Abstract
African swine fever virus (ASFV), the causative agent of one of the most devastating swine diseases, has been considered exclusively cytoplasmic, even though some authors have shown evidence of an early stage of nuclear replication. In the present study, an increment of lamin A/C phosphorylation was observed in ASFV-infected cells as early as 4 h postinfection, followed by the disassembling of the lamina network close to the sites where the viral genome starts its replication. At later time points, this and other nuclear envelope markers were found in the cytoplasm of the infected cells. The effect of the infection on the cell nucleus was much more severe than previously expected, since a redistribution of other nuclear proteins, such as RNA polymerase II, the splicing speckle SC-35 marker, and the B-23 nucleolar marker, was observed from 4 h postinfection. All this evidence, together with the redistribution, dephosphorylation, and subsequent degradation of RNA polymerase II after ASFV infection, suggests the existence of sophisticated mechanisms to regulate the nuclear machinery during viral infection.
INTRODUCTION
Viruses are obligate intracellular parasites that have evolved many diverse strategies to remodel the infected cell, thus providing an ideal environment for their replication and optimal virus production.
The nucleus of the infected cell plays an essential role during most viral infections. While some viruses, such as retroviruses, replicate entirely within the nucleus (42), some others are considered nucleocytoplasmic viruses due to the fact that they have an early stage of nuclear replication (20). New evidence demonstrates the relevance of the nucleus and/or its components, even for viruses traditionally considered cytoplasmic (19, 29, 41). This is the case for one of the most complex viruses found in the animal kingdom, African swine fever virus (ASFV). Despite ASFV being the sole member of the family Asfarviridae (9), it has been phylogenetically incorporated within the nucleocytoplasmic large DNA virus clade, together with iridoviruses, phycodnaviruses, mimiviruses, and poxviruses, forming an individual lineage with poxviruses (20, 21). ASFV and poxviruses share several characteristics that have caused them to be considered purely cytoplasmic viruses, including their capacity to encode a wide range of enzymes that could allow self-replication and transcription, theoretically without needing the cell machinery. In spite of the above-mentioned data, there is some evidence indicating an early stage of nuclear replication during ASFV infection (13, 30, 35, 40).
In situ hybridization and autoradiography experiments revealed ASF viral DNA in the nuclei of infected cells (macrophages and Vero cells) at early times of viral DNA synthesis (13), confirming largely ignored pioneer studies done 20 years ago showing, for the first time, ASFV DNA within the nuclei of infected macrophages (40) or the incapability of ASFV to replicate in enucleated cells (30). Today, we know that small DNA fragments are synthesized intranuclearly in proximity to the nuclear membrane at an early time, whereas at later times, larger fragments and mature cross-linked ASFV DNA are found in the cytoplasm, suggesting some kind of egress from the nuclear membrane (13, 35).
Little is known about the pathway/s of viral and/or ASFV DNA entrance within the nucleus and about the mechanism(s) of its egress from the nucleus to the cytoplasm. Considering the large size of the ASFV genome, ranging between 170 and 193 kbp (5), an active transport process of the ASFV DNA associated with shuttling viral and cellular proteins has been proposed, with the structural p37 protein playing a key role in this process (11). Although a similar mechanism could be hypothesized to explain the transport of the ASFV DNA to the cytoplasm, some evidence pointed toward a budding-like process through the nuclear envelope (NE) being responsible for DNA egress (13).
In the present study, we investigated the dramatic modifications suffered by the host nucleus early after ASFV infection using a combination of three-dimensional (3D) immunofluorescence in situ hybridization (immuno-FISH) experiments imaged by confocal microscopy and biochemical assays. On one hand, a disruption of peripheral lamin A/C, underlying the inner nuclear membrane (INM), was evident as early as 6 h postinfection (p.i.) close to the sites where the newly synthesized ASFV DNA is found. Helping to explain the mechanisms of lamin disruption, an increment of lamin A/C phosphorylation was observed in ASFV-infected cells as early as 4 h p.i. At later times postinfection, lamin A/C and nucleoporin p62, a nuclear pore marker, were found within the cytoplasm and viral factories.
On the other hand, the concomitant reorganization of the nucleoplasmic lamin A/C, the B23 nucleolar marker, and the SC35 splicing speckle marker, together with the redistribution, dephosphorylation, and subsequent degradation of RNA polymerase II (RNA Pol II), indicates an early impairment of the cell cycle regulation, including cellular transcription (8, 18, 38, 44). The disruption and reorganization of nuclear components during the initial steps of ASFV infection indicate a more serious involvement of the nucleus during ASFV infection than was previously believed.
MATERIALS AND METHODS
Cell culture and virus.
Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% fetal calf serum at 37°C in a 5% CO2 humidified atmosphere.
Infections were carried out using Percoll-purified ASFV (strain BA71V) adapted to grow in Vero cells (10). Percoll purification has been described previously (7).
ASFV-specific DNA probe and nuclear markers used for immunofluorescence (IF).
The probe used for DNA FISH was obtained from the viral DNA (strain E75L) isolated from purified virions (3). The probe was labeled with biotin using the BioPrime DNA labeling system (Invitrogen, Barcelona, Spain) and purified with the Qiagen nucleotide removal kit (Qiagen, Barcelona, Spain).
Nuclear proteins were detected using mouse monoclonal antibodies against lamin A/C (1:20), nucleoporin p62 (1:10) (Santa Cruz Biotechnology, Inc.), RNA Pol II H5 (1:50) (Covance, CA), SC-35 (1:500) (Sigma, Steinheim, Germany), and nucleophosmin/B23 (1:300) (Invitrogen, Barcelona, Spain). Polyclonal rabbit antibody against the structural protein p54 (1:100) was used to detect the viral factories (1).
3D immuno-FISH.
Vero cells (250,000 cells/ml suspension) were cultured on glass coverslips and mock infected or infected with 5 PFU per cell of purified BA71V. In order to preserve the cellular 3D structure, a previously described protocol was performed (2). Briefly, mock-infected and ASFV-infected Vero cells were fixed at different times postinfection (4, 6, 8, 12, and 18 h p.i.) with 4% paraformaldehyde (PFA) for 20 min. The cells were then permeabilized in 0.5% Triton X-100 for 25 min at room temperature (RT) and incubated in 0.1 N HCl for 5 min at RT, and the RNA was removed with 200 μg/ml RNase in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C for 30 min. The cells were kept in 50% formamide-2× SSC until hybridization. Three hundred nanograms of DNA probe was precipitated with 3 μg salmon sperm DNA and resuspended in 18 μl of hybridization mix. The probe was denatured at 75°C for 10 min and preannealed at 37°C for 90 min. The cells were then denatured at 75°C for 3 min, and hybridization was performed at 37°C for 24 h. Next, the cells were washed once in 50% formamide-2× SSC at 45°C for 3 min, four times in 2× SSC at 45°C for 3 min, and four times in 0.1× SSC at 60°C for 3 min and then blocked twice in 0.12% BSA-1× phosphate-buffered saline (PBS)-0.1% Tween 20 at RT for 10 min.
To complete the FISH, detection was performed with either streptavidin-fluorescein isothiocyanate (FITC) (1:250) (BD Biosciences, NJ) or streptavidin-tetramethyl rhodamine isocyanate (TRITC) (1:500; Zymed Laboratories, CA) for 1 h at 37°C.
After the FISH protocol, the cells were washed three times in 1× PBS for 5 min and blocked with 3% bovine serum albumin (BSA)-1× PBS for 30 min. The cells were then incubated with the primary antibodies in the blocking solution for 1 h at RT. After three washes (5 min each) with 1× PBS, incubation with the secondary antibodies was performed for 1 h at RT. Different fluorescein-conjugated anti-mouse antibodies were used throughout this study: Cy2 (1:100), Cy3 (1:500), Cy5 (1:500), and FITC (1:100) (Jackson Immunoresearch Europe Ltd.) and anti-rabbit-FITC (1:500) (Sigma). Finally, after three washes with 1× PBS, nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (1 μg/ml) and mounted with Vectashield.
Immunofluorescence microscopy.
Vero cells, mock infected or infected with 5 PFU per cell, were fixed with 4% PFA and permeabilized with 0.5% Triton X-100 as described above for the FISH protocol. Then, the cells were blocked with 3% BSA-1× PBS for 1 h and incubated with the primary antibodies for 1 h in the blocking solution at RT. After three washes with 1× PBS (5 min each), the cells were incubated with secondary antibodies for 1 h in blocking solution at RT. Finally, nuclei were counterstained with DAPI (1 μg/ml) and mounted with Vectashield.
Image acquisition and processing.
Fluorescence images were viewed on a Nikon eclipse 90i epifluorescence microscope equipped with a DXM 1200F camera (Nikon Corporation, Japan). Image stacks were captured using a Leica TCS SP5 confocal microscope (×40, 1.25-numerical-aperture [NA] and ×63, 1.40-NA objectives). Z stack images were acquired at intervals of 0.3 to 0.4 μm. The images were processed by using the LAS AF Lite program from Leica and Image J v1.44e software (http://rsb.info.nih.gov/ij).
Western blot analysis.
Western blot analysis was performed using cell extracts from either uninfected or infected Vero cells at different times postinfection. Samples were washed with 1× PBS and then lysed in cold buffer containing 50 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM EDTA, 10% Triton X-100, and a protease inhibitor cocktail (Roche). Cell extracts were quantified, and equal quantities of protein from each sample were resolved in 6% or 12% SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with the following primary antibodies: a rabbit polyclonal antibody against lamin A/C (1:1,000) from Cell Signaling; the monoclonal antibodies H5 and 8WG16 (1:500 in both cases) from Covance, recognizing the hyperphosphorylated form at serine 2 and the hypophosphorylated form of RNA Pol II, respectively; the polyclonal antibody N-20 (1:500) from Santa Cruz, recognizing the N-terminal part of the largest RNA Pol II subunit; a mouse monoclonal antibody against β-actin (1:5,000) from Sigma; and a rabbit polyclonal antibody against α-tubulin (1:1,000) from Sigma. Protein detection was performed using peroxidase-conjugated antibodies and an ECL system (GE Healthcare Life Sciences).
Metabolic labeling of proteins with [32P]orthophosphate.
Vero cells were mock infected or infected with ASFV at a multiplicity of infection (MOI) of 5. After 1 h of adsorption, the inoculum was removed and the cells were incubated with medium containing 100 μCi/ml of [32P]orthophosphate (Perkin Elmer). At 4 h p.i., the medium was removed and the cells were washed with 1× PBS and disrupted in lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM EDTA, 10% Triton X-100) in the presence of the complete Mini EDTA-free protease inhibitor (Roche) and phosphatase inhibitors (500 μM sodium orthovanadate, 500 μM β-glycerophosphate, and 500 μM sodium molybdate). After centrifugation at 12,000 × g for 15 min at 4°C, the supernatants were collected and immunoprecipitation was performed. Briefly, labeled cell lysates were incubated overnight at 4°C in a rotator with a rabbit anti-lamin A/C polyclonal antibody (1:100), and next, 30 μl of protein A/G Plus-agarose (Santa Cruz Biotechnology, Inc.) was added for 2 additional hours at 4°C. Finally, the immune complexes were spun down by centrifugation at 13,000 × g for 30 s at 4°C, and after three washes in lysis buffer, the pellets were resuspended in 1× Laemmli sample buffer, loaded in 12% SDS-polyacrylamide gels, and processed for autoradiography. Densitometry of the labeled bands was performed using QuantityOne software (Bio-Rad), and the increase of lamin phosphorylation was corrected for the total amount of lamin A and C present in the cell extracts (measured by Western blotting).
RESULTS
The nuclear lamina is phosphorylated and disassembled during the initial phase of ASFV replication.
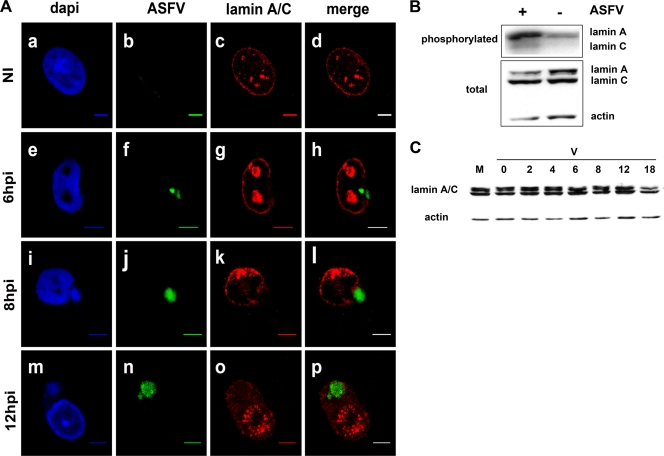
To characterize the integrity of the NE, 3D immuno-FISH experiments were performed on uninfected and infected Vero cells harvested at different times postinfection, using a DNA probe to specifically detect the ASFV nucleic acid, followed by staining with a specific antibody against lamin A/C. Viral DNA was detectable from 4 h p.i. and was mainly found at that time as faint dots within the cell cytoplasm in proximity to the nuclear envelope (data not shown). At this time point, lamin A/C showed the same distribution as in uninfected cells (data not shown). In comparison with uninfected cells (Fig. 1A, a to d), the nuclear distribution of lamin A/C was clearly disrupted at 6 h p.i. in close proximity to the sites where the viral DNA was located (Fig. 1A, e to h). The intensities of both the DAPI counterstaining and the ASFV-specific probe labeling showed an exponential accumulation of nascent viral DNA at 6 h p.i. (Fig. 1A, e and f, respectively), coinciding with the initial phase of viral DNA replication (13, 35). Supporting the involvement of the nucleus in the early events of ASFV infection, a very evident phosphorylation of lamin A and C was found as early as 4 h p.i., increasing 4.9 and 2.9 times, respectively, compared with mock-infected Vero cells (Fig. 1B). Nuclear lamin disruption was still evident at 8 h p.i. (Fig. 1A, k), and at 12 h p.i., lamin A/C staining was also found diffusely distributed within the cytoplasm of the infected cells (Fig. 1A, o). As expected, ASFV DNA accumulation reached its maximum in the cytoplasm at late times postinfection (Fig. 1A, m and n), where the final stage of replication and ASFV DNA maturation takes place (35). ASFV DNA was undetectable before 4 h p.i. and after 18 h p.i., coinciding with the detection of the major capsid protein p72 (data not shown), most probably indicating its inclusion in fully assembled viral particles.
Fig. 1.
Peripheral lamin A/C phosphorylation and disruption at early times in ASFV infection. (A) 3D immuno-FISH of Vero cells uninfected (NI) and infected at 6, 8, and 12 h p.i. with 5 PFU of the Ba71V strain of ASFV. Nuclei were counterstained with DAPI (blue signal in a, e, i, and m), labeled with the FITC-conjugated ASFV genome probe (green signal in b, f, j, and n), and stained with a monoclonal antibody against lamin A/C (red signal in c, g, k, and o). Scale bars, 5 μm. (B) ASFV infection induces increased phosphorylation of lamin A and C in Vero cells. (Top) Autoradiography showing the immunoprecipitated 32P-labeled lamin A/C proteins obtained from mock-infected (−) and ASFV-infected (+) Vero cells at 4 h p.i. (Bottom) Western blot analysis showing the total amounts of lamin A/C and β-actin contained in each cell extract. (C) Degradation of nuclear lamin A/C at late times after ASFV infection. Western blot analysis of cell extracts from mock-infected (M) and ASFV-infected (V) Vero cells at different times postinfection (0, 2, 4, 6, 8, 12, and 18 h p.i.) using a polyclonal antibody against nuclear lamin A/C and an anti-β-actin antibody as an endogenous control.
Nuclear membrane markers are found in ASFV factories late after infection.
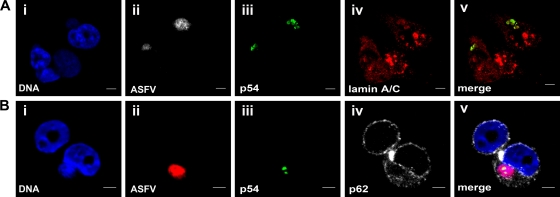
To confirm the presence of nuclear lamin A/C in the viral factories at late times postinfection, FISH (Fig. 2A, ii) combined with double IF was performed using an anti-lamin A/C antibody (panel iv in Fig. 2A) and a polyclonal antibody against p54 (Fig. 2A, iii), a late structural ASFV protein that specifically stains the viral factories (34). p54 showed perfect colocalization with lamin A/C at late times postinfection (Fig. 2A, v). However, no direct interaction between the two proteins was demonstrable in coimmunoprecipitation studies using cell extracts from ASFV-infected cells at any of the times postinfection tested (data not shown). Interestingly, lamin A/C staining covers larger regions of the cytoplasm (Fig. 2A, iv) than the ASFV DNA (Fig. 2A, ii) or the p54 protein (Fig. 2A, iii), clearly demonstrating that this NE marker can be found outside the viral factories within the cytoplasm of infected cells.
Fig. 2.
Localization of nuclear envelope markers in viral factories at 12 h p.i. with ASFV. Shown is 3D double immuno-FISH of Vero cells infected at 12 h p.i. with 5 PFU of the BA71V strain of ASFV. FISH was performed using the ASFV probe, followed by double IF using antibodies against nuclear envelope proteins and the ASFV protein p54 to label viral factories. (A) Lamin A/C localization in viral factories. (i) DAPI-counterstained DNA is shown in blue. (ii) ASFV DNA was visualized with streptavidin-TRITC, shown in gray. (iii and iv) 3D double immunofluorescence was developed using a polyclonal antibody against the ASFV marker p54, revealed with anti-rabbit-FITC (green signal in iii) and a monoclonal antibody against lamin A/C revealed with anti-mouse-Cy5 (red signal in iv). (v) Colocalization of both markers (yellow signal). (B) Nucleoporin p62 localization in viral factories. (i) DAPI-counterstained DNA is shown in blue. (ii) ASFV DNA was visualized with streptavidin-TRITC, shown in red. (iii and iv) 3D double immunofluorescence was developed using a polyclonal antibody against the ASFV marker p54, revealed with anti-rabbit-FITC (green signal in iii) and a monoclonal antibody against nucleoporin p62 revealed with anti-mouse-Cy5 (gray signal in iv). (v) Merged image. Scale bars, 5 μm.
The egress of ASFV DNA from the nucleus could imply sequestering of other components of the NE to the viral factories, where ASFV-morphogenesis takes place. To confirm this hypothesis, FISH experiments (Fig. 2B, ii) were followed by double IF using anti-p54 (Fig. 2B, iii) and a monoclonal antibody against nucleoporin p62 (Fig. 2B, iv), a specific marker of NE. In contrast with lamin A/C staining, p62 did not colocalize with p54, showing a diffused cytoplasmic labeling around the viral factories (Fig. 2B, v).
Disruption of nuclear organization during the initial phase of ASFV infection.
Lamin A/C is a multifunctional protein found not only in the nuclear periphery, but also in the nucleoplasm, participating in chromatin organization (8). In the present study, lamin A/C was also found in the nucleoplasm of infected cells. In contrast to uninfected cells (Fig. 1A, c), enlarged foci of lamina aggregates were localized throughout the nonnucleolar regions of the nucleoplasm of infected cells at early times postinfection (Fig. 1A, g and k). At later times postinfection (≥12 h p.i.), a more punctate labeling of lamina aggregates was found throughout the nucleus (Fig. 1A, o), corresponding to areas without DAPI staining (Fig. 1A, m). This nuclear lamina redistribution (both perinuclear and nucleoplasmic) was not accompanied by an increment of expression, as revealed by the Western blot analysis performed in Vero cells infected at different times postinfection (Fig. 1C). Conversely, and only at very late time points postinfection, a specific degradation of lamin A/C was observed (Fig. 1C).
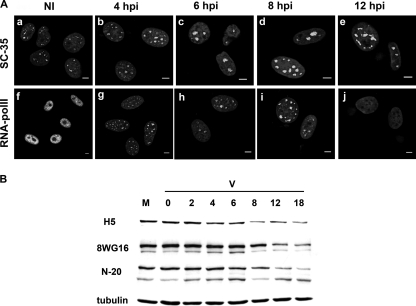
Due to the fact that similar lamin A/C aggregation in nucleoplasmic foci has been associated with a redistribution of transcriptionally related markers (22, 23, 38, 39), comparative IF experiments were performed on uninfected and ASFV-infected cells using antibodies against SC-35 (a marker specifically labeling the splicing speckles) and RNA Pol II H5 (a marker specifically labeling only the active, hyperphosphorylated form of polymerase II at serine 2).
The SC35 marker showed a distribution similar to that of lamin A/C, with ASFV-infected cells showing enlarged splicing speckles (Fig. 3A, b to e), most probably reflecting the inhibition of cellular transcription (6, 23, 39, 44). This hypothesis was confirmed with the results obtained using an antibody against the active form of RNA Pol II. While noninfected cells showed the typical diffused distribution of RNA Pol II (Fig. 3A, f), the enzyme was immediately recruited to very well defined areas of the nucleoplasm at early time points of infection (Fig. 3A, g to i), to finally disappear from 12 h p.i. (Fig. 3A, j).
Fig. 3.
(A) Redistribution of transcriptionally related nuclear markers during ASFV infection. Intranuclear detection of splicing speckles (SC-35) and RNA Pol II in Vero cells uninfected (NI) and infected at 4, 6, 8, and 12 h p.i. with 5 PFU of BA71V. Specific monoclonal antibodies and Cy2- or FITC-conjugated anti-mouse antibodies were used for the IF in all cases. Scale bars, 5 μm. (B) Dephosphorylation and degradation of RNA Pol II during ASFV infection. Vero cells were mock infected (M) or infected with the BA71V strain of ASFV (V). At different times postinfection, cell extracts were analyzed by Western blotting with antibodies against RNA Pol II Ser-2P (H5), hypophosphorylated RNA Pol II (8WG16), and the N-terminal part of the largest RNA Pol II subunit (N-20). Anti-α-tubulin was used as an endogenous control.
Western blot analysis with antibodies that recognize different forms of RNA Pol II allowed us to confirm that during ASFV infection, an early hypophosphorylation of RNA Pol II occurred, followed by the degradation of the enzyme at late time points of infection (Fig. 3B). Thus, at 4 and 6 h p.i., coinciding with the nuclear redistribution of RNA Pol II, there was a reduction in the hyperphosporylated form of RNA Pol II (recognized by the H5 antibody) and a proportional increase in the hypophosphorylated band of RNA Pol II (a smaller band detected with the 8WG16 antibody) (32). The dramatic reduction of all forms of RNA Pol II was evident from 8 h p.i., as shown with the N-20 RNA Pol II polyclonal antibody (Fig. 3B).
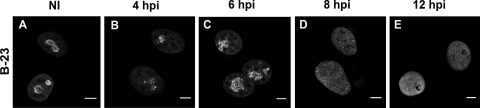
Apart from the above-described effects, ASFV infection severely affected other essential nuclear structures, such as the nucleolus, a nuclear structure playing a key role in many cell-regulatory processes, including ribosome biogenesis and the cell cycle (18). As for the rest of the markers studied, a very clear redistribution was found for the B23 nucleolar marker in ASFV-infected cells. While one or two well-defined nucleoli were visualized in uninfected cells (Fig. 4A), they lost definition as early as 4 h p.i., with B23 staining being found forming much smaller foci than in uninfected cells (Fig. 4B). From 6 h p.i., the B23-specific staining started to be found dispersed within the nucleus, finally showing a diffuse nucleoplasmic pattern (Fig. 4C and D) that at 12 h p.i. excluded the nucleoli of ASFV-infected cells (Fig. 4E).
Fig. 4.
Nucleolar disruption during ASFV infection. Intranuclear detection of the nucleolar B-23 marker in Vero cells uninfected (NI) and infected at 4, 6, 8, and 12 h p.i. with 5 PFU of BA71V. A specific monoclonal anti-B23 antibody and a Cy2-conjugated anti-mouse antibody were used for immunofluorescence. Scale bars, 5 μm.
DISCUSSION
Our results clearly demonstrate an early disorganization of nuclear components during ASFV infection, probably reflecting the viral strategies to control the cell cycle, from the disruption of the NE to nucleoplasmic reorganization of essential components for cellular transcription and replication.
The involvement of the NE in viral replication has been previously demonstrated for many other viruses (43). In particular, the nuclear lamina, a proteinaceous mesh described as essential to maintain the integrity of the NE (27), has also been shown to play a key role during infection with different RNA and DNA viruses (37, 41). The multifunctional role of lamin A/C protein during herpesvirus infection has been well characterized. Lamin A/C seems to play a key role during lytic herpesvirus infection by recruiting viral DNA and chromatin-modifying enzymes at the nuclear periphery in order to maintain chromatin in an active conformational stage (37). Additionally, herpesviruses can also provoke the recruitment of cellular and viral kinases to phosphorylate lamin A/C, producing its disassembly (16, 24, 25, 26, 28). In a similar way, the early phosphorylation and disassembly of lamin A/C might facilitate the interaction of ASFV DNA with nuclear proteins required for proper viral DNA conformation and/or initiation of the replication process. The presence of a serine protein kinase in the ASF viral particle (4) allows us to speculate about early phosphorylation of lamin A/C immediately after virus uncoating, as soon as the viral nucleoprotein complex reaches the nucleus. We are currently addressing this specific issue.
At later time points, both lamin A/C and nucleoporin p62 were found in the cytoplasm of the infected cells. Although far from conclusive, our results seem to support the hypothesis of a budding-like process through the NE being responsible for DNA egress (13). This result was also supported by experimental data showing that chemical inhibition of the CRM1-mediated nuclear export pathway did not affect the kinetics of ASFV replication (11). To our knowledge, this is the first study in which these NE proteins have been found in the cytoplasm of infected cells, although other nuclear proteins have been found in the cytoplasm of cells infected with vaccinia virus (29) or ASFV (15). Attempts to describe direct interactions between lamina and/or nucleoporin p62 and viral components (coimmunoprecipitation and electron microscopy studies) have failed or produced inconclusive results (data not shown), even though such interactions cannot be ruled out. Further studies are needed to elucidate the functional role of lamin A/C and nucleoporin p62 during ASFV infection.
Apart from maintaining the integrity of the NE, the lamina has also been implicated in DNA replication and chromatin organization (for reviews, see references 8 and 27). A reorganization of nucleoplasmic lamin A/C in large nucleoplasmic foci was observed from very early after infection. A similar picture has been previously described during herpesvirus infections (24, 25), in the presence of transcription inhibitors (23) or hypertonic conditions (12). Nucleoplasmic lamin A/C redistribution has been correlated with both enlargement of the splicing speckles and the inhibition of RNA Pol II-dependent transcription (23, 39), providing evidence for an important role of this protein in the organization of transcriptional and splicing events (23). Our results clearly demonstrate that early during ASFV infection, intranuclear lamin A/C, SC-35, and RNA Pol II are reorganized in enlarged nucleoplasmic foci, suggesting a possible mechanism to block cellular transcription early during viral infection. This hypothesis seems to reinforce previous experiments in which a sudden decrease in the incorporation of [3H]uridine was observed at 4 h p.i. in cells infected with ASFV compared with mock-infected cells (M. L. Salas, unpublished data). Following nuclear reorganization, RNA Pol II dephosphorylation and degradation were produced, thereby confirming that ASFV does not need the enzyme to replicate (36) and contributing to the cellular shutoff described during ASFV infection (33).
To our knowledge, this is the first report showing the inactivation of RNA Pol II in ASFV-infected cells. Whether these events are directly driven by viral or cellular proteins is not fully known, although structural proteins and/or polypeptides expressed very early after the infection might be involved in these processes.
There are several viral structural proteins with the potential to play a role in the above-described events. The structural protein p37 has already been suggested to participate in the nuclear transport of viral DNA immediately after virus uncoating (11). As with p37, many other proteins might also be transported as nucleoprotein complexes together with DNA, including the above-described serine-protein kinase (4) or p30 and p54, two of the best-characterized ASFV structural antigens. On one hand, p30, an early phosphoprotein that binds the heterogeneous nuclear ribonucleoprotein K within the nuclei of infected cells, might be involved in downregulation of cellular mRNA translation (17). On the other hand, p54 is a multifunctional protein that mediates the transport of ASFV to the microtubular organizing center (MTOC) (perhaps in the form of a multiprotein-DNA complex) through its binding to the light chain of dynein (1). In silico studies clearly demonstrated the presence of a nuclear localization signal (RKKK) in p54 (reliability, 76.7) (31), supporting the possibility that the protein might also play a role in the transport of DNA to the nucleus. Similarly, viral proteins containing strong nucleolar targeting signals (14) might be involved in nucleolar disorganization during ASFV infection. Redistribution of nucleolar markers has been reported before for other viral infections as altering the normal function of the cell and tightly regulating the cellular cycle during viral infection (18).
In summary, our results clearly demonstrate that nuclear interactions early during infection with ASFV are more important than previously believed. For the first time, clear phosphorylation and disassembly of lamin A/C was observed early after infection close to the sites where the viral genome starts its replication. On the other hand, disruption of both NE components and intranuclear structures was shown, leading to the inactivation of RNA Pol II through its recruitment to enlarged transcription speckles and ulterior dephosphorylation and degradation. Further studies will be necessary to understand the molecular mechanisms governing all these cellular modifications. These discoveries might have important implications when searching for antiviral strategies. Lessons learned from other intranuclear or nucleocytoplasmic viruses might be useful to fight against ASF, a disease that is currently causing real economic problems in many sub-Saharan African countries.
ACKNOWLEDGMENTS
We thank G. Andrés, A. Bensaid, R. Cordón, M. J. Bustos, H. Domingo, C. Kress, A. D. Sánchez, A. Olvera, and M. del Rosal for their technical help and support and A. Nieto and C. Alonso for providing RNA Pol II and anti-p54 antibodies, respectively. The manuscript was edited by Kevin Dalton.
M.B. and F.R. are financially supported by contracts from the Juan de la Cierva and I3 programs, respectively, and from the Spanish Ministry of Science and Innovation, which also financed the CONSOLIDER-Porcivir CDS2006-00007, AGL2007-66441-C03-01/GAN, and AGL2010-22229-C03-01 research projects. Work at the Centro de Biología Molecular Severo Ochoa was supported by grants from the Wellcome Trust (075813/C/04/Z) and the Spanish Ministerio de Ciencia e Innovación (BFU2007-61647) and by an institutional grant from Fundación Ramón Areces.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Alonso C., et al. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 75:9819–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballester M., et al. 2008. The nuclear localization of WAP and CSN genes is modified by lactogenic hormones in HC11 cells. J. Cell. Biochem. 105:262–270 [DOI] [PubMed] [Google Scholar]
- 3. Ballester M., et al. 2010. Intranuclear detection of African swine fever virus DNA in several cell types from formalin-fixed and paraffin-embedded tissues using a new in situ hybridisation protocol. J. Virol. Methods 168:38–43 [DOI] [PubMed] [Google Scholar]
- 4. Baylis S. A., Banham A. H., Vydelingum S., Dixon L. K., Smith G. L. 1993. African swine fever virus encodes a serine protein kinase which is packaged into virions. J. Virol. 67:4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blasco R., de la Vega I., Almazán F., Aguero M., Viñuela E. 1989. Genetic variation of African swine fever virus: variable regions near the ends of the viral DNA. Virology 173:251–257 [DOI] [PubMed] [Google Scholar]
- 6. Bregman D. B., Du L., van der Zee S., Warren S. L. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carrascosa A. L., del Val M., Santerén J. F., Viñuela E. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dechat T., Adam S. A., Goldman R. D. 2009. Nuclear lamins and chromatin: when structure meets function. Adv. Enzyme Regul. 49:157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dixon L. K., et al. 2005. Asfarviridae, p. 135–143 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom [Google Scholar]
- 10. Enjuanes L., Carrascosa A. L., Moreno M. A., Viñuela E. 1976. Titration of African swine fever (ASF) virus. J. Gen. Virol. 32:471–477 [DOI] [PubMed] [Google Scholar]
- 11. Eulálio A., et al. 2007. African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times. Virus Res. 130:18–27 [DOI] [PubMed] [Google Scholar]
- 12. Favale N. O., Sterin Speziale N. B., Fernandez Tome M. C. 2007. Hypertonic-induced lamin A/C synthesis and distribution to nucleoplasmic speckles is mediated by TonEBP/NFAT5 transcriptional activator. Biochem. Biophys. Res. Commun. 364:443–449 [DOI] [PubMed] [Google Scholar]
- 13. García-Beato R., Salas M. L., Viñuela E., Salas J. 1992. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology 188:637–649 [DOI] [PubMed] [Google Scholar]
- 14. Goatley L. C., et al. 1999. Nuclear and nucleolar localization of an African swine fever virus protein, I14L, that is similar to the herpes simplex virus-encoded virulence factor ICP34.5. J. Gen. Virol. 80:525–535 [DOI] [PubMed] [Google Scholar]
- 15. Granja A. G., et al. 2004. Modulation of p53 cellular function and cell death by African swine fever virus. J. Virol. 78:7165–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamirally S., et al. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernáez B., Escribano J. M., Alonso C. 2008. African swine fever virus protein p30 interaction with heterogeneous nuclear ribonucleoprotein K (hnRNP-K) during infection. FEBS Lett. 582:3275–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiscox J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hiscox J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iyer L. M., Aravind L., Koonin E. V. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iyer L. M., Balaji S., Koonin E. V., Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184 [DOI] [PubMed] [Google Scholar]
- 22. Jagatheesan G., et al. 1999. Colocalization of intranuclear lamin foci with RNA splicing factors. J. Cell Sci. 112:4651–4661 [DOI] [PubMed] [Google Scholar]
- 23. Kumaran R. I., Muralikrishna B., Parnaik V. K. 2002. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 159:783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C. P., et al. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 82:11913–11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marschall M., et al. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357–33367 [DOI] [PubMed] [Google Scholar]
- 26. Milbradt J., Webel R., Auerochs S., Sticht H., Marschall M. 2010. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285:13979–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moir R. D., Spann T. P. 2001. The structure and function of nuclear lamins: implications for disease. Cell. Mol. Life Sci. 58:1748–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 29. Oh J., Broyles S. S. 2005. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 79:12852–12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ortín J., Viñuela E. 1977. Requirement of cell nucleus for African swine fever virus replication in Vero cells. J. Virol. 21:902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reinhardt A., Hubbard T. 1998. Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res. 26:2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodríguez A., Pérez-Gónzalez A., Nieto A. 2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 81:5315–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodríguez J. M., Salas M. L., Santarén J. F. 2001. African swine fever virus-induced polypeptides in porcine alveolar macrophages and in Vero cells: two-dimensional gel analysis. Proteomics 1:1447–1456 [DOI] [PubMed] [Google Scholar]
- 34. Rodríguez J. M., García-Escudero R., Salas M. L., Andrés G. 2004. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 78:4299–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rojo G., García-Beato R., Viñuela E., Salas M. L., Salas J. 1999. Replication of African swine fever virus DNA in infected cells. Virology 257:524–536 [DOI] [PubMed] [Google Scholar]
- 36. Salas J., Salas M. L., Viñuela E. 1988. Effect of inhibitors of the host cell RNA polymerase II on African swine fever virus multiplication. Virology 164:280–283 [DOI] [PubMed] [Google Scholar]
- 37. Silva L., Cliffe A., Chang L., Knipe D. M. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spann T. P., Moir R. D., Goldman A. E., Stick R., Goldman R. D. 1997. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136:1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spann T. P., Goldman A. E., Wang C., Huang S., Goldman R. D. 2002. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156:603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tabarés E., Sánchez-Botija C. 1979. Synthesis of DNA in cells infected with African swine fever virus. Arch. Virol. 61:49–59 [DOI] [PubMed] [Google Scholar]
- 41. Uchil P. D., Kumar A. V., Satchidanandam V. 2006. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 80:5451–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whittaker G. R., Kann M., Helenius A. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627–651 [DOI] [PubMed] [Google Scholar]
- 43. Zebovitz E., Leong J. K., Doughty S. C. 1974. Involvement of the host cell nuclear envelope membranes in the replication of Japanese encephalitis virus. Infect. Immun. 10:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng C., Kim E., Warren S. L., Berget S. M. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]