Abstract
There are 9 serotypes of neuraminidase (NA) from influenza A virus (N1 to N9), which are classified into two groups based on primary sequences (groups 1 and 2). The structural hallmark of the two groups is the presence or absence of an extra 150-cavity (formed by the 150-loop) in the active site. Thus far, structures of NAs from 6 out of the 9 serotypes have been solved. Here, we solved the N5 structure, the last unknown structure group 1 serotype with a unique Asn147 residue in its 150-loop, demonstrating that it has an extended 150-cavity that closes upon inhibitor binding.
TEXT
Influenza virus is the causative agent of seasonal and, occasionally, pandemic flu (23). The outbreak of the swine origin influenza A virus (S-OIV) H1N1 pandemic in 2009 has led to serious economic losses and public panic, which illustrates the importance of flu prevention and control. The neuraminidase (NA) of influenza A virus is a surface glycoprotein that cleaves terminally linked sialic acid from hemagglutinin (HA) binding receptors to facilitate the release of progeny virions from infected cells and prevent the aggregation of virus particles (5). NA is the most efficient drug target for treatment of influenza infection so far (7, 21). Currently, there are four approved antiflu drugs targeting NA: oseltamivir (10), zanamivir (22), peramivir (3), and laninamivir (25), as well as some others undergoing clinical trials. Due to the appearance of drug-resistant strains and pandemics, there is always an urgent need for the development of new drugs. Structure-based rational drug design is always ideal, and NA-targeting drugs are good examples in this regard.
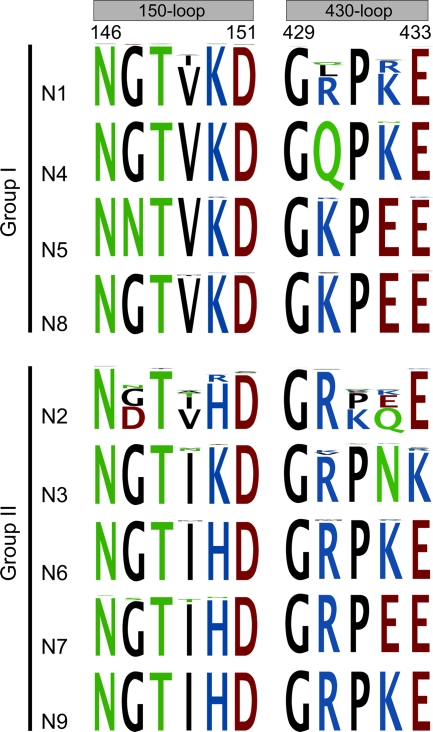
There are 9 serotypes of NA, named N1 to N9. Based on their primary nucleotide sequences, NAs can be divided into two groups, group 1 (N1, N4, N5, and N8) and group 2 (N2, N3, N6, N7, and N9) (2). Zanamivir and oseltamivir were initially designed to mimic the binding of the transition state analogue Neu5Ac2en to the NA active site based on group 2 NA structures (6, 20, 21). Later, an extra cavity in the active site, called the 150-cavity and formed by 150-loop amino acids, was identified in group 1 NAs and proposed as a new drug target (17). Specifically targeting this cavity, a novel inhibitor, 3-(p-tolyl)allyl-Neu5Ac2en, has recently been confirmed to selectively inhibit group 1 NAs, as well as 2009-pandemic H1N1 neuraminidase (09N1) (16). Although our recent work showed that 09N1 has no 150-cavity, which is determined and affected by the 150-loop and 430-loop (12) (Fig. 1), we hypothesize that the atypical group 1 09N1 may possess a 150-loop for the binding of group 1-specific inhibitors that is more easily opened than those of group 2 NAs. This can also explain the more efficient inhibition of 3-(p-tolyl)allyl-Neu5Ac2en to 09N1 than to group 2 NAs. This raised questions as to whether or not (i) the 150-cavity is really conserved for all group 1 members and (ii) a new drug specifically designed targeting the 150-cavity can work for all group 1 NAs. Although the structures for 3 out of 4 group 1 NA serotypes have been solved so far (17, 24), and they all (with the exception of 09N1) have the 150-cavity, with unique sequences in the 150-loop and 430-loop, the crystal structure of group 1 member N5 remains elusive; its 150-loop has a special amino acid at position 147 (Asn instead of a conserved Gly) (Fig. 1), which might have some effect on the 150-cavity formation. Therefore, we set up experiments to produce soluble N5 in a baculovirus expression system and obtain enough protein for crystallization.
Fig. 1.
Comparative sequence alignments of the 150-loop and 430-loop in NAs from 9 different serotypes using CLC Main Workbench software. One-third of the N1 sequences and one-half of the N2 to N9 sequences (a total of 9,020 sequences) available in GenBank were randomly selected for alignment. Comparison of the unique N5 Asn147 with the highly conserved Gly147 from other group 1 serotypes is clearly shown in the alignment. Typical group 1 NA structures with a 150-cavity contain Val149, and typical group 2 structures without a 150-cavity contain Ile149. The Tokyo and Memphis N2 structures contain Val at position149; however, they are still unable to form a 150-cavity due to a salt bridge between Asp147 and His150, which holds the 150-loop in a closed conformation.
Highly stable and pure N5 protein was prepared based on previous methods (24, 26), with modification using a Bac-to-Bac baculovirus expression system (Invitrogen). cDNA encoding residues 83 to 471 (N2 numbering) of the A/duck/Alberta/60/1976 (H12N5) virus NA was cloned into pFastBac1 (Invitrogen), with a GP67 signal peptide, a 6×His tag, a tetramerizing sequence, and a thrombin cleavage site at the N terminus. The recombinant baculovirus was prepared based on the manufacturer's protocol (Invitrogen).
N5 protein was obtained from infected Sf9 insect cells using the purification methods of our previous work on 09N1 (12). Crystallization conditions were screened using the hanging-drop vapor diffusion method with commercial kits (Hampton Research). N5 crystals were obtained with 0.1 M HEPES (pH 7.5), 12% (wt/vol) polyethylene glycol 3,350 at 18°C. X-ray diffraction data were collected at 100° K at beamline NE3A of the Photon Factory, Tsukuba, Japan. The data were processed and scaled by using the HKL-2000 software program (14).
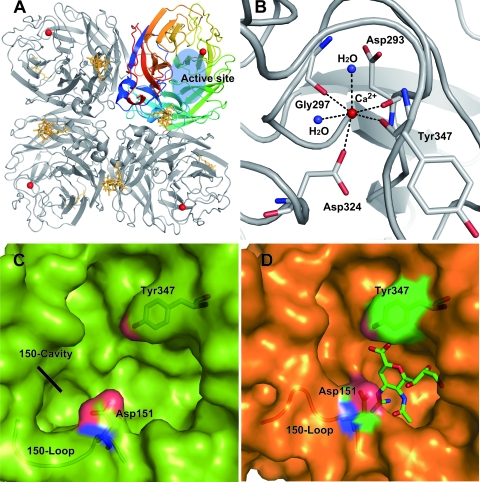
The structure of N5 and its complex with zanamivir (through crystal soaking) were solved by the molecular replacement method using Phaser (15) from the CCP4 program suite (4), with the structure of 09N1 (Protein Data Bank [PDB] identifier, 3NSS) as the search model. Model building and refinement were performed as described previously (12) using COOT (8), REFMAC5 (13), and the PHENIX package (1). The stereochemical quality of the final models was assessed with the program PROCHECK (11). The N5 structures were refined to crystallographic Rwork/Rfree values of 0.126/0.164 and 0.121/0.157, respectively (Table 1) (R = Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|, where Rfree is calculated for a randomly chosen 5% of reflections and Rwork is calculated for the remaining 95% of reflections used for structure refinement). The overall N5 structure shows a typical “box-shaped” tetramer observed in other influenza NAs (5). Each monomer (all of which are identical) contains a propeller-like arrangement of six four-stranded, antiparallel sheets (Fig. 2 A).
Table 1.
Data collection and refinement statisticsa
| Parameter | Value(s) for: |
|
|---|---|---|
| Native N5 | N5-zanamivir | |
| Data collection | ||
| Space group | P4 | P4 |
| Cell dimensions | ||
| a, b, c (Å) | 111.51, 111.51, 66.47 | 112.11, 112.11, 66.77 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.00–1.50 (1.55–1.50) | 50.00–1.60 (1.66–1.60) |
| Rmerge | 0.083 (0.544) | 0.068 (0.345) |
| Ι/σI | 21.9 (2.2) | 20.8 (3.6) |
| Completeness (%) | 99.7 (98.0) | 99.9 (100.0) |
| Redundancy | 6.4 (4.5) | 4.4 (4.3) |
| Refinement | ||
| Resolution (Å) | 37.17–1.50 | 35.45–1.60 |
| No. of reflections | 125,014 | 105,577 |
| Rwork/Rfree | 0.1260/0.1640 | 0.1205/0.1572 |
| No. of atoms | ||
| Protein | 6,270 | 6,243 |
| Ligand/ion | 2 | 48 |
| Water | 949 | 1,138 |
| B factors | ||
| Protein | 15.2 | 13.2 |
| Ligand/ion | 11.3 | 9.2 |
| Water | 32.6 | 34.1 |
| RMSDs | ||
| Bond length (Å) | 0.004 | 0.005 |
| Bond angle (°) | 0.949 | 1.040 |
| Ramachandran plot | ||
| Most favored (%) | 85.9 | 86.4 |
| Additionally favored (%) | 13.6 | 13.2 |
| Generally allowed (%) | 0.4 | 0.4 |
| Disallowed (%) | 0 | 0 |
Values in parentheses are for the highest-resolution shell. Rmerge = Σhkl Σi|Ii(hkl) − 〈I(hkl)〉|/Σhkl ΣiIi(hkl), where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections; I, the intensity of a reflection; σI, the standard deviation of intensity; B factor, a mix of thermal displacement and disorder.
Fig. 2.
Overall structure of N5 showing the calcium binding site, N-linked glycosylation site, and an enzymatic active site with and without zanamivir binding. (A) Overall structure of the N5 homotetramer. The active site is located on top of the molecule and is highlighted in blue. Calcium ions are shown as red spheres, and N-linked glycans are gold. One monomer is colored using a rainbow gradient to illustrate the canonical propeller arrangement, with six four-stranded, antiparallel sheets. (B) There is only one calcium ion binding site in each N5 monomer. A single calcium ion is coordinated by Asp293, Gly297, Asp324, Tyr347, and two water molecules, which are shown as blue spheres. (C) Native, uncomplexed N5 structure, with the open conformation of the 150-loop revealing the existence of a 150-cavity. (D) Zanamivir-complexed N5 structure, in which the 150-loop moves proximal to the zanamivir binding site and induces the closure of the 150-cavity. The binding also changes the orientation of the Asp151 side chain, which moves 2.04 Å (Cα) closer to the zanamivir 4-guanidino group. Binding of zanamivir to N5 is highly similar to that of the previously reported group 1 NA structures.
Uncomplexed N5 is strikingly similar to other typical group 1 NAs, as it indeed contains a 150-cavity (Fig. 2C). After zanamivir soaking, the 150-loop adopts a closed conformation, which is an induced conformational change shared by all other typical group 1 members, indicating the flexibility of the 150-loop and its involvement in ligand binding (Fig. 2D). Superimposition of the Cα atoms of the 150-loop of native N5 and its complex with zanamivir reveals a root mean square deviation (RMSD) of 2.04 Å. In the N5-zanamivir complex (Fig. 2D), Asp151 approaches zanamivir to hydrogen bond with its 4-guanidino group and shifts 2.04 Å (Cα atom) toward the inhibitor binding site, compared with uncomplexed N5. In accordance with the N8-zanamivir and VN04N1 (neuraminidase from Vietnam 2004 H5N1)-zanamivir complexes, Tyr347 of N5 is located on the top of zanamivir's carboxylate group and hydrogen bonds with it. It can be inferred that Tyr347 of N5 might also interact with the oseltamivir carboxylate group, which may lower the probability of developing oseltamivir resistance for N5 (17, 24). We then found that Tyr347 is highly conserved in group 1 avian influenza NAs, including three NAs mentioned above, whereas for human virus NAs, the residue at position 347 is not Tyr but commonly Glu, Asp, His, or Asn. Furthermore, based on the recent simulation study on the substrate binding of H5N1 NA, we speculate that the highly conserved Tyr347 among group 1 avian virus NAs might play an important role in recognizing the α-2,3-linked sialic acid avian type receptor (9). More experiments are clearly needed to support this hypothesis. Taken together, these results demonstrate that N5 possesses the common characteristics of the reported typical group 1 NAs.
In addition, regarding the calcium ion binding site, two calcium ions were found in each monomer of the tetramer in most of the known NA structures (18, 19), except 18N1 and 09N1, which contain three (24). However, only one calcium ion binding site was observed in N5, which is formed with the aid of Asp293, Gly297, Asp324, Tyr347, and two water molecules (Fig. 2B); this difference of N5 from other known structure NAs might be attributed to our crystallization condition in which there is no calcium ion. This site has been unambiguously resolved in most of the known structures and was suggested to be important for the enzyme activity and stability of NA (18). Furthermore, two N-linked glycosylation sites in the globular head region, Asn93 and Asn146, were identified; however, N5 lacks N-linked glycans at Asn88 and Asn234, which contrasts with avian N1, which has three N-linked glycans at Asn88, Asn146, and Asn234 at the same region (24).
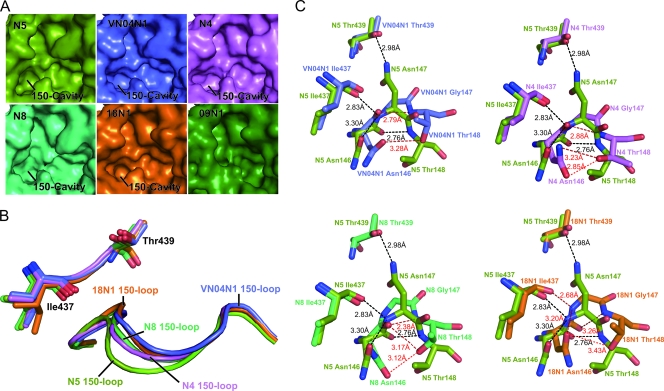
Upon closer comparison of the uncomplexed N5 active site with those of all other known structure group 1 NAs, it is clear that the N5 150-cavity is extended relative to those of all other group 1 structures (Fig. 3 A). Superimposition of 150-loops from all available NAs with an open conformation indicates that the unusual 150-cavity of N5 is attributed mainly to a more extended 150-loop (Fig. 3B). Based upon detailed comparison (Fig. 3C), the 150-loops of VN04N1, N4, N8, and 18N1 (1918 pandemic H1N1 neuraminidase) with Gly147 were found to adopt a conformation much closer to that of the center of the active site. For VN04N1, N4, and N8, the side chains of Asn146 and Thr148 are able to hydrogen bond with each other and thus help to maintain the conformation of their 150-loops. Furthermore, there is a hydrogen bond between the Asn146-Gly147 peptide carbonyl and Gly147-Thr148 peptide nitrogen, which further stabilizes the 150-loop conformation of VN04N1, N5, and N8.
Fig. 3.
Comparison of the unique conformation of the N5 (splitpea) 150-cavity and 150-loop to those of VN04N1 (tv_blue), N4 (violet), N8 (lime green), and 18N1 (orange). (A) Comparison of molecular surfaces from all currently available group 1 NA structures, in which N5 displays an extended 150-cavity relative to those of other known structure group 1 members. (B) Comparison of N5's 150-loop with those of N4, N8, 04VN1, and 18N1. The N5 150-loop adopts an extended conformation relative to those of 04VN1, N4, N8, and 18N1. (C) Comparison of detailed polar interactions of residues 146, 147, and 148 of N5 with those of N4, N8, 04VN1, and 18N1. N5 hydrogen bonds are colored black, and N4, N8, 04VN1, and 18N1 hydrogen bonds are colored red. The upper left panel displays superimposition of residues 146 to 148 from N5 and VN04N1, the upper right displays superimposition of residues 146 to 148 from N5 and N4, the lower left displays superimposition of residues 146 to 148 from N5 and N8, and the lower right displays superimposition of residues 146 to 148 from N5 and 18N1. The presence of Asn147 in N5 promotes a further distance from Thr439, which is a key factor in the extension of the N5 150-loop relative to those in VN04N1, N4, N8, and 18N1. The distance between the Asn146 and Thr148 side chains is also further for N5 than it is for VN04N1, N4, N8, and 18N1. Furthermore, the Asn146-Asn147 peptide bond in N5 and 18N1 is rotated relative to those of VN04N1, N4, and N8, which creates an altered hydrogen bond network.
In the case of N5 (Fig. 3C), Asn147 allows for a unique hydrogen bond with Thr439. The lifting of the N5 150-loop away from the active site relative to the other group 1 structures is necessary for the Asn147 and Thr439 hydrogen bond to form at a distance of 2.98 Å. In N5, the Asn146-Asn147 peptide bond is in a rotated conformation relative to that of N4, N8, and VN04N1. This orientation leads to an altered hydrogen bond network in N5. Notably, the Asn146-Asn147 peptide nitrogen of N5 is able to form an extra hydrogen bond with the Ile437-Trp438 peptide carbonyl in addition to the hydrogen bond between the Asn146-Asn147 peptide carbonyl and the Asn147-Thr148 peptide nitrogen. Moreover, the distance between the Asn146 and Thr148 side chains is greater in the extended N5 150-loop than in VN04N1, N4, N8, and 18N1. These interactions, taken together, may explain why the N5 150-loop adopts a more extended conformation.
In conclusion, we have successfully solved the crystal structure of the last group 1 serotype, N5, and discovered some unique characteristics especially regarding a special, extended 150-cavity, due to the polymorphic amino acid Asn147. The common characteristic, especially the open 150-loop like those of other typical group 1 NAs, indicates that any new drugs targeting the group 1-specific 150-cavity should work for N5.
Acknowledgments
This work was supported by the China Ministry of Science and Technology (MOST, 973, project grant 2011CB504703). G.F.G. is a leading principal investigator of the Innovative Research Group of National Natural Science Foundation of China (NSFC grant 81021003).
We thank Yi Shi, Di Liu, Enguang Feng, Hong Liu, and Hualiang Jiang for discussions and reagents.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Air G. M., Laver W. G. 1989. The neuraminidase of influenza virus. Proteins 6:341–356 [DOI] [PubMed] [Google Scholar]
- 3. Babu Y. S., et al. 2000. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 43:3482–3486 [DOI] [PubMed] [Google Scholar]
- 4. Collaborative Computational Project, number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 5. Colman P. M. 1994. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 3:1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colman P. M., Varghese J. N., Laver W. G. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41–44 [DOI] [PubMed] [Google Scholar]
- 7. Das K., Aramini J. M., Ma L. C., Krug R. M., Arnold E. 2010. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 17:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 9. Jongkon N., Sangma C. 27 April 2011. Receptor recognition mechanism of human influenza A H1N1 (1918), avian influenza A H5N1 (2004), and pandemic H1N1 (2009) neuraminidase. J. Mol. Model. [Epub ahead of print.] doi:10.1007/s00894-011-1071-4 [DOI] [PubMed] [Google Scholar]
- 10. Kim C. U., et al. 1997. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 119:681–690 [DOI] [PubMed] [Google Scholar]
- 11. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 12. Li Q., et al. 2010. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat. Struct. Mol. Biol. 17:1266–1268 [DOI] [PubMed] [Google Scholar]
- 13. Murshudov G. N., Vagin A. A., Dodson E. J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 14. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 15. Read R. J. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373–1382 [DOI] [PubMed] [Google Scholar]
- 16. Rudrawar S., et al. 2010. Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat. Commun. 1:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Russell R. J., et al. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49 [DOI] [PubMed] [Google Scholar]
- 18. Smith B. J., et al. 2006. Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding. Acta Crystallogr. D Biol. Crystallogr. 62:947–952 [DOI] [PubMed] [Google Scholar]
- 19. Taylor G., Garman E., Webster R., Saito T., Laver G. 1993. Crystallization and preliminary X-ray studies of influenza A virus neuraminidase of subtypes N5, N6, N8 and N9. J. Mol. Biol. 230:345–348 [DOI] [PubMed] [Google Scholar]
- 20. Varghese J. N., Laver W. G., Colman P. M. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303:35–40 [DOI] [PubMed] [Google Scholar]
- 21. von Itzstein M. 2007. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6:967–974 [DOI] [PubMed] [Google Scholar]
- 22. von Itzstein M., et al. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418–423 [DOI] [PubMed] [Google Scholar]
- 23. Wright P. F., Neumann G., Kawaoka Y. 2007. Orthomyxoviruses, p. 1691–1740 In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 24. Xu X., Zhu X., Dwek R. A., Stevens J., Wilson I. A. 2008. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 82:10493–10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamashita M., et al. 2009. CS-8958, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob. Agents Chemother. 53:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang W., et al. 2010. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell 1:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]