdown resulted in a reduction of the expression of the proteins produced from the viral polyprotein. The observed decreased production of viral protein in nucleolin siRNA-transfected cells was not due to any gross effects on host protein synthesis, as the levels of hnRNP A1 were unaffected (Fig. 7A and B), as was cell viability (Fig. 7D).
Nucleolin is required for efficient FCV replication.
Since the reduction of nucleolin using siRNAs had a significant effect on the NS6/7 levels produced during FCV infection, the consequences of nucleolin knockdown on FCV replication were evaluated by monitoring the levels of infectious virus produced. Nucleolin knockdown was found to reduce the FCV yield by at least 1.5 log after 4 h and 6 h of infection (Fig. 8 A). Nucleolin siRNAs has no gross effect on FCV binding, as similar levels of infectious virus were found bound to cells at time zero. In addition, although virus entry was not monitored directly, the duration of the eclipse phase would indicate that it was also largely unaffected. In agreement with the observed effect on viral titer, cells transfected with the nucleolin siRNA displayed reduced and delayed cytopathic effect compared that observed in the nontarget siRNA-treated cells (Fig. 8B). As previously stated, the viability of cells was unaffected by siRNA treatment (Fig. 7B); therefore, the observed decreased replication of FCV in nucleolin siRNA-transfected cells was specific and not due to an indirect effect resulting in gross changes in cellular metabolism. These results taken together indicate that nucleolin is required for FCV replication.
Fig. 8.
FCV replication is inhibited by nucleolin siRNAs. (A) One-step growth curve analysis of FCV in CrFK cells treated with either nontargeting or nucleolin-specific siRNAs. CrFK cells treated with either nontargeting or nucleolin-specific siRNAs were infected with FCV (MOI 10), and the virus yield was determined at 0, 2, 4, and 6 hpi by plaque assay. Infections were performed a minimum of 5 times in duplicate, with one representative data set shown. Error bars represent standard deviations. *, P < 0.005, and **, P < 0.001, by one-way ANOVA. (B) Phase-contrast microscopy of FCV-infected cells transfected with nucleolin or nontargeting siRNAs. Note the delayed and reduced cytopathic effect in nucleolin-treated cells.
DISCUSSION
Positive-strand RNA viral genomes often contain RNA elements that can vary greatly in their primary sequence but which maintain relatively consistent structures within virus families. Studies with numerous RNA viruses have highlighted that these RNA elements are extensively implicated in viral replicative cycle regulation (3, 9, 16, 18, 20, 33, 49, 62). These structured elements have been predicted to be formed within the genomes of several members of the Caliciviridae family (31, 54), and their functional role in viral replication has been confirmed for the MNV (6, 54). RNA stem-loop structures are targets for various cytoplasmic and nuclear factors that may function to regulate aspects of viral replication. Several cellular proteins, such as La, PABP, PTB, and PCBP-2, interact with the 5′ and 3′ ends of NV genomic RNA (30, 31). In addition, we have recently highlighted the role of PTB as a negative regulator of FCV translation, possibly contributing to the switch from translation to replication (6, 41). In this study, we describe that nucleolin, a ubiquitous, multifunctional 105-kDa nucleolar shuttling phosphoprotein (25, 47), binds to the NV and FCV 3′ UTR in vitro. This interaction was also found to occur during FCV replication in cell culture. Nucleolin is a multifunctional cellular nucleolar protein that is implicated in cell proliferation and growth, including functions during rRNA processing from rRNA gene transcription to the assembly of preribosomal particles (25, 47, 56). In recent years, the role of nucleolin in viral infections has been widely established and supported by its colocalization with many viral components. In particular, nucleolin binds to several viral UTRs and participates in the replication of RNA viruses (34, 39, 60).
Nucleolin interacts with the 3′ UTR of wild-type PV but not the 3′ UTR of a replication-defective mutant, and this interaction plays a role in viral genome amplification during the early stages of the viral life cycle (60). The putative nucleolin binding site within the 3′ end of the PV genome has been identified as a CAUUUUAGU sequence, located in a loop of a predicted pseudoknot structure (60). This sequence is very similar to the nucleolin binding site present in the amyloid precursor protein mRNA, which consists of a CAUUUUGGU sequence (63). The NV 3′ UTR contains a CAUUUAAU sequence that may represent a partial consensus binding site; however, bioinformatic analysis indicates that it is not located in a loop structure (Fig. 2B). Although no related sequences are present in the FCV 3′ UTR, a previously identified nucleolin binding site in pre-rRNA (U/A)CCCG(A/G) (24, 26, 40) appears to be at least partially conserved in FCV (Fig. 2A). A very similar sequence (ACCCCA) is present within the stem region of the FCV 3′ UTR (Fig. 2A). Although exact copies of the consensus nucleolin binding motifs were not present within the NV and FCV 3′ UTRs, the protein can bind to both regions. It is also possible that nucleolin binding to the calicivirus 3′ UTR relies on the interaction with a novel, as-yet-unidentified binding site, as has been recently proposed for the tombusvirus 3′ UTR-nucleolin interaction (39). Since comparable stem-loop RNA structures are present in both the NV and FCV 3′ UTRs, it is likely that the nucleolin binding depends not only on the primary sequence but also on the secondary (or tertiary) structure of the RNA, as has been reported for other cellular factors (32).
The expression and subcellular localization of cellular proteins implicated in viral replication are often modified during the course of infection. In this work, the levels and integrity of nucleolin RNA and protein were not altered; however, FCV infection clearly resulted in the redistribution of nucleolin from the nucleoli to the nucleoplasm, as well as to the surrounding perinuclear area during the latter stages of virus replication. This type of nucleolin relocalization is also reported to occur as a result of various stress-related stimuli, such as exposure to ionizing radiation, heat shock, oxidative shock, and camptothecin, as well as a consequence of viral infection (47). Proteins from several viruses are known to colocalize with nuclear factors, such as nucleolin, B23, and fibrillarin, resulting in their subsequent relocalization to play a role in the viral replication process (35, 36). Human adenovirus and herpes simplex virus 1 (HSV-1) both interact with nuclear factors such as nucleolin (8, 12, 45, 48), and during HSV-1 infection, the viral protein UL24, required for efficient viral replication, induces the dispersion of nucleolin. In cells infected with human cytomegalovirus, an increased level of nucleolin and redistribution throughout the nucleus were observed (57). It has been suggested that nucleolin localization depends on its phosphorylation state (25); therefore, the possibility that the phosphorylation or dephosphorylation of nucleolin could be associated with its relocalization during FCV infection remains to be determined.
Our coprecipitation assay using recombinant NV NS6/7 protein showed that this protein and nucleolin can associate to form a complex, although whether this is a direct protein-protein interaction or mediated by an unidentified intermediary is not clear. The analogous interaction in FCV was further confirmed by immunoprecipitation assays with antisera to the FCV NS6/7 protein and extracts from FCV-infected cells. The association of nucleolin with elements that are essential for the replication of the viral negative-strand RNA, such as the genomic 3′ UTR and the NS6/7 protease-polymerase protein, suggests a role of this protein during calicivirus replication. Indeed, the function of nucleolin in the various steps of the replication cycle of other viruses is well documented: it has been reported that nucleolin stimulates IRES-mediated translation (38), and it plays a role during HSV-1 egress (45), as well as in the budding and assembly of retrovirus virions (5, 58). Nucleolin has also been implicated in PV replication because of its ability to interact with the 3′ UTR of the genomic RNA (60). Furthermore, nucleolin has been described as an indispensable factor for HCV replication due to its association with the replicase NS5B (34, 43, 52). In contrast, nucleolin binds to the 3′ UTR of the tombusvirus genomic RNA and plays an inhibitory role during the early stages of tombusvirus replication, possibly via interference with the recruitment of viral RNA at the replication site (39).
The results of confocal laser scanning microscopy indicated that colocalization of the FCV NS6/7 and nucleolin occurs in the perinuclear area. Similar colocalization has been reported during HCV replication where the NS5B replicase-nucleolin association causes a change in nucleolin distribution from nucleoli to the perinuclear area (34). It is within these perinuclear sites that HCV-specific plus-strand synthesis is reported to take place, in replication complexes derived from the membranes of the ER (27). In CrFK cells infected with FCV, NS6/7 localization was reported predominantly in membrane-associated replication complexes in the cytoplasm (28). Recent reports point to an ER-derived origin of membranous vesicles induced during FCV and MNV infection (7, 37). In particular, over the course of MNV infection, double-stranded RNA and NS7 were observed to proliferate from punctuate foci located in the perinuclear region (37). In agreement with this information, we found that nucleolin, FCV NS6/7, and PDI, a protein that has been reported as an ER marker, are present in partially purified membrane-bound replication complexes.
Given that in FCV-infected CrFK cells, the colocalization of the protease-polymerase NS6/7 with nucleolin was observed in the perinuclear area and that both proteins are present within RCs and associate with the viral RNA, a role for nucleolin in the calicivirus life cycle was expected. This was confirmed subsequently by the use of siRNAs specific for nucleolin, which displayed a significant inhibitory effect on FCV NS6/7 production and virus replication. Furthermore, the reduced and delayed cytopathic effect, as well as the reduction in virus yield observed in cells transfected with the nucleolin siRNA compared to the yield in the nontarget siRNA-treated cells, indicates that nucleolin, a nucleolar host cell protein, is required for efficient FCV replication. Although not measured directly, since the growth characteristics and the viability of the cells were largely unaffected when transfected with nucleolin siRNAs, we can conclude that there was no gross effect on cell cycle progression that could affect the replication of the virus in an indirect manner.
Because both protease and polymerase activities are present in the NS6/7 protein, it is possible that a protease activity could also take place in the perinuclear area. In regard to this possibility, a nuclear-cytoplasmic relocalization of host cell proteins after FCV infection has been reported recently (6). This finding may be the result of the alteration of nucleopore complex composition by the FCV protease activity, as reported during PV infection (29).
Here, we report that nucleolin, a nucleolar host cell protein, binds to the 3′ UTR and the NS6/7 protease-polymerase, relocates from the nucleoli to the nucleoplasm, and acts as a positive regulator of FCV replication. It is likely that nucleolin functions either directly or indirectly as an RNA chaperone promoting the formation of a complex suitable for translation and/or replication. Although the precise role of nucleolin in the calicivirus life cycle remains to be determined, this work identifies a cellular factor critical for the calicivirus life cycle and, thus, adds to the insights into the life cycle of this family of poorly understood pathogens.
ACKNOWLEDGMENTS
We thank Rosa M. del Angel and Juan Ludert for many helpful suggestions and for critical comments on the manuscript, Fernando Medina for the cell cultures, and Eduardo Carrillo for technical assistance.
This work was supported by grants 43788-Q from Consejo Nacional de Ciencia y Tecnología and ICYTDF/247 from Instituto de Ciencia y Tecnología del Distrito Federal, México, D. F., to A.L.G.-E. and by a grant from the Wellcome Trust to I.G. I.G. is a Wellcome Trust Senior Fellow.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Agis-Juárez R. A., et al. 2009. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J. Gen. Virol. 90:2893–2901 [DOI] [PubMed] [Google Scholar]
- 2. Ahlquist P., Noueiry A. O., Lee W. M., Kushner D. B., Dye B. T. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarez D. E., Filomatori C. V., Gamarnik A. V. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′ UTRs. Virology 375:223–235 [DOI] [PubMed] [Google Scholar]
- 4. Andino R., Boddeker N., Silvera D., Gamarnik A. V. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76–82 [DOI] [PubMed] [Google Scholar]
- 5. Bacharach E., Gonsky J., Allin K., Oriova M., Goff S. P. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domains of retroviral gag proteins and inhibits virion assembly. J. Virol. 74:11027–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey D., et al. 2010. Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J. Virol. 84:2859–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bailey D., et al. 2010. Feline calicivirus p32, p39 and p30 proteins localize to the endoplasmic reticulum to initiate replication complex formation. J. Gen. Virol. 91:739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertrand L., Leiva-Torres G. A., Hyjazie H., Pearson A. 2010. Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J. Virol. 84:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bode J. G., Brenndörfer E. D., Karthe J., Häussinger D. 2009. Interplay between host cell and hepatitis C virus in regulating viral replication. Biol. Chem. 290:1031–1032 [DOI] [PubMed] [Google Scholar]
- 10. Bradford M. M. 1976. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 11. Bugler B., Caisergues-Ferrer M., Bouche G., Bourbon H., Amalric F. 1982. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur. J. Biochem. 128:475–480 [DOI] [PubMed] [Google Scholar]
- 12. Callé A., et al. 2008. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82:4762–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen C. M., Chiang S. Y., Yeh N. H. 1991. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J. Biol. Chem. 266:7754–7758 [PubMed] [Google Scholar]
- 14. De Nova-Ocampo M., Villegas-Sepúlveda N., del ÁAngel R. M. 2002. Translation elongation-1a, La, and PTB interact with the untranslated region of dengue 4 virus RNA. Virology 295:337–347 [DOI] [PubMed] [Google Scholar]
- 15. Domitrovitch A. M., Diebel K. W., Ali N., Sarker S., Siddiqui A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335:72–82 [DOI] [PubMed] [Google Scholar]
- 16. Dreher T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151–174 [DOI] [PubMed] [Google Scholar]
- 17. Elbashir S. M., Martinez J., Patkaniowska A., Lendecke W., Tusch T. 2001. Functional anatomy of siRNA for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Enjuanes L., Almazan F., Solá I., Zuñiga S. 2006. Biochemical aspects of coronavirus replication and virus-host interactions. Annu. Rev. Microbiol. 60:211–230 [DOI] [PubMed] [Google Scholar]
- 19. Fankhauser R. L., et al. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1–7 [DOI] [PubMed] [Google Scholar]
- 20. Filomatori C. V., et al. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20:2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22. Franze de Fernandez M. T., Eoyang L., August J. T. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588–590 [DOI] [PubMed] [Google Scholar]
- 23. Gamarnik A., Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghisolfi-Nieto L., Joseph G., Puvion-Dutilleu F., Amalic F., Bouvet P. 1996. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-rRNA. J. Mol. Biol. 206:34–53 [DOI] [PubMed] [Google Scholar]
- 25. Ginisty H., Sicard H., Roger B., Bouvet P. 1999. Structure and function of nucleolin. J. Cell Sci. 112:761–772 [DOI] [PubMed] [Google Scholar]
- 26. Ginisty H., et al. 2000. Interaction of nucleolin with an evolutionarily conserved pre-rRNA sequence is required for the assembly of the primary processing complex. J. Biol. Chem. 275:18845–18850 [DOI] [PubMed] [Google Scholar]
- 27. Gosert R., et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green K. Y., et al. 2002. Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J. Virol. 76:8582–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gustin K. E., Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gutiérrez-Escolano A. L., Uribe-Brito Z., del ÁAngel R. M., Jiang X. 2000. Interaction of cellular proteins with the 5′ end of Norwalk virus genomic RNA. J. Virol. 74:8558–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gutiérrez-Escolano A. L., Vázquez-Ochoa M., Escobar-Herrera J., Hernández-Acosta J. 2003. La, PTB, and PAB proteins bind to the 3′ untranslated region of Norwalk virus genomic RNA. Biochem. Biophys. Res. Commun. 311:759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hellen C. U., et al. 1993. A cytoplasmic 57 kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:7642–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herold J., Andino R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirano M., et al. 2003. Direct interaction between nucleolin and hepatitis C virus NS5B*. J. Biol. Chem. 278:5109–5115 [DOI] [PubMed] [Google Scholar]
- 35. Hiscox J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hiscox J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nature Rev. 5:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hyde J. L., et al. 2009. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 83:8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Izumi R. E., Valdez B., Banerjee R., Srivastava M., Dasgupta A. 2001. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 76:17–29 [DOI] [PubMed] [Google Scholar]
- 39. Jiang Y., Zhenghe L., Nagy P. D. 2010. Nucleolin/Nsr1 binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology 396:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johansson C., et al. 2004. Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target. J. Mol. Biol. 337:799–816 [DOI] [PubMed] [Google Scholar]
- 41. Karakasiliotis I., Chaudhry Y., Roberts L. O., Goodfellow I. G. 2006. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 87:3339–3347 [DOI] [PubMed] [Google Scholar]
- 42. Karakasiliotis I., et al. 2010. Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PLoS One 5:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kusakawa T., Shimakami T., Kaneko S., Yoshioka K., Murakami S. 2007. Functional interaction of hepatitis C virus NS5B with nucleolin GAR domain. J. Biochem. 141:917–927 [DOI] [PubMed] [Google Scholar]
- 44. Li W., et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lymberopoulos M. H., Pearson A. 2007. Involvement of UL24 in herpes-simplex-1-induced dispersal of nucleolin. Virology 363:397–409 [DOI] [PubMed] [Google Scholar]
- 46. Mas A., Alves-Rodrigues I., Noueiry A., Ahlquist P., Díez J. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Masiuk M. 2008. Nucleolin—characteristics of protein and its role in biology of cancers and viral infections. Adv. Cell Biol. 35:1–19 [Google Scholar]
- 48. Matthews D. A. 2001. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 75:1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Minkyung Y., Lemon S. M. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oka T., et al. 2007. Highly conserved configuration of catalytic amino acid residues among calicivirus-encoded proteases. J. Virol. 81:6798–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scheller N., et al. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimakami T., et al. 2006. Effect of hepatitis C virus (HCV) NS5B-nucleolin interaction on HCV replication with HCV subgenomic replicon. J. Virol. 80:3332–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 54. Simmonds P., et al. 2008. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 36:2530–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sosnovtsev S. V., Prikhodko E. A., Belliot G., Cohen J. I., Green K. Y. 2003. Feline calicivirus replication induces apoptosis in cultured cells. Virus Res. 94:1–10 [DOI] [PubMed] [Google Scholar]
- 56. Srivastava M., Pollard H. B. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13:1911–1922 [PubMed] [Google Scholar]
- 57. Strang B. L., Boulant S., Cohen D. M. 2010. Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. J. Virol. 84:1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ueno T., et al. 2004. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type 1 (HIV-1). Microbiol. Immunol. 48:111–118 [DOI] [PubMed] [Google Scholar]
- 59. Vashist S., Bailey D., Putics A., Goodfellow I. 2009. Model systems for the study of human norovirus biology. Future Virol. 4:353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Waggoner S., Sarnow P. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walter B. L., Parsley T. B., Ehrenfeld E., Semler B. L. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yi M., Lemon S. M. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zaidi S. H., Malter J. S. 1994. Amyloid precursor protein mRNA stability is controlled by a 29-base element in the 3′-untranslated region. J. Biol. Chem. 269:24007–24013 [PubMed] [Google Scholar]
- 64. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]