Abstract
The head of bacteriophage T4 is decorated with 155 copies of the highly antigenic outer capsid protein (Hoc). One Hoc molecule binds near the center of each hexameric capsomer. Hoc is dispensable for capsid assembly and has been used to display pathogenic antigens on the surface of T4. Here we report the crystal structure of a protein containing the first three of four domains of Hoc from bacteriophage RB49, a close relative of T4. The structure shows an approximately linear arrangement of the protein domains. Each of these domains has an immunoglobulin-like fold, frequently found in cell attachment molecules. In addition, we report biochemical data suggesting that Hoc can bind to Escherichia coli, supporting the hypothesis that Hoc could attach the phage capsids to bacterial surfaces and perhaps also to other organisms. The capacity for such reversible adhesion probably provides survival advantages to the bacteriophage.
INTRODUCTION
Bacteriophage T4 is a large, double-stranded DNA virus that belongs to the family Myoviridae of the order Caudovirales and infects Escherichia coli. The T4 virion consists of a prolate head and a contractile tail, terminating in a baseplate to which are attached six long tail fibers (LTFs). The T4 virus initiates infection of a host cell by reversible binding of the LTFs to lipopolysaccharide molecules on the bacterial surface or to the outer membrane protein OmpC (50). Upon binding of the LTFs, a recognition signal is transmitted to the phage baseplate (16), causing the short tail fibers around the periphery of the baseplate to unravel and bind irreversibly to the lipopolysaccharides on the cell surface, thus securely anchoring the phage tail to the cell (36). The binding of the short tail fibers is followed by contraction of the tail sheath (3, 24), penetration of the bacterial membrane by the tail tube, and ejection of the viral DNA into the bacterium (27, 28, 37).
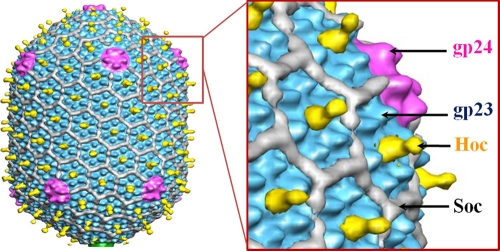
The prolate T4 head, encapsidating a ∼171-kbp DNA genome, has a length of ∼1,200 Å and a width of ∼860 Å. The head has icosahedral ends and an elongated midsection (Fig. 1). The major capsid protein, the product of gene 23 (gp23), is organized into a hexagonal lattice characterized by triangulation numbers of 13 (laevo) for the icosahedral ends and 20 for the midsection (13). One vertex of the T4 head is the portal for DNA packaging during virus assembly (43), tail attachment, and DNA ejection through the tail during infection. The portal vertex is occupied by a dodecamer of gp20 (9), whereas the other 11 head vertices are occupied by pentamers of the special vertex protein gp24 (13, 19). gp24 and gp23 have a fold similar to that of the capsid protein of bacteriophage HK97 (48) with an additional “insertion” domain that forms characteristic protrusions on the T4 capsid surface (Fig. 1) (14).
Fig. 1.
Cryo-EM structure of the phage T4 head (13). The major capsid protein gp23, which forms a hexagonal lattice, is shown in cyan, the vertex protein gp24 in magenta, Hoc in the center of each gp23 hexameric capsomer in yellow, and Soc in white. The figure is modified from reference 13.
Two accessory proteins, Soc (small outer capsid protein; 9 kDa) and Hoc (highly immunogenic outer capsid protein, 40 kDa), decorate the outer surface of the T4 capsid (18). Both Hoc and Soc are dispensable for capsid assembly and bind to the assembled capsid in the last stage of its maturation (18). These proteins have been used as a platform for display of pathogenic antigens on the capsid surface (12, 20, 29, 35, 40). Soc helps to stabilize the T4 capsid against extremes of pH and temperature (18, 34, 42). There are 870 Soc molecules attached to the T4 capsid surface at the interfaces between adjacent gp23 hexameric capsomers, serving as molecular glue (34). The capsid is further reinforced by trimerization of adjacent Soc molecules on the head surface. Analogous glue proteins have also been found in other phages and viruses (34).
Hoc-like proteins, on the other hand, have been found only in T4-like phages. Unlike Soc, Hoc has only a marginal effect on capsid stability, and its function had not been established. There are 155 Hoc molecules per T4 capsid, and they bind near the centers of gp23 hexamers. In the cryoelectron microscopy (cryo-EM) reconstruction of the T4 head (13), the density corresponding to Hoc molecules has the shape of a dumbbell (Fig. 1). The Hoc molecules do not bind to the pentameric gp24 vertices of the wild-type T4 capsid, nor do they attach to the pentameric gp23 “gp24-bypass” mutant vertices (11). The ability of Hoc to bind to gp23 hexamers but not to gp23 pentamers suggests that more than one gp23 molecule contributes to the Hoc binding site. Alternatively, Hoc may attach to one gp23 molecule, but the binding site might be inaccessible in the gp23 pentamers.
Analysis of the sequence suggested that the Hoc protein contains four domains, of which domains 1, 2, and 3 have an immunoglobulin-like fold (5, 39). The Hoc proteins of other T4-like bacteriophages have 1 to 5 domains (Table 1). The sequence of the C-terminal domain is better conserved than those of the other domains among the T4-like phages; thus, the C-terminal domain is likely to be involved in capsid binding. Mutagenesis data suggested that the C-terminal sequence, Glu-Ser-Arg-Asn-Gly, which is strictly conserved in all known T4-like phages, is likely required for the binding of Hoc to the capsid. Based on biochemical and cryo-EM data, it was suggested that the T4 Hoc molecule might have two functional modules: a capsid binding module containing domains 1 and 4 and a solvent-exposed module containing domains 2 and 3 (39). A model for the Hoc molecule that has a horseshoe-like shape, with domain 1 interacting with domain 4, has been proposed (39).
Table 1.
Hoc proteins of T4-like bacteriophages
| Bacteriophage name | No. of residues in the Hoc protein | Sequence identity (%) with T4 Hoc | Probable no. of domains |
|---|---|---|---|
| T4 | 376 | 100.0 | 4 |
| RB51 | 376 | 90.0 | 4 |
| RB32 | 376 | 88.0 | 4 |
| RB14 | 376 | 86.0 | 4 |
| RB30 | 472 | 77.0 | 5 |
| AR1 | 474 | 76.0 | 5 |
| Acj61 | 106 | 68.0 | 1 |
| SP18 | 282 | 58.0 | 3 |
| JS98 | 282 | 57.0 | 3 |
| RB69 | 471 | 52.0 | 5 |
| IME08 | 377 | 53.0 | 4 |
| JS10 | 469 | 50.0 | 5 |
| CC31 | 232 | 45.0 | 2 |
| vB_EcoM_VR7 | 367 | 44.0 | 4 |
| IME08 | 288 | 43.0 | 3 |
| RB16 | 167 | 39.0 | 2 |
| KP15 | 91 | 36.0 | 1 |
| Acj9 | 178 | 34.0 | 2 |
| KP15 | 177 | 32.0 | 2 |
| 65 | 273 | 29.0 | 3 |
| 31 | 180 | 29.0 | 2 |
| 44RR2.8t | 180 | 29.0 | 2 |
| JS10 | 286 | 28.0 | 3 |
| φAS4 | 178 | 25.0 | 2 |
| 25 | 177 | 24.0 | 2 |
| JSE | 404 | 22.0 | 4 |
| RB49 | 404 | 22.0 | 4 |
| Phi1 | 404 | 22.0 | 4 |
Bacteriophage RB49 is a T4-like virus that uses E. coli as a host. The Hoc protein of bacteriophage RB49, like the Hoc protein of T4, consists of four domains. The RB49 and T4 Hoc proteins have only 22% sequence identity (Fig. 2), whereas the major capsid proteins of these phages have 67% sequence identity. RB49 Hoc binds to the T4 phage and saturates its capsid surface, although RB49 Hoc has a slightly lower binding affinity to the T4 capsid (39). Here we report a crystal structure of Hoc domains 1, 2, and 3 from bacteriophage RB49. In addition, we present biochemical data suggesting that Hoc can bind to the cell surface of E. coli, and we discuss possible functions of the Hoc protein.
Fig. 2.
Alignment of bacteriophage T4 and RB49 Hoc sequences. β-Strands and α-helices are marked with the letters s and h, respectively. Regions corresponding to domains 1, 2, 3, and 4 of RB49 Hoc are shown in cyan, yellow, gray, and green, respectively. Since the structure of domain 4 of Hoc was not available, the secondary structure for this domain was predicted using the JPRED3 server (http://www.compbio.dundee.ac.uk/www-jpred) (7). The conserved motif in domain 4 is shown in boldface.
MATERIALS AND METHODS
Purification of recombinant Hoc proteins.
RB49 Hoc, T4 Hoc, and a T4 Hoc domain(s) fused to the C terminus of HIV p24 were cloned into pET expression vectors, expressed, and purified as described previously (39). For crystallizations, hexahistidine-tagged Hoc purified by HisTrap chromatography (AKTA Prime; GE Healthcare) was cleaved by thrombin (Novagen, San Diego, CA) to remove the histidine tag. The sample was then reapplied to the HisTrap column, and the flowthrough fractions containing no hexahistidine tag were concentrated and further purified by size exclusion chromatography using a Superdex-200 (Hi-Load 16/16, prep-grade) gel filtration column (AKTA FPLC; GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 8.0) and 30 mM NaCl. The sample was concentrated by Amicon Ultra-4 centrifugal filtration.
Protein crystallization and structure determination.
Attempts at crystallizing the complete Hoc molecule of T4 were unsuccessful. Subsequently, it was found to be possible to crystallize bacteriophage RB49 Hoc. Initial screening for crystallization conditions for RB49 Hoc was performed using the sitting drop vapor diffusion method. Drops containing 1 μl of the protein at a concentration of ∼6 mg/ml were mixed with 1 μl of reservoir solutions using various commercial crystallization screens and were equilibrated against 100 μl of the reservoir solution. RB49 Hoc crystals were obtained in 20% polyethylene glycol 8000 (PEG 8000), 200 mM MgCl2, and 100 mM Tris-HCl (pH 8.5). Crystals were detected 6 weeks after the crystallization trays were set up. Later it was found that C-terminal domain 4 of the crystallized protein had been cleaved off by an unidentified protease present in the protein solution. This process had occurred not only in the sample set up for crystallization but also in the sample kept at 4°C in 20 mM Tris-HCl (pH 8.0) and 30 mM NaCl buffer. Thus, to avoid having to wait for the cleavage process, crystallization conditions were repeated using the cleaved protein. These crystallization experiments produced crystals in 4 days. The crystals belonged to space group C2, with one molecule in the asymmetric unit (Table 2). Platinum derivatives were obtained by soaking crystals for 2.5 h in a solution containing 10 mM K2PtCl4.
Table 2.
Crystal parameters and diffraction data statistics
| Crystal | RB49 Hoc native data set 1 | RB49 Hoc platinum derivative (MAD data) | RB69 Soc native data set 2 | ||
|---|---|---|---|---|---|
| Space group | C2 | C2 | C2 | ||
| Unit cell parameters (Å) | a = 210.97, b = 36.23, c = 62.48, β = 98.71 | a = 212.19, b = 36.79, c = 62.99, β = 98.85 | a = 213.52, b = 36.73, c = 62.69, β = 99.28 | ||
| Wavelength (Å) | 1.03328 | 1.07219 (peak) | 1.07246 (inflection point) | 1.06333 (high-energy remote) | 0.97950 |
| No. of images | 180 | 360 | |||
| Oscillation angle (°) | 1.0 | ||||
| Resolution (Å) | 3.0 | 2.7 | 1.95 along a*/|a*| + c*/|c*|; 2.80 along a*/|a*| − c*/|c*|; 2.7 along b* | ||
| No. of unique reflections | 9,629 | 12,881 | 12,713 | 13,040 | 18,095 |
| Redundancy, ratio | 3.5 | 6.6 | 6.7 | 6.9 | 6.2 |
| Completeness (%) | 99.7 | 96.6 | 94.0 | 97.6 | 80.8 (resolution range from infinity to 2.45 Å); 50.9 (resolution range from infinity to 1.95 Å) |
| Rsym (%) | 9.9 | 7.8 | 8.4 | 7.4 | 6.3 |
| Mosaicity (°) | 2.1 | 0.7 | 0.6 | ||
| 〈I〉/〈σ(I)〉 | 12.3 | 23.0 | 21.5 | 27.0 | 23.14 |
| Last resolution shell (Å) | 3.11-3.0 | 2.8-2.7 | 2.8-2.7 | 2.8-2.7 | 1.98-1.95 |
| Rsym, last spherical resolution shell (%) | 61.6 | 30.2 | 33.3 | 27.8 | 29.1 |
| 〈I〉/〈σ(I)〉, outermost shell | 1.71 | 2.66 | 2.45 | 4.66 | 4.24 |
Data collection and structure determination.
X-ray data (Table 2) were collected at GM/CA-CAT beamline 23-ID-D of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Before data collection, crystals were placed in a cryoprotectant solution for a few seconds and were flash-frozen in the nitrogen stream. The cryoprotectant solution was obtained from the crystal growth solution by adding ethylene glycol to obtain a 10% (vol/vol) concentration.
Initially, a native data set was collected to 3 Å resolution, and a multiwavelength anomalous dispersion (MAD) data set from a platinum derivative was collected to 2.7 Å resolution. The data sets were processed using the HKL2000 program (33). The diffraction data were anisotropic. The diffraction was stronger in the direction corresponding to the vector a*/∣a*∣ + c*/∣c*∣ in the reciprocal space than in the other orthogonal directions determined by the vectors a*/∣a*∣ − c*/∣c*∣ and b*. Structure was determined using the PHENIX software suite (1) with the help of the AutoSol wizard (45). Correction for the diffraction anisotropy was performed using the XTRIAGE program (51). Determination of the platinum positions and subsequent phasing using the SIRAS (single isomorphous replacement with anomalous scattering) method were performed using the SOLVE program (46). The phases were improved by density modification with the RESOLVE program (44). An atomic model was built into the electron density using the COOT program (10). The model was subjected to several rounds of crystallographic refinement using the PHENIX.REFINE program (2) with manual rebuilding in COOT (10) and automatic rebuilding using PHENIX AutoBuild (47). The refinement was initially performed using the structure factor amplitudes corresponding to the remote wavelength of the platinum derivative data set. Later, a higher-resolution data set was collected from a native crystal. The maximum resolution of the data set (limited by the crystal-to-detector distance) was 1.95 Å. In the worst diffraction direction, defined by the vector a*/∣a*∣ − c*/∣c*∣, 〈I〉/〈σ(I)〉 was less than 2 at resolutions higher than 2.8 Å, whereas in the best direction, determined by the vector a*/∣a*∣ + c*/∣c*∣, 〈I〉/〈σ(I)〉 was ∼8 even at 1.95 Å resolution. In the third direction, defined by the vector b*, 〈I〉/〈σ(I)〉 fell below 2 at a resolution of 2.7 Å. The data were truncated using an ellipsoid that had its principal axes along these vectors. The lengths of the principal axes were determined by the resolution limits of 1.6, 2.8, and 2.7 Å. All reflections that were outside this ellipsoid and had an I/σ(I) of <2.0 were rejected. The structure factors were modified by applying anisotropic scaling to scale up reflections located in the weak directions of reciprocal space. The components of the B-factor scaling tensor corresponding to the principal axes of the ellipsoid were 0, −60, and −35 Å2, respectively. The refinement of the structure was continued using the anisotropically scaled structure amplitudes. This anisotropy correction substantially improved the robustness of refinement and the quality of the electron density maps. Final refinement statistics are presented in Table 3.
Table 3.
Crystallographic refinement statistics for RB49 Hoc
| Statistic | Value |
|---|---|
| No. of protein nonhydrogen atoms | 2,334 |
| No. of solvent atoms | 90 |
| R factor (working set) | 0.208 |
| Rfree (test set, 10% of the data) | 0.268 |
| Last resolution shell | 2.02–1.95 |
| R factor in the last resolution shell | 0.233 |
| Rfree in the last resolution shell | 0.307 |
| Avg B factor (Å2) | 59 |
| RMSD | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.175 |
| Dihedral angles (°) | 14.3 |
| Planarity (Å) | 0.005 |
| Chirality (Å3) | 0.086 |
| Peptide bond ω angle (°) | 5.9 |
| % of residues in different regions of the Ramachandran plota | |
| Most favored regions | 93.5 |
| Additional allowed regions | 6.2 |
| Generously allowed regions | 0.4 |
| Disallowed regions | 0 |
Residues except Gly and Pro.
The homology model of T4 Hoc based on the RB49 Hoc structure was constructed using the MODELLER program (38). Figures were prepared using the MOLSCRIPT (25) and Raster3D (31) programs.
Binding of Hoc to E. coli.
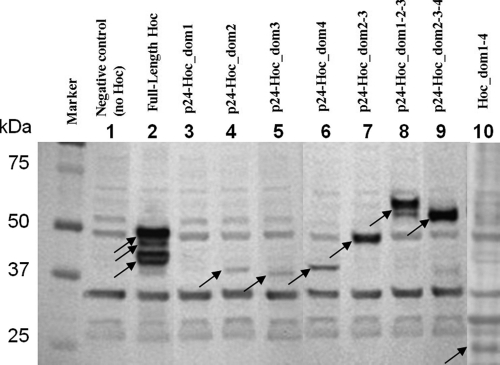
The full-length T4 Hoc protein and the domains of Hoc fused to the C terminus of HIV-1 p24 were added to freshly cultured E. coli P301. As discussed previously, fusion to p24 was necessary because it increased the size of ∼10-kDa Hoc domains to a high molecular mass range (>35 kDa), allowing unambiguous identification of these bands by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (39). About 108 cells were centrifuged at 2,300 × g for 3 min at 4°C in a low-bind Eppendorf tube. The sedimented cells were resuspended in phosphate-buffered saline (PBS). The purified Hoc proteins were centrifuged at 20,000 × g for 45 min at 4°C to remove any aggregated protein. The supernatant was then added to E. coli cells at a ratio of 2 × 106 Hoc molecules per cell in a reaction volume of 150 μl. This ratio was selected based on optimization experiments in which the ratios were varied over a wide range, and the results showed that binding reached saturation at this ratio. The samples were incubated at room temperature for 45 min, and the E. coli cells were sedimented by centrifugation at 2,300 × g for 3 min at 4°C and were washed twice with 1 ml of PBS. The final pellet was resuspended in 10 μl of PBS and was transferred to a new tube, and 10 μl of 2× SDS-PAGE sample buffer was added. The proteins were separated by electrophoresis, transferred to a polyvinylidene difluoride (PVDF) membrane, and incubated with chicken polyclonal anti-Hoc antibodies (a kind gift from Lindsay Black, University of Maryland Medical School, Baltimore) to detect bound Hoc proteins. Alkaline phosphatase-conjugated rabbit anti-chicken IgG was used as secondary antibody, and the bands were visualized by adding a chromogenic substrate (Western blot kit; Invitrogen) (Fig. 3). Each experiment included two negative controls: (i) a control where E. coli cells were omitted, which showed no bands, and (ii) a control where Hoc was omitted, which showed several background bands due to nonspecific interaction of the Hoc antibodies with E. coli proteins (Fig. 3, lane 1). Each reaction was carried out in duplicate.
Fig. 3.
Binding of T4 Hoc and of T4 Hoc fragments to E. coli. The binding assays were performed using 108 E. coli cells and 2 × 106 Hoc molecules per cell (see Materials and Methods). Bound Hoc was detected by Western blotting using polyclonal anti-Hoc antibodies. Lane 1 represents the negative control in which the Hoc protein was omitted. It shows several bands corresponding to E. coli proteins that interact with anti-Hoc antibodies. These background bands are present in all lanes. Arrows indicate the bands corresponding to the Hoc proteins bound to E. coli.
Protein Data Bank accession number.
The atomic coordinates and structure factors have been deposited with the Protein Data Bank under PDB accession number 3SHS.
RESULTS
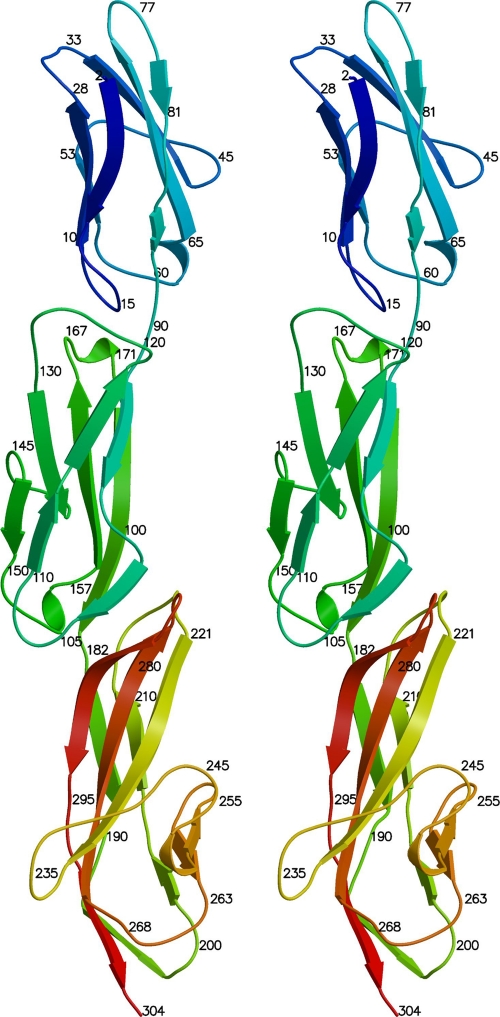
The structures of three N-terminal domains of Hoc from bacteriophage RB49 were determined by X-ray crystallography. The diffraction of the crystals was anisotropic (see Materials and Methods). The resolutions of the best datasets along the three principal mutually perpendicular directions were 1.95, 2.8, and 2.7 Å, respectively. The final atomic model contained residues 2 to 304, corresponding to about three-fourths of the 404 residues in RB49 Hoc. The molecule has an elongated shape with an approximately linear arrangement of domains, contrary to the horseshoe-like model proposed previously for T4 Hoc (39). Consistent with bioinformatics predictions, all three domains have an immunoglobulin-like fold. Domains 1 and 2 are similar in size, containing 90 and 91 residues, respectively, whereas domain 3 is larger and consists of 123 residues.
The structures of domains 1 and 2 of RB49 Hoc are very similar to each other, with 23% sequence identity and a 1.7-Å root mean square deviation (RMSD) between 80 equivalenced Cα atoms. This suggests that these domains evolved recently from a common ancestor by gene duplication. Comparison of the individual RB49 Hoc domains with known protein structures using the Dali algorithm (17) shows their similarity to various eukaryotic molecules involved in protein-protein interaction and cell adhesion (Table 4). Each Hoc domain has a large number of structural relatives with Z-scores higher than 9. Domains 1 and 2 of Hoc belong to the I-set family of the immunoglobulin superfamily, as classified in the Structural Classification of Proteins (SCOP) (32). Domain 1 of Hoc (residues 1 to 90) is structurally closely similar to extracellular domains of the immunoglobulin Fc receptors (30). These receptors bind Fc fragments of antibodies to initiate cellular responses against antigens. The structure of Hoc domain 2 (residues 91 to 181) is closely similar to that of the third immunoglobulin-like domain of Perlecan, a multifunctional protein from the basement membrane (26), and to the N-terminal domain of the neural cell adhesion molecule (22). Domain 3 of RB49 Hoc (residues 182 to 304) belongs to the V-set family of the immunoglobulin superfamily. The V-set domains resemble the antibody variable domain. The closest structural relative of domain 3 from RB49 Hoc is the variable region of the T-cell receptor α chain (8). T-cell receptors are located on the surfaces of T lymphocytes and recognize antigens bound to the major histocompatibility complex.
Table 4.
Some structural relatives of the individual domains of RB49 Hoc
| Molecule name | PDB IDa | Dali Z-score | RMSD between equivalence Cα atoms (Å) | No. of equivalenced residues | Sequence identity (%) |
|---|---|---|---|---|---|
| Domain 1 of RB69 Hoc | |||||
| Fcγ receptor | 2fcg | 12.2 | 1.8 | 82 | 16 |
| High-affinity immunoglobulin epsilon receptor | 1j88 | 11.9 | 1.9 | 82 | 17 |
| Low-affinity immunoglobulin gamma receptor | 1e4j | 11.9 | 1.8 | 82 | 17 |
| Collagen binding domain of human glycoprotein IV | 2gi7 | 11.3 | 1.6 | 80 | 18 |
| Immunoglobulin alpha Fc receptor | 1uct | 10.7 | 2.4 | 84 | 11 |
| Primordial avian classical Fc receptor | 2vsd | 10.7 | 1.9 | 82 | 16 |
| Leukocyte immunoglobulin-like receptor | 2d3v | 10.7 | 1.7 | 80 | 13 |
| Neural cell adhesion molecule | 1epf | 10.4 | 2.2 | 81 | 22 |
| Domain 2 of RB69 Hoc | |||||
| Perlecan (third immunoglobulin-like domain) | 1gl4 | 10.5 | 1.9 | 79 | 16 |
| Neural cell adhesion molecule | 1epf | 10.1 | 1.7 | 79 | 19 |
| Neural cell adhesion molecule 2 | 2v44 | 10.1 | 1.5 | 75 | 16 |
| Vascular cell adhesion protein 1 | 1ij9 | 9.9 | 1.9 | 78 | 14 |
| Mucosal addressin cell adhesion molecule 1 | 1gsm | 9.7 | 2.5 | 79 | 6 |
| Programmed cell death protein 1 | 3bp6 | 9.7 | 2.0 | 76 | 13 |
| Cervical EMMPRIN | 3b5h | 9.6 | 1.6 | 75 | 11 |
| Domain 3 of RB49 Hoc | |||||
| T-cell receptor α chain variable domain | 2e7l | 13.0 | 2.2 | 100 | 23 |
| Matuzumab Fab light chain | 3c09 | 12.4 | 1.9 | 99 | 16 |
| T-cell surface glycoprotein CD1D1 | 3huj | 12.2 | 2.5 | 103 | 17 |
| Amyloid lambda-6 light chain variable region PIP | 3bdx | 11.6 | 2.2 | 99 | 20 |
| Lutheran blood group glycoprotein N-terminal domain | 2pf6 | 11.6 | 2.0 | 98 | 15 |
| Extracellular domain of CRIgb protein from the complement system | 2icc | 11.3 | 2.7 | 103 | 17 |
PDB, Protein Data Bank; ID, identification code.
CRIg, complement receptor of the immunoglobulin superfamily.
Since the Hoc domains share similarity to molecules involved in protein binding and surface adhesion, it is possible that one of the functions of Hoc is to attach the virus reversibly to E. coli surface molecules. This hypothesis was tested by mixing purified T4 Hoc or individual T4 Hoc domains with E. coli and analyzing for the presence of bound Hoc by Western blotting using polyclonal anti-Hoc antibodies. The results (Fig. 3) showed that all individual Hoc domains bind to E. coli, although the binding of domain 1 was very weak. The binding of Hoc appears to increase synergistically with increasing numbers of domains. For example, the Hoc protein that contains domains 2 and 3 bound more strongly than the individual domains 2 and 3 combined. Full-length Hoc, containing all four domains, showed the strongest binding.
DISCUSSION
Here we report the structure of three of the four domains of Hoc, a virus decoration protein present only on T4-like bacteriophage heads. Only the first three domains could be crystallized, possibly because the linker between domains 3 and 4 is flexible. The C-terminal domain (residues 305 to 404 in RB49 Hoc) is involved in capsid binding (39) and is well conserved among T4-like bacteriophages. This domain has no detectable sequence similarity to any nonbacteriophage protein. The secondary-structure predictions (7) suggest with high probability that there are three α-helices in domain 4 of RB49 Hoc (Fig. 2). Therefore, it is unlikely that the C-terminal domain of Hoc proteins has an immunoglobulin-like fold.
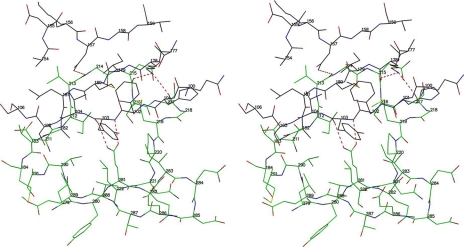
The crystal structure shows that RB49 Hoc has short linkers between domains, resulting in extensive interactions between domains 2 and 3 (Fig. 4 and 5). This is inconsistent with the previously proposed model of the T4 Hoc protein (39), which has a sharp bend between domains 2 and 3, producing a horseshoe-like shape that brings domains 1 and 4 into proximity with each other. Therefore, it is unlikely that the sharp bend between domains 2 and 3, responsible for the horseshoe-like shape in the model, can occur in RB49 Hoc. Homology modeling (38) of T4 Hoc, based on the RB49 Hoc crystal structure, suggests that the linkers between domains 2 and 3 in T4 Hoc are also short. Thus, the T4 Hoc molecule probably also has an elongated shape. However, because of the low sequence similarity between the T4 and RB49 Hoc proteins (22% sequence identity), the position and the length of the linkers in T4 Hoc are uncertain. Therefore, the possibility that T4 Hoc may adopt a horseshoe-like shape cannot be completely excluded.
Fig. 4.
Ribbon stereodiagram showing the structures of the three RB49 Hoc N-terminal domains. Colors range from blue at the amino end to red at the carboxy end of the polypeptide chain.
Fig. 5.
Stereodiagram showing the interactions between domains 2 and 3 of RB49 Hoc. Oxygen atoms are shown as red, nitrogen atoms as blue, and sulfur atoms as orange. Carbon atoms are shown as black in domain 2 and green in domain 3.
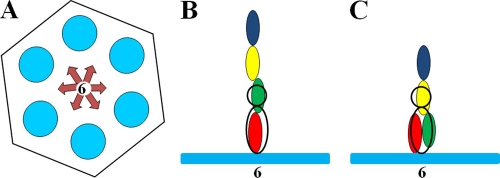
In the cryo-EM reconstruction of the T4 capsid (13), the density corresponding to the Hoc molecules is located at the centers of the gp23 hexamers and has the shape of a dumbbell. However, this cryo-EM density is the result of the averaging between many phage particles during the reconstruction process. Most likely, a Hoc molecule can bind to a gp23 hexameric capsomer in any of six possible orientations related by rotation around the quasi-6-fold axis of the gp23 capsomer (Fig. 6A). However, some of these orientations might occur less frequently due to small differences in the conformation or orientation of the gp23 molecules related by the quasisymmetry. Since the bound Hoc molecules are not necessarily aligned with the axes of the gp23 hexamers, and since the orientation of the Hoc molecules may be different in different phage particles, the dumbbell-shaped density may correspond to an average density of many Hoc molecules attached to the capsid in different orientations. If this is the case, the base of the dumbbell may correspond only to the C-terminal domain 4 of Hoc, interacting with the T4 capsid. The distal part of the dumbbell would correspond to domain 3, and the density of domains 1 and 2 would be completely absent (Fig. 6B). Alternatively, both domains 3 and 4 may contribute to the density of the base of the dumbbell (Fig. 6C).
Fig. 6.
Schematic representation of Hoc bound to a gp23 hexamer. (A) The gp23 capsomer is represented by a hexagon. Six blue circles represent the gp23 monomers. The six possible orientations of the Hoc molecule with respect to the gp23 capsomer are represented by arrows. (B and C) The gp23 capsomer is represented by the blue bar. Domains 1, 2, 3, and 4 of Hoc are represented by blue, yellow, green, and red ellipses, respectively. The dumbbell-shaped density of Hoc, observed in the cryo-EM reconstruction, is represented by the black ellipse and circle. (B) Elongated conformation of the Hoc molecule. Domains 1 and 2 are presumably not visible in the cryo-EM density, in part because of a likely different displacement of the Hoc domains from the 6-fold axis of the gp23 hexamer in different particles used to generate the cryo-EM density. (C) Alternative structure of Hoc in which domains 2, 3, and 4 contribute to the observed cryo-EM density.
The horseshoe-like model of Hoc would imply that the N terminus of the protein is located on the virus close to the major capsid protein shell. However, when bulky antigens, such as the 88-kDa anthrax protective antigen, were fused to the N terminus of Hoc, there was no change in the affinity of Hoc binding to the capsid, whereas fusion of antigens to the C terminus caused a 13- to 400-fold decrease in affinity (40, 41). This is consistent with the notion that the elongated structure of Hoc has its C-terminal domain bound to the capsid.
The density corresponding to Hoc molecules cannot be easily distinguished in raw EM images of the native T4 virion. However, the shape and size of the horseshoe-like Hoc structure would be comparable to those of an antibody's Fab fragment. Experience with virus-Fab complexes (see, for example, reference 23) has shown that the density of Fab bound to a virus is usually detectable in raw EM images. Thus, in the absence of detectable Hoc density, the elongated model of the Hoc protein would be favored.
Depending on ambient conditions, a virus might have a survival advantage by staying close to a host without infecting it, but when the conditions become more favorable for infection, the virus can detach from the cell and start the infection process. The T4 virus starts the infection of an E. coli bacterium by attaching to its surface using the LTFs. Each of six LTFs can assume two different states. In the “up” position, the LTFs are positioned along the tail, whereas in the “out” position, the LTF are released from the tail and can oscillate freely while searching for cell surface receptors (24). The release of LTFs is controlled by ambient conditions, such as pH and temperature. Therefore, depending on the ambient conditions, the T4 bacteriophage can switch from the “waiting” mode to the “infection” mode. In the “waiting” mode, it would be beneficial for T4 to stay close to E. coli cells by attaching itself to the bacterial surface. Biochemical experiments showed that the T4 Hoc protein can bind to E. coli cells. In addition, Hoc might allow the phage to use a bacterium (which is not necessarily its host), as a “vehicle” for travel to different locations. Since bacteria use chemotaxis to direct their motion to places with favorable chemical environments, traveling on a bacterial surface may be a more effective way to find new hosts than random diffusion. The binding of Hoc to a host cell may also be beneficial to the bacteriophage during the infection process, because it may allow the virus to stay attached to the cell while its tail fibers find their receptors.
T4 Hoc− mutant phages tend to aggregate at low ionic strength (in the absence of divalent ions), whereas the native T4 phages do not aggregate under the same conditions (49), presumably because of the repulsive negative charge on the Hoc molecules (the T4 Hoc has a charge of −13 electrons at neutral pH). Therefore, another function of Hoc might be to disperse the virus particles, preventing aggregation.
Because the first three Hoc domains have many traits similar to each other and to those of other cell binding immunoglobulin domains, it is more probable that these domains and the eukaryotic immunoglobulin domains have evolved from a common ancestor than that they have converged to a similar structure. Since the immunoglobulin-like domains play essential roles in surface interactions between cells (4, 6, 21), the presence of a large number of immunoglobulin-like domains on the T4 capsid surface may allow the phage particles to interact with the surfaces of eukaryotic cells. This may be beneficial for the bacteriophage when it propagates on symbiotic bacteria (8a).
Phages and viruses encode “nonessential” proteins to expand their survival advantage to environmental conditions. Decorating the virion with long fibers consisting of domains derived from recent hosts gives the virus options to evolve survival advantages, such as the ability to accumulate near the host, sensing of the environmental conditions for infection, transportation to favorable environments, and dispersion of particles to maximize productivity. Although such mechanisms have been well documented in bacteria and higher organisms, the Hoc protein of T4-like bacteriophages might represent the first example of a viral protein entirely dedicated to such roles. This might be widespread in phages and other viruses, because insertion of “foreign” domains (such as Ig domains) on exposed regions of the virus has been found frequently in the sequences of phages and viral genomes (15).
ACKNOWLEDGMENTS
We thank Sheryl Kelly for help in the preparation of the manuscript.
This work was supported by a National Science Foundation grant (MCB-0443899) to M.G.R., a National Institutes of Health grant (NIAID; AI082086) to V.B.R., and a National Institutes of Health grant (NIAID; R01AI081726) to M.G.R. and V.B.R.
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. 2005. The Phenix refinement framework. CCP4 Newsl. 42:contribution 8 [Google Scholar]
- 3. Aksyuk A. A., et al. 2009. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bateman A., Eddy S. R., Chothia C. 1996. Members of the immunoglobulin superfamily in bacteria. Protein Sci. 5:1939–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bateman A., Eddy S. R., Mesyanzhinov V. V. 1997. A member of the immunoglobulin superfamily in bacteriophage T4. Virus Genes 14:163–165 [DOI] [PubMed] [Google Scholar]
- 6. Bork P., Holm L., Sander C. 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242:309–320 [DOI] [PubMed] [Google Scholar]
- 7. Cole C., Barber J. D., Barton G. J. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Colf L. A., et al. 2007. How a single T cell receptor recognizes both self and foreign MHC. Cell 129:135–146 [DOI] [PubMed] [Google Scholar]
- 8a. Dabrowska K., Switala-Jelen K., Opolski A., Gorski 2006. Possible association between phages. Hoc protein, and the immune system. Arch. Virol. 151:209–215 [DOI] [PubMed] [Google Scholar]
- 9. Driedonks R. A., Engel A., ten Heggeler B., van Driel R. 1981. Gene 20 product of bacteriophage T4. Its purification and structure. J. Mol. Biol. 152:641–662 [DOI] [PubMed] [Google Scholar]
- 10. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 11. Fokine A., Battisti A. J., Kostyuchenko V. A., Black L. W., Rossmann M. G. 2006. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: the evolution of pentameric vertex proteins in icosahedral viruses. J. Struct. Biol. 154:255–259 [DOI] [PubMed] [Google Scholar]
- 12. Fokine A., et al. 2007. Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins. Virology 367:422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fokine A., et al. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U. S. A. 101:6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fokine A., et al. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 102:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fraser J. S., Yu Z., Maxwell K. L., Davidson A. R. 2006. Ig-like domains on bacteriophage: a tale of promiscuity and deceit. J. Mol. Biol. 359:496–507 [DOI] [PubMed] [Google Scholar]
- 16. Goldberg E., Grinius L., Letellier L. 1994. Recognition, attachment, and injection, p. 347–356 In Karam J. D., et al. (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC [Google Scholar]
- 17. Holm L., Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishii T., Yanagida M. 1977. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid: their properties, isolation and characterization of defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 109:487–514 [DOI] [PubMed] [Google Scholar]
- 19. Iwasaki K., et al. 2000. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology 271:321–333 [DOI] [PubMed] [Google Scholar]
- 20. Jiang J., Abu-Shilbayeh L., Rao V. B. 1997. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 65:4770–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jing H., et al. 2002. Archaeal surface layer proteins contain β propeller, PKD, and β helix domains and are related to metazoan cell surface proteins. Structure 10:1453–1464 [DOI] [PubMed] [Google Scholar]
- 22. Kasper C., et al. 2000. Structural basis of cell-cell adhesion by NCAM. Nat. Struct. Biol. 7:389–393 [DOI] [PubMed] [Google Scholar]
- 23. Kaufmann B., et al. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 103:12400–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kostyuchenko V. A., et al. 2005. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12:810–813 [DOI] [PubMed] [Google Scholar]
- 25. Kraulis P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946–950 [Google Scholar]
- 26. Kvansakul M., Hopf M., Ries A., Timpl R., Hohenester E. 2001. Structural basis for the high-affinity interaction of nidogen-1 with immunoglobulin-like domain 3 of perlecan. EMBO J. 20:5342–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leiman P. G., Chipman P. R., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. 2004. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419–429 [DOI] [PubMed] [Google Scholar]
- 28. Leiman P. G., Kanamaru S., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60:2356–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q., Shivachandra S. B., Leppla S. H., Rao V. B. 2006. Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes. J. Mol. Biol. 363:577–588 [DOI] [PubMed] [Google Scholar]
- 30. Maxwell K. F., et al. 1999. Crystal structure of the human leukocyte Fc receptor, FcγRIIa. Nat. Struct. Biol. 6:437–442 [DOI] [PubMed] [Google Scholar]
- 31. Merritt E. A., Bacon D. J. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505–524 [DOI] [PubMed] [Google Scholar]
- 32. Murzin A. G., Brenner S. E., Hubbard T., Chothia C. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536–540 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 34. Qin L., Fokine A., O'Donnell E., Rao V. B., Rossmann M. G. 2010. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J. Mol. Biol. 395:728–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren Z. J., et al. 1996. Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 5:1833–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Riede I. 1987. Receptor specificity of the short tail fibres (gp12) of T-even type Escherichia coli phages. Mol. Gen. Genet. 206:110–115 [DOI] [PubMed] [Google Scholar]
- 37. Rossmann M. G., Mesyanzhinov V. V., Arisaka F., Leiman P. G. 2004. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 14:171–180 [DOI] [PubMed] [Google Scholar]
- 38. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23:318–326 [DOI] [PubMed] [Google Scholar]
- 39. Sathaliyawala T., et al. 2010. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 77:444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sathaliyawala T., et al. 2006. Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: a novel in vitro approach to construct multicomponent HIV vaccines. J. Virol. 80:7688–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shivachandra S. B., et al. 2007. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 25:1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steven A. C., Greenstone H. L., Booy F. P., Black L. W., Ross P. D. 1992. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, cooperativity, and super-stabilization by soc binding. J. Mol. Biol. 228:870–884 [DOI] [PubMed] [Google Scholar]
- 43. Sun S., et al. 2008. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135:1251–1262 [DOI] [PubMed] [Google Scholar]
- 44. Terwilliger T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56:965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Terwilliger T. C., et al. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65:582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terwilliger T. C., Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 55:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terwilliger T. C., et al. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wikoff W. R., et al. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133 [DOI] [PubMed] [Google Scholar]
- 49. Yamaguchi Y., Yanagida M. 1980. Head shell protein hoc alters the surface charge of bacteriophage T4. Composite slab gel electrophoresis of phage T4 and related particles. J. Mol. Biol. 141:175–193 [DOI] [PubMed] [Google Scholar]
- 50. Yu F., Mizushima S. 1982. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 151:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zwart P. H., Grosse-Kunstleve R. W., Adams P. D. 2005. Xtriage and Fest: automatic assessment of X-ray data and substructure structure factor estimation. CCP4 Newsl. 43:contribution 7 [Google Scholar]