Abstract
Respiratory syncytial virus (RSV) G protein deletion mutants replicate effectively in vitro but have not been detected in nature. Subtyping of RSV strains in hospitalized children in South Africa identified G protein PCR amplicons significantly reduced in size in 2 out of 209 clinical specimens screened over 4 years. Sequence analysis revealed subtype B strains lacking nearly the entire G protein ectodomain in one HIV-positive and one HIV-exposed child hospitalized with pneumonia. The association of clinical strains lacking most of the G protein with lower respiratory tract infection in immunocompromised children may have implications for RSV vaccine development.
TEXT
Respiratory syncytial virus (RSV), a Pneumovirus, family Paramyxoviridae, is a major cause of pneumonia in children. Two antigenic subgroups (A and B) exist with partial cross-protection (1). Neutralizing antibodies are directed against the F and G surface glycoproteins. Major antigenic differences are features of the G protein, a type II transmembrane glycoprotein that mediates viral attachment. The G protein has an ectodomain consisting of a central region of four conserved cysteines and a putative receptor binding site, flanked by two hypervariable regions (5). Several G protein genotypes have been documented within each subgroup based predominantly on the second hypervariable region (13, 21, 24).
Evolutionary studies of subgroup B strains have described major differences in G protein length due to alternative termination codon usage, premature stop codons, and in-frame duplications, deletions, and insertions (10, 22). G proteins of especially subtype B isolates have been identified that are truncated by 30 bp due to frameshift mutations (11). The central region remained conserved in all sequences, with an absolute conserved region between amino acid positions 164 and 187 (26).
Cell surface glycosaminoglycans (GAGs) are responsible for the majority of RSV attachment to cultured cells leading to infection (3). RSV virions containing the F protein as the sole surface protein bind to GAGs as well as another unidentified molecule, suggesting that the F protein may have an auxiliary role as an attachment protein (16).
Variation in neutralizing epitopes in the hypervariable region of the G protein suggests that immune selection of new variants may contribute to generation of human RSV diversity (20). A cold-passaged subtype B mutant containing large deletions spanning most of the coding sequences of the small hydrophobic (SH) and attachment (G) proteins was generated in vitro. This virus replicated efficiently in Vero cells but was found to be overattenuated in RSV-seronegative infants and children (6). Deletion of the central conserved domain and cysteine noose was shown to not affect virus growth in vitro or in vivo (in mice) (17), although no record exists of strains that lack this in humans with clinical disease. Augmentation of the Th2 immune response by the G protein and antigenic variation in G make delta G mutants attractive vaccine candidates (2, 18).
As part of a molecular epidemiology investigation of a nosocomial outbreak of RSV in a ward for premature infants in Kalafong Hospital in Pretoria, South Africa, in 2006, RSV-positive nasopharyngeal aspirates (NPA) from patients in the general pediatric ward were investigated. The HIV-1 seroprevalence among mothers of these infants was 52.6%, suggesting a high level of perinatal exposure (25). Here, we describe identification of G protein deletion mutants that were significantly reduced in size in comparison to that of prototype controls in children with pneumonia. To investigate this phenomenon, these strains were amplified and sequenced with primers that stretch from the start of the G protein across to the F protein. In total, 209 children were screened that were <1 year of age and diagnosed with RSV between February 2006 and May 2009 with the immunofluorescence respiratory panel 1 immunofluorescence assay (IFA) that detects RSV, parainfluenza viruses 1 to 3, adenovirus, and influenza A and B viruses (Chemicon, Hampshire, United Kingdom) or with the Directigen RSV rapid test (Becton Dickinson Microbiology Systems, Franklin Lakes, NJ) at the Department of Medical Virology, University of Pretoria, Tshwane Academic Division, National Health Laboratory Service (NHLS). NHLS serves three secondary and tertiary hospitals in the Pretoria region: Kalafong Secondary Hospital, Steve Biko Academic Hospital, and the 1 Military Hospital.
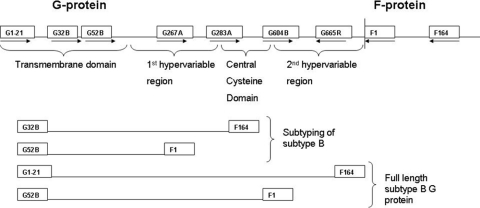
RNA was extracted directly from RSV-positive NPAs with the QIAamp viral RNA minikit (Qiagen, Valencia, CA) and subtyped with multiplex nested reverse transcription (RT)-PCR that distinguishes subtypes A and B by size (9, 23, 24). Positive specimens resulted in PCR fragments of 950 bp for RSV subtype A and 1,200 bp for subtype B strains in the first round and 700 and 950 bp, respectively, for the nested PCR corresponding with the RSV prototype strains A2 and B1. Figure 1 indicates a diagram of the different primers used.
Fig. 1.
Diagram of primers used. G32B, G267A, and F164, first-round multiplex RT-PCR; G52B, G283A, and F1, nested multiplex PCR for subtyping; G1-21 and F164, first-round RT-PCR; G32B and F1, nested PCR for full-length G protein amplification; G32B, G604B, F1, and G665R, sequencing primers.
Two clinical specimens were identified with amplicons that were significantly reduced in size in comparison to that of the prototype controls. Sequence analysis identified G protein deletion mutant subtype B strains that lacked most of the ectodomain, including the conserved cysteine noose. These deletion mutant strains as well as one representative strain of each subtype B genotype were selected for full-length G protein sequencing. Furthermore, any specimens that had G protein subtyping amplicons smaller than the expected size were selected for sequencing, as was a truncated strain previously identified in South Africa, AgJ15_99 (23). The full-length G protein was amplified with primers G1-21 (22) and F164 (Fig. 1) with the Titan one-step RT-PCR system (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's recommendations with the following cycle: 50°C for 30 min; 94°C for 2 min; 35 cycles of 94°C for 10 s, 53°C for 30 s, and 68°C for 1 min; and 68°C for 7 min. This was followed by nested PCR with primers G32B (15) and F1 (14) as described before (23). PCR products were analyzed on a 1.5% agarose gel, against a 100-bp molecular weight marker (Roche Diagnostics, Mannheim Germany).
Nucleotide sequencing was carried out using the BigDye version 3.0 kit on an ABI 3130 sequencer (Applied Biosystems, Foster City, CA) using primers G32B, G604B (5′AAACCAACCATCAAACCCACA3′), F1, and G665R (5′TTTTGGGGCTCTTTTGTTTG3′) (24). Sequence alignments were performed with Clustal X version 1.81 (19) and analyzed with BioEdit Sequence Alignment Editor version 7.0.4.1 (4).
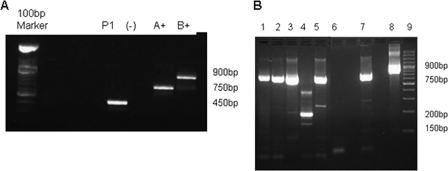
The first specimen with a suspicious G protein amplicon (SA367859Pt06) was from an 8-month-old HIV-positive child hospitalized with pneumonia that tested positive for RSV by the RSV rapid antigen kit. This child had symptoms of breathing distress and tachypnea, was very underweight (close to marasmic), and was on continued oxygen treatment. His CD4 count was 1,038 (27.5%). The percentage of CD4 cells is the best indicator of immunodeficiency in children, although guidelines exist only for infants older than 12 months. The immune systems of infants <1 year of age are still immature, and HIV-infected infants are at serious risk of morbidity and mortality (12). The subtyping multiplex nested PCR resulted in a fragment of approximately 450 bp (Fig. 2 A). Sequencing and blast search analysis identified close homology to the RSV G protein. PCR amplification and sequencing from the start of the G protein through to the F protein and alignment to other RSV strains identified a subtype B strain for which amino acids 101 to 297 (Fig. 3) were deleted, corresponding to the BA1428/99B (AY333364) reference strain used (21).
Fig. 2.
G protein-specific subtyping multiplex nested RT-PCR. (A) Amplicon from patient 1 (P1) (SA367859Pt06) next to a negative control (–) and the prototype subtype A- and B-positive-control strains. (B) Multiplex nested PCR products: lanes 1 to 5, clinical specimens; lane 6, negative control; lane 7, positive-control RSV A; lane 8, positive-control RSV B; lane 4, deletion mutant 2 (SA374801Pt06).
Fig. 3.
Complete amino acid alignment of the three deletion mutants in comparison to the full G protein subtype B sequences of each genotype. The first three sequences are for deletion mutants followed by the full G protein of each genotype. The genotype name is indicated in brackets after the strain name. Accession numbers: HQ711840 to HQ711842 (deletion mutant strains SA367859Pt06, SA374801Pt06a, and SA374801Pt06b), AF065250 (GB1), AF065251 (GB2), JF704217 (GB3), JF704214 (GB4), JF704213 (SAB1), JF704218 (SAB2), JF704215 (truncated SAB3), JF704216 (SAB3), AY333364 (BA), and JF704219 (South African BA).
One further specimen (SA374801Pt06) was identified that had 3 amplicons of significantly reduced size (Fig. 2B, lane 4). This specimen was from an HIV-exposed male, 2 months and 13 days old, hospitalized for 7 days for gastroenteritis and severe dehydration. He developed pneumonia in the ward and tested positive for RSV 5 days after admission. The child and mother live in an informal settlement with limited resources, and the infant was extremely malnourished upon admission. HIV PCR was performed due to the HIV-seropositive status of the mother as part of the antiretroviral treatment program for prevention of mother-to-child transmission. Follow-up testing for HIV would be carried out up to 18 months as per protocol for HIV-exposed children. Sequence analysis and BLAST searches confirmed the 200- and 150-bp bands to have homology to RSV subgroup B, while the larger band was nonspecific.
In both RSV deletion mutant species identified in this patient, nearly the entire ectodomain of the G protein gene was deleted. The smaller of the two RSV-specific amplicons had a deletion from amino acid 31 to 299, while amino acids 87 to 280 were deleted in the larger of the two amplicons (Fig. 3). The amplicon from patient 1 and both amplicons from patient 2 lacked the conserved cysteine region. To our knowledge, this is the first time that strains with such extreme deletions in the G glycoprotein have been identified in children with lower respiratory tract infections, which shows the flexibility of the G protein to tolerate significant antigenic changes in nature. No amplicons were visible at the expected position for full-length G proteins, and no other coinfections were identified. SA374801Pt06 was negative for human metapneumovirus, bocavirus, parainfluenza viruses 1, 2, and 3, influenza viruses A and B, adenovirus, and coronaviruses 229E, OC43, HKU1, and NL63 by multiplex real-time PCR (7). SA367859Pt06 was negative for other viruses detected by the IFA, but insufficient material was available for additional PCRs. Attempts to culture virus from these specimens was not successful, probably due to low concentrations present and repeat freeze-thawing of specimens that had been screened retrospectively.
These findings suggest that despite being attenuated in immunocompetent children, the G protein might not be necessary for infection and replication in immunosuppressed individuals. These viruses may not be pathogenic or able to replicate in immunocompetent individuals and therefore were not detected previously in patients. The F protein might be sufficient to play the role of the auxiliary attachment protein in such cases. It is unclear if the deletion mutants evolved in these children or if they were transmitted. Lazar et al. (8) identified a premature stop codon in the G glycoprotein gene when sequencing strains at three different time points from a patient with severe combined immune deficiency syndrome. The RSV G gene encoded a truncated G glycoprotein lacking 42 carboxy-terminal amino acids but still had the conserved central cysteine noose. The investigators hypothesized that these mutations developed during prolonged infection due to severe immune deficiency in these patients and immunologic pressure as a result of the monthly treatment with intravenous immunoglobulin.
To conclude, data presented here suggest that RSV clinical strains lacking most of the G protein gene, including the central conserved cysteine noose, may occur in immunocompromised patients with lower respiratory tract infections. Reduced immune pressure in these patients may allow these strains to utilize other proteins for binding and replication. This may have implications for the utilization of certain attenuated strains as vaccine candidates in immunocompromised children.
Nucleotide sequence accession numbers.
HQ711840 to HQ711842 (deletion mutants) and JF704213 to JF704219 (full G proteins) were deposited in GenBank.
Acknowledgments
This study was reviewed, approved, and monitored by the human ethics committee, University of Pretoria (25/2006), and funded by the Poliomyelitis Research Foundation.
Work was done at the Department of Medical Virology, University of Pretoria, Pretoria, South Africa.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Anderson L. J., et al. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626–633 [DOI] [PubMed] [Google Scholar]
- 2. Collins P. L., Murphy B. R. 2002. Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296:204–211 [DOI] [PubMed] [Google Scholar]
- 3. Feldman S. A., Audet S., Beeler J. A. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 5. Johnson P. R., Spriggs M. K., Olmsted R. A., Collins P. L. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karron R. A., et al. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. U. S. A. 94:13961–13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lassauniere R., Kresfelder T., Venter M. 2010. A novel multiplex real-time RT-PCR assay with FRET hybridization probes for the detection and quantitation of 13 respiratory viruses. J. Virol. Methods 165:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lazar I., Canaan A., Weibel C., Kahn J. S. 2006. Novel mutations in the respiratory syncytial virus G gene identified in viral isolates from a girl with severe combined immune deficiency treated with intravenous immune globulin. J. Clin. Virol. 37:168–173 [DOI] [PubMed] [Google Scholar]
- 9. Madhi S. A., Venter M., Madhi A., Petersen M. K., Klugman K. P. 2001. Differing manifestations of respiratory syncytial virus-associated severe lower respiratory tract infections in human immunodeficiency virus type 1-infected and uninfected children. Pediatr. Infect. Dis. J. 20:164–170 [DOI] [PubMed] [Google Scholar]
- 10. Martinez I., et al. 1999. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J. Gen. Virol. 80(Pt. 1):125–130 [DOI] [PubMed] [Google Scholar]
- 11. Melero J. A., Garcia-Barreno B., Martinez I., Pringle C. R., Cane P. A. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78(Pt. 10):2411–2418 [DOI] [PubMed] [Google Scholar]
- 12. National Department of Health South Africa 2010. Guidelines for the management of HIV in children. National Department of Health South Africa, Pretoria, South Africa: www.hiv911.org.za/wp-content/uploads/2010/04/2010-Paediatric-Guidelines.pdf [Google Scholar]
- 13. Peret T. C., et al. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891–1896 [DOI] [PubMed] [Google Scholar]
- 14. Peret T. C., Hall C. B., Schnabel K. C., Golub J. A., Anderson L. J. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221–2229 [DOI] [PubMed] [Google Scholar]
- 15. Sullender W. M., Sun L., Anderson L. J. 1993. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J. Clin. Microbiol. 31:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Techaarpornkul S., Collins P. L., Peeples M. E. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296–304 [DOI] [PubMed] [Google Scholar]
- 17. Teng M. N., Collins P. L. 2002. The central conserved cystine noose of the attachment G protein of human respiratory syncytial virus is not required for efficient viral infection in vitro or in vivo. J. Virol. 76:6164–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teng M. N., Whitehead S. S., Collins P. L. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283–296 [DOI] [PubMed] [Google Scholar]
- 19. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trento A., et al. 2010. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J. Virol. 84:7500–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trento A., et al. 2003. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 84:3115–3120 [DOI] [PubMed] [Google Scholar]
- 22. Trento A., et al. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J. Virol. 80:975–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Venter M., Collinson M., Schoub B. D. 2002. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: comparison of viruses and genotypes responsible for different disease manifestations. J. Med. Virol. 68:452–461 [DOI] [PubMed] [Google Scholar]
- 24. Venter M., Madhi S. A., Tiemessen C. T., Schoub B. D. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 82:2117–2124 [DOI] [PubMed] [Google Scholar]
- 25. Visser A., Delport S., Venter M. 2008. Molecular epidemiological analysis of a nosocomial outbreak of respiratory syncytial virus associated pneumonia in a kangaroo mother care unit in South Africa. J. Med. Virol. 80:724–732 [DOI] [PubMed] [Google Scholar]
- 26. Zlateva K., Lemey P., Moes E., Vandamme A., Ranst M. V. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157–9167 [DOI] [PMC free article] [PubMed] [Google Scholar]