Abstract
The analysis of HIV-1 envelope carbohydrates is critical to understanding their roles in HIV-1 transmission as well as in binding of envelope to HIV-1 antibodies. However, direct analysis of protein glycosylation by glycopeptide-based mass mapping approaches involves structural simplification of proteins with the use of a protease followed by an isolation and/or enrichment step before mass analysis. The successful completion of glycosylation analysis is still a major analytical challenge due to the complexity of samples, wide dynamic range of glycopeptide concentrations, and glycosylation heterogeneity. Here, we use a novel experimental workflow that includes an up-front complete or partial enzymatic deglycosylation step before trypsin digestion to characterize the glycosylation patterns and maximize the glycosylation coverage of two recombinant HIV-1 transmitted/founder envelope oligomers derived from clade B and C viruses isolated from acute infection and expressed in 293T cells. Our results show that both transmitted/founder Envs had similar degrees of glycosylation site occupancy as well as similar glycan profiles. Compared to 293T-derived recombinant Envs from viruses isolated from chronic HIV-1, transmitted/founder Envs displayed marked differences in their glycosylation site occupancies and in their amounts of complex glycans. Our analysis reveals that the glycosylation patterns of transmitted/founder Envs from two different clades (B and C) are more similar to each other than they are to the glycosylation patterns of chronic HIV-1 Envs derived from their own clades.
INTRODUCTION
The systematic mapping and characterization of protein glycosylation provide a wealth of molecular information that is crucial for understanding a wide variety of biochemical and cellular processes. However, comprehensive analysis of protein glycosylation has proven to be difficult due to the wide dynamic range of glycopeptide concentrations and immense structural diversity of glycans. Glycan modifications on proteins undergo a series of glycan processing steps from the endoplasmic reticulum (ER) to the Golgi apparatus with a diverse array of glycan processing enzymes that compete for available substrate, resulting in multiple glycosylation patterns for each glycosylation site for a given protein and variation in glycosylation site occupancy (35, 52, 53). Moreover, protein glycosylation varies significantly across different cell types, cell states, tissues, and organisms (3, 12, 16, 34, 35, 53, 56, 62). Despite these challenges, recent advances in proteomics have accelerated the pace of the development of efficient methods and technologies that can be tailored for the analysis of protein glycosylation (6, 51, 66). To date, the analysis of protein glycosylation by mass spectrometry (MS) is underpinned by an array of sample preparation methods that include affinity/enrichment schemes (6, 18, 26), modern chromatographic methods (31, 50, 54), and remarkable improvements in mass spectrometry instrumentation (47, 48, 51). When used effectively, these methods provide a means to qualitatively and quantitatively profile the global glycosylation of a given glycoprotein sample.
An essential requirement to globally profile protein glycosylation and maximize the information content of a given glycosylation site is the characterization of glycosylation in a site-specific fashion. A typical analysis employs a bottom-up proteomics approach wherein glycoproteins are enzymatically digested using a specific or nonspecific protease to generate the peptide/glycopeptide mixture that is subsequently analyzed by a mass spectrometry (MS) platform in tandem with the chromatographic separation of choice (2, 11, 14, 18, 29, 48, 51, 66). Following data acquisition, all possible glycopeptides are identified by peaks that are separated by monosaccharide units and/or by peaks of characteristic glycopeptide-marker ions in the MS1 and tandem-MS (MS/MS) data (9, 21, 66). The data sets that are generated are then analyzed with downstream software tools to deduce the glycopeptide compositions, thereby elucidating glycosylation site occupancy and the glycan motif at a given glycosylation site.
While this approach has proven to be useful, the high complexity of glycoprotein samples imposes restrictions on the detection sensitivity of glycopeptides. Aside from having low ionization efficiency compared to coeluting peptides, ionization of glycopeptides is also obscured by ion suppression effects and competition for ionization with coeluting glycopeptides in complex biological milieu (11, 68). As a result, glycopeptides that are present in relatively low abundance are detected with reduced sensitivity or may be not detected at all. Herein, we demonstrate an alternative approach to achieve a more comprehensive glycopeptide profile involving the use of glycosidases, wherein glycopeptides are partially or completely deglycosylated prior to mass analysis. This approach affords the benefits of accurate identification of glycosylation site occupancy and the identification of the type of glycan populating a particular site, and it facilitates the characterization of glycopeptides that poorly ionize in a given elution window or are not detected at all when other glycopeptides are present, thus maximizing glycosylation coverage. We have used this technology to perform glycosylation site occupancy analysis and glycopeptide profiling of proteins of great biological interest—recombinant HIV-1 Env oligomers derived from transmitted/founder viruses.
A single transmitted/founder virus quasispecies has been found to be involved in 70 to 80% of heterosexually transmitted infections, but the characteristics of transmitted/founder HIV-1 strains that permit transmission are not known (8, 13, 32, 58). The types of glycans on recombinant viral Env are beginning to be characterized (5, 34), and the carbohydrates on recombinantly expressed envelopes may not be fully representative of those glycans on virion Env spikes (15). Nonetheless, comparison of recombinant transmitted/founder and chronic HIV-1 Env glycosylation with new carbohydrate analysis technology is an important initial analysis, primarily because of the anticipated use of recombinant transmitted/founder Envs in human immunogenicity trials. The RV144 Thai HIV-1 vaccine efficacy trial used a canarypox virus expressing envelope as a vector prime followed by a bivalent Env gp120 protein boost (57), and new efficacy trials will almost certainly also contain Env protein boosts. Moreover, transmitted/founder viruses differ from chronic viruses in their biological functions, including infectivity of macrophages (58), and in their ability to bind to the integrin homing receptor α4β7 (49). In particular, the ability of recombinant transmitted/founder Envs to bind to α4β7 while chronic Envs bind less well may in part be due to their glycan profile (49).
If, when using new technology, site-specific carbohydrates vary between transmitted/founder and chronic Envs, then it will additionally warrant the even more difficult studies of site-specific carbohydrates of transmitted/founder and chronic virion trimers on CD4 T-cell-derived virions, to determine if Env carbohydrates contribute to transmitted/founder virus success in establishing the initial productive infection.
In the present study, we describe the utility of glycosidases to characterize the glycosylation profiles of two recombinant HIV-1 transmitted/founder Env oligomers based on the envelope proteins (Envs) derived from a clade B transmitted/founder virus, B.700010040.C9 (33), and from a clade C transmitted/founder virus, C.1086 (1). HIV-1 Env is heavily glycosylated, with at least 24 N-linked glycosylation sites (10, 17, 39, 45, 67, 69). Glycosylation profiles of HIV-1 Env have been characterized by structural analysis of chemically or enzymatically released glycans (45, 46, 56) or by glycopeptide-based analysis (10, 17, 19, 20, 29, 39, 65, 68, 69). In this study, we employed a glycopeptide-based mass mapping approach that includes a sequential digestion with glycosidases and trypsin prior to liquid chromatography-electrospray ionization-Fourier transform ion cyclotron resonance MS (LC/ESI-FTICR MS) analysis to maximize glycosylation information and determine the N-glycosylation profiles inherent to two transmitted/founder Envs, where “transmitted/founder” Envs are defined as Envs derived from the early env gene sequences of acutely infected individuals that coalesce to a consensus env gene sequence of the transmitted virus (1, 33). Our results show that both transmitted/founder Envs had a similar degree of glycosylation site occupancy, and >50% of the sites are either variably utilized or completely nonutilized. When glycosylation sites are utilized, 20% and 30% of glycosylation sites on B.700010040.C9 gp140 ΔC and C.1086 gp140 ΔC, respectively, are populated exclusively with high-mannose glycans. Five conserved glycosylation sites in both transmitted/founder Envs are populated with high-mannose glycans. Compared to the glycosylation profiles of the Envs derived from viruses isolated from chronic HIV-1 infections, B.JR-FL and C.97ZA012 (19, 20), the transmitted/founder Envs differed in their glycosylation patterns in terms of site occupancy and regarding levels of complex glycans present compared to those present on chronic HIV-1 recombinant Envs.
MATERIALS AND METHODS
Reagents.
Ammonium bicarbonate, Trizma hydrochloride, Trizma base, EDTA, high-performance liquid chromatography (HPLC)-grade acetonitrile (CH3CN) and methanol (CH3OH), urea, dithiothreitol (DTT), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), iodoacetamide (IAM), ammonium hydroxide, ammonium acetate, glacial acetic acid, and formic acid were purchased from Sigma (St. Louis, MO). Water was purified using a Millipore Direct-Q3 water purification system (Billerica, MA). Sequencing-grade trypsin was obtained from Promega (Madison, WI). Glycerol-free peptide-N-glycosidase F (PNGase F) cloned from Flavobacterium meningosepticum and endo-β-N-acetylglucosaminidase H (Endo H) cloned from Streptomyces plicatus, were purchased from New England BioLabs (Ipswich, MA). Sialidase A cloned from Arthrobacter ureafaciens and endo-β-N-acetylglucosaminidase F3 (Endo F3) cloned from Elizabethkingia meningoseptica, were obtained from Prozyme (Hayward, CA) and EMD Biosciences (Gibbstown, NJ), respectively.
Expression and purification of transmitted/founder HIV-1 subtype B and C envelope proteins.
B.700010040.C9 and C.1086 envelope proteins were constructed (41). Acute HIV-1 envelope gene sequences of B.700010040.C9 and C.1086 were isolated from individuals with acute subtype B and subtype C HIV-1 infections, respectively, by single genome amplification (SGA) (1, 58). To produce recombinant soluble gp140 oligomer proteins, gp140 ΔC env genes were designed by introducing a stop codon before the membrane-spanning domain, mutating two critical Arg residues in the gp120-gp41 cleavage site, and preserving all other regions of the extracellular domain of the env gene. The B.700010040.C9 and C.1086 gp140 env genes were codon optimized by converting amino acid sequences to nucleotide sequences employing the codon usage of highly expressed human housekeeping genes (4, 41), de novo synthesized (Blue Heron, Bothell, WA), and cloned into pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA). The resulting recombinant plasmids were linearized by digestion with restriction endonuclease SspI (Invitrogen, Carlsbad, CA) and used to generate stably transfected 293T cells (ATCC, Bethesda, MD) under selection of hygromycin B (Sigma, St. Louis, MO) at 200 μg/ml. Recombinant HIV-1 gp140ΔC Envs were purified from serum-free supernatant of the stably transfected 293T cells by lectin agarose beads (Galanthus nivalis; Vector Laboratories, Burlingame, CA) (41) and stored at −80°C until use. Prior to any mass spectrometric experiments, Env samples were thawed and aliquoted into separate tubes containing ∼75 μg of the Env sample and stored at −80°C until analysis.
Glycosidase digestion of envelope proteins.
Deglycosylation of Envs using PNGase F was performed by incubating ∼75 μg of the Env sample (protein concentration of >4 mg/ml) with 1 μl of PNGase F solution (500,000 U/ml) for a week at 37°C at pH 8.5. Endo H and Endo F3 deglycosylation experiments were performed by incubating ∼75 μg of the Env (protein concentration of >4 mg/ml) with the enzyme solutions. For the Endo H deglycosylation experiment, samples were denatured with 2 M urea in 100 mM Tris buffer (pH 5.5), followed by the addition of 2 μl of Endo H (≥5 U/ml). After thorough mixing, the reaction mixture was incubated for 48 h at 37°C. In the Endo F3 deglycosylation experiment, samples were deglycosylated by addition of 2 μl of Endo F3 (≥5 U/ml) to a reaction mixture containing 75-μg Env samples in 50 μM NH4C2H2O2 (pH 4.5). The reaction mixture was incubated for 2 weeks at 37°C. As a quality control experiment, the Endo F3 and Endo H studies were first conducted on the HIV-1 Env B.JR-FL. The results from these experiments indicated that site occupancy analysis with Endo H and Endo F3 produced results identical to those that occurredwhen the site occupancy was determined by PNGase F. Desialylation with sialidase A was performed by adding 2 μl of sialidase A (≥5 U/ml) to 75-μg Env samples at pH 6.5, with the mixture incubated overnight at 37°C. Prior to tryptic digestion, the pH of the deglycosylated Env samples was adjusted to 8.5 with 300 mM NH4OH. Deglycosylated Env samples were digested with trypsin as described below.
Digestion of envelope proteins.
The total protein concentration, which was measured by absorbance, was >4 mg/ml, and samples containing 75 μg of the HIV-1 Envs were used for protein digestion. Proteins were denatured with 6 M urea in 100 mM Tris buffer (pH 8.5) containing 3 mM EDTA and were fully reduced using either 15 mM DTT or 5 mM TCEP at room temperature for 1 h. TCEP was used for samples treated with glycosidases due to its stability and efficiency at breaking disulfide bonds at low pH (25). Both TCEP- and DTT-reduced samples were alkylated with 20 mM IAM at room temperature for another hour in the dark. Excess IAM in both samples was quenched by adding DTT to a final concentration of 25 mM, and the mixture was incubated for 20 min at room temperature. The reduced and alkylated Env samples were digested with trypsin (30:1 protein/enzyme ratio) at 37°C and incubated overnight, followed by a second trypsin addition under the same conditions. The resulting HIV-1 envelope protein digest was either directly analyzed by LC/ESI-FTICR MS or stored at −20°C until further analysis. To ensure reproducibility of the method, protein digestion was performed at least three times on different days with Env samples obtained from the same batch and analyzed with the same experimental procedure.
Mass spectrometry.
LC/ESI-FTICR MS experiments were performed using a hybrid linear ion-trap (LIT) Fourier transform (FT) ion cyclotron resonance mass spectrometer (LTQ-FT; ThermoScientific, San Jose, CA) directly coupled to the Dionex UltiMate capillary LC system (Sunnyvale, CA) equipped with a FAMOS well plate autosampler. Mobile phases utilized for the experiment consisted of solvent A (99.9% deionized H2O plus 0.1% formic acid) and solvent B (99.9% CH3CN plus 0.1% formic acid). Five microliters of the sample (∼7 μM) was injected onto a C18 PepMap 300 column (300-μm inside diameter [i.d.] by 15 cm, 300 Å; LC Packings, Sunnyvale, CA) at a flow rate of 5 μl/min. The following CH3CN/H2O multistep gradient was used: 5% solvent B for 5 min followed a linear increase to 40% solvent B in 50 min, and then a linear increase to 90% solvent B in 10 min. The column was held at 95% solvent B for 10 min for reequilibration. A short wash and blank run were performed between every sample to ensure there was no sample carryover. The ESI source was operated under the following conditions: source voltage of 2.8 kV, capillary temperature of 200°C, and capillary offset voltage of 44 V. Data were collected in the positive-ion mode in a data-dependent fashion in which the five most intense ions in an FT scan at resolution (R)=25,000 at m/z 400 were sequentially and dynamically selected for subsequent collision-induced dissociation (CID) in the LTQ linear ion trap using a normalized collision energy of 30% and a 3-min dynamic exclusion window followed by two neutral loss scans when a loss of a monosaccharide unit was detected in the MS2 scan. (Note that under these conditions, the resolution, R, at m/z 1,000 is 10,000, and at m/z 1,500, R is 6,700.)
Glycopeptide identification.
Compositional analyses were performed using GlycoPep DB (21), GlycoPep ID (28), and GlycoMod (9). Details of the analysis for glycopeptides have been described previously (19, 20, 29). Briefly, for glycopeptides with a singly utilized glycosylation site, compositional analysis was performed using the MS2 data. First, the peptide portion of a glycopeptide was identified in the tandem-MS spectrum. The peptide sequence was elucidated from the characteristic fragment of the glycosidic cleavage ion, Y1, using GlycoPep ID. Once the peptide sequence was identified, plausible glycopeptide compositions were obtained using the high-resolution MS data and GlycoPep DB, and the putative glycan candidate was confirmed manually by identifying the Y1 ion and inspecting the glycan fragmentation pattern from the tandem-MS data. For glycopeptides with multiply utilized glycosylation sites, experimental masses of glycopeptide ions from the high-resolution MS data were converted to singly charged masses and submitted to GlycoMod. This program calculates plausible glycopeptide compositions from the set of experimental mass values entered by the user, compares these mass values with theoretical mass values, and then generates a list of plausible glycopeptide compositions within a specified mass error. Plausible glycopeptide compositions in GlycoMod were deduced by providing the mass of the singly charged glycopeptide ion, enzyme, protein sequence, cysteine modification, mass tolerance, and possible types of glycans present in the glycopeptide. Plausible glycopeptide compositions obtained from the analysis were manually confirmed and validated from MS2 data.
Peptide identification.
Deglycosylated peptides were identified by searching raw data acquired on the hybrid LTQ FTICR mass spectrometer against a custom HIV database with 107 protein entries, obtained from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/content/), using Mascot, version 2.2.04 (Matrix Science, London, United Kingdom). The peak list was extracted from raw files using BioWorksBrowser, version 3.5 (Thermo Electron Corporation). DTA files were searched specifying the following parameters: (i) enzyme, trypsin; (ii) missed cleavage, 2; (iii) fixed modification, carbamidomethyl; (iv) variable modification, methionine oxidation, carbamyl, HexNAc, and dHexNAc; (v) peptide tolerance, 0.8 Da; and (vi) MS/MS tolerance of 0.4 Da. Peptides identified from the Mascot search were manually validated from MS2 data to ensure major fragmentation ions (b and y ions) were observed, especially for peptides generated from PNGase F-treated Envs that contain N-to-D conversions.
Hierarchical clustering of Envs based on glycosylation site occupancy.
Clustering analysis was performed for comparison of glycosylation site occupancy of the two acute and chronic Envs. The protein sequences of the Envs were aligned using ClustalW2 (37) to determine fixed, shifting, and missing glycosylation sites. Note that missing glycosylation sites are due to mutation or deletion. A table for glycosylation occupancy was generated, with each row corresponding to the site occupancy of each glycosylation site arranged according to the Env sequence position. The following values were used for site occupancy: 1 for a fully utilized site; 0 for an unutilized or missing site; and between 0 and 1 for a variably utilized site, specifically 0.25, 0.33, 0.5, and 0.67. These values were deduced depending on the number of potential N-glycosylation (PNG) sites for a given tryptic peptide and the degree of site occupancy. For tryptic peptides with (i) one PNG site and site occupancy of 0 and 1, the value is 0.5, and for those with (ii) two PNG sites and site occupancy of 0, 1, and 2, the values are 0.25, 0.33, 0.5, or 0.67, depending on how they are occupied. Heirarchical clustering was performed using the R language (version 2.11.1, 2010; R Foundation for Statistical Computation) (27). The hclust function was used for clustering the column by the Ward method for linkage. A heat map with a column dendrogram was generated using the heatmap.2 function.
RESULTS
Characteristics of HIV-1 Env from transmitted/founder viruses.
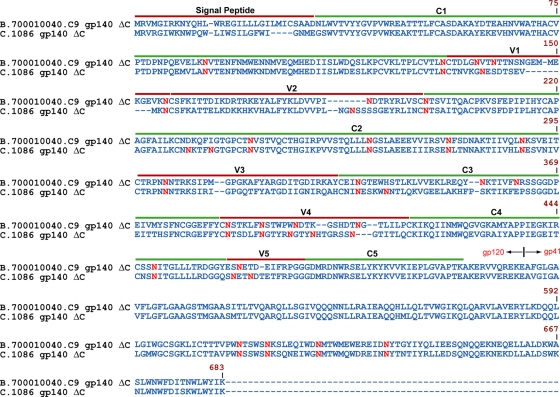
The transmitted/founder Envs used in this study were expressed in 293T stably transfected cell lines (7) and were derived from a clade B transmitted/founder virus (B.700010040.C9) (31) and a clade C transmitted/founder virus (C.1086) (1) with the deletion of the cleavage site (C) in the gp140 sequence. The full sequence alignment of the two transmitted/founder Envs showing the five conserved regions, the five hypervariable regions, and the gp41 region is shown in Fig. 1. Sequence alignment analysis using ClustalW2 (37) revealed that the protein sequences of the transmitted/founder clade B and C Envs were 71% identical. Potential N-linked glycosylation (PNG) sites are shown in red (Fig. 1), and overall, there were 25 and 27 PNG sites for B.700010040.C9 gp140 and C.1086 gp140, respectively. Comparison of the PNG sites of the transmitted/founder Envs revealed that 17 of the PNG sites were conserved. These sites are in the conserved Env regions C1 (N88 and N130), C2 (N197, N241, N262, N276, and N289), C3 (N334), and C4 (N448) and the hypervariable Env regions V1 (N156), V3 (N301), and V4 (N386 and N392), as well as in the gp41 region (N611, N616, N625, and N637). Numbering of glycosylation sites was based on the reference HIV-1 strain, HXB2 (UniProtKB/Swiss-Prot accession no. P04578) (67).
Fig. 1.
Sequence alignment of the early transmitted Env immunogens B.700010040.C9 gp140 ΔC and C.1086 gp140 ΔCF. Dashes indicate gaps in amino acid sequence and the location of the variable (V1 to V5) and conserved (C1 to C5) regions, with potential glycosylation sites in red. To standardize the sequence positions, the numbering was based on that of the reference HIV strain HXB2.
Site-specific glycosylation analysis of Envs derived from transmitted/founder viruses.
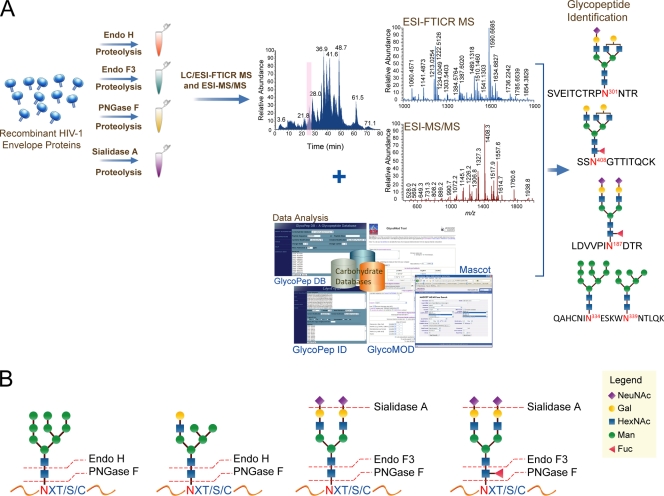
We implemented an experimental workflow for the characterization of the glycosylation of the transmitted/founder Envs, as shown schematically in Fig. 2A. This workflow included an up-front complete and partial enzymatic deglycosylation step using the endoglycosidases PNGase F, Endo H, and Endo F3 and an exoglycosidase, sialidase A, on separate aliquots of Env samples followed by an in-solution trypsin digestion. The resulting Env digests were analyzed by LC/ESI-FTICR MS, and glycopeptide compositions were deduced using the software tools Mascot, GlycoPep DB, GlycoMod, and GlycoPep ID (9, 21, 28), as shown in Fig. 2A. The inclusion of complete and partial deglycosylation in the sample preparation step generated complementary MS data sets from four separate glycosidase-treated Env samples that were subsequently integrated to obtain the overall glycosylation profiles of the transmitted/founder Envs. Our initial efforts to characterize the glycosylation of these Envs without the use of the glycosidases resulted in <40% of the putative glycosylation sites detected with <30% of the glycan compositions detected. Therefore, the newly implemented approaches were designed to increase the glycosylation coverage and maximize glycosylation information. Specifically, we obtained complete coverage of the glycosylation sites and a more than 2-fold increase in the number of glycan compositions detected.
Fig. 2.
(A) Experimental workflow for the glycopeptide-based mass mapping approach that includes a sequential digestion with endoglycosidase and trypsin. Glycopeptide compositions and site occupancy are determined using software-based analysis tools. (B) Glycan specificities of the glycosidases used to cleave glycans from substrate.
The glycosidases shown in Fig. 2B that were used in these experiments have the following glycan specificities. (i) PNGase F catalyzes the complete cleavage of glycans attached to the glycosylated asparagine (N) residue in the consensus sequence NXT/S/C, where X is any amino acid except proline, resulting in the conversion of N to D. (ii) Endo H and Endo F3 catalyze the cleavage of glycosidic bond between the two N-acetylglucosamine (HexNAc) residues in the chitobiose core, generating N-glycosylation sites with HexNAc or fucosylated HexNAc residues. Endo H cleaves only high-mannose and hybrid glycans, while Endo F3 cleaves complex glycans with specificity for bianternnary and trianternnary complex glycans (43, 61). (iii) Sialidase A catalyzes the cleavage of sialic acid residues. The use of these glycosidases followed by LC/ESI-FTICR analysis provided complementary information on the degree and type of glycosylation on B.700010040.C9 gp140 and C.1086 gp140 due to their selective glycan specificities. For instance, Env samples treated with Endo H permitted analysis of glycopeptides with complex glycans, while Env samples treated with Endo F3 permitted analysis of glycopeptides with high-mannose and hybrid glycans. In addition to the glycopeptide species, Endo H- and Endo F3-treated samples generated peptides with HexNAc or fucosylated HexNAc residues, thereby providing information on site occupancy.
Degree of glycosylation site occupancy.
Using the experimental workflow described in the previous section, we have identified all of the 25 and 27 potential N-linked glycosylation (PNG) sites from the tryptic digest of B.700010040.C9 gp140 and C.1086 gp140 Envs, respectively. Table 1 shows the identified glycopeptides with their corresponding glycosylation site occupancy. The mapping of the 25 and 27 PNG sites from B.700010040.C9 gp140 and C.1086 gp140, respectively, was accomplished by ∼20 tryptic peptides with single and multiple PNG sites per Env that were detected from the LC/ESI-FTICR MS and MS/MS analyses of endoglycosidase-treated Env samples. Deglycosylated peptides generated from endoglycosidase-treated Env samples would either have corresponding N-to-D conversion when PNGase F is used, or HexNAc/fucosylated HexNAcs are attached to glycosylated asparagine in the consensus sequence after treatment with Endo H or Endo F3. We relied mostly on the data obtained from Endo H- and Endo F3-treated Env samples to discern glycosylation site occupancy due the fact that a larger mass shift of 203.0793 Da (when a single HexNAc remains attached to N) or 349.1373 Da (when a fucosylated HexNAc remains attached to N) is observed for deglycosylated peptides, thus permitting the most conclusive assignments. With PNGase F, assignment of glycosylation site occupancy is not always straightforward due to a relatively small mass shift (0.9840 Da) when both deglycosylated and nonglycosylated forms of the same peptide coelute and the fact that potential deamidation artifacts could occur during digestion, resulting in false-positive identification. Accordingly, data obtained from PNGase F-treated Env samples were used only to determine glycosylation site occupancy of glycopeptides that were not detected from Endo H- and Endo F3-treated samples.
Table 1.
Glycopeptides detected from LC/ESI-FTICR MS analysis
| Glycopeptide sequencea | No. of potential glycosylation sites | Sites occupied |
|---|---|---|
| B.700010040.C9 gp140 ΔC | ||
| N88VTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | 1 | 1 |
| LTPLCVTLN130CTDLGN136VTN139TTNSNGEMMEK | 3 | 2 and 3 |
| N156CSFK | 1 | 0 and 1 |
| LDVVPIN186DTR | 1 | 1 |
| LVSN197TSVITQAPCK | 1 | 0 and 1 |
| QFIGTGPCTN241VSTVQCTHGIRPVVSTQLLLN262GSLAEEEVVIR | 2 | 2 |
| SVN276FSDNAK | 1 | 0 and 1 |
| TIIVQLN289K | 1 | 0 and 1 |
| SVEITCTRPNN301NTR | 1 | 0 and 1 |
| AYCEIN334GTEWHSTLK/KAYCEIN334GTEWHSTLK | 1 | 0 and 1 |
| EQYN356K/LREQYN356K | 1 | 1 |
| TIVFN362R | 1 | 0 and 1 |
| SSGGDPEIVMYSFNCGGEFFYCN386STK | 1 | 1 |
| LFN392STWPWN398DTK | 2 | 1 |
| GSHDTN409GTLILPCK | 1 | 0 and 1 |
| CSSN448ITGLLLLR | 1 | 0 and 1 |
| DGGYESN463ETDEIFRPGGGDMR | 1 | 0 and 1 |
| LICTTTVPWN611TSWSN616K | 2 | 0, 1, and 2 |
| SLEQIWN625MTWMEWER | 1 | 1 |
| EIDN637YTGYIYQLEIEESQNQQEK | 1 | 0 |
| C.1086 gp140 ΔC | ||
| EVHNVWATHACVPTDPNPQEMVLAN88VTENFNMWK | 1 | 0 and 1 |
| LTPLCVTLN130CTNVK | 1 | 0 and 1 |
| GN137ESDTSEVMK | 1 | 0 and 1 |
| N156CSFK | 1 | 0 and 1 |
| LDVVPLNGN184SSSSGEYR | 1 | 1 |
| LINCN197TSAITQAPCK | 1 | 0 and 1 |
| CNN230K | 1 | 1 |
| TFN234GTGPCR | 1 | 0 and 1 |
| N241VSTVQCTHGIKPVVSTQLLLN262GSLAEEEIIIR | 2 | 0, 1, and 2 |
| SEN276LTNNAK | 1 | 0 and 1 |
| TIIVHLN289ESVNIVCTRPNN301NTR | 2 | 0, 1, and 2 |
| QAHCNIN334ESK | 1 | 0 and 1 |
| WN339NTLQK | 1 | 0 and 1 |
| GEFFYCN386TSDLFN392GTYR | 2 | 0, 1, and 2 |
| N397GTYN401HTGR | 2 | 1 |
| SSN408GTITLQCK/SSN408GTITLQCKIK | 1 | 0 and 1 |
| AIYAPPIEGEITCNSN448ITGLLLLR | 1 | 0 and 1 |
| DGGQSN462ETN465DTETFRPGGGMR | 2 | 0 and 1 |
| LICTTAVPWN611SSWSN616K | 2 | 0, 1, and 2 |
| SQNEIWGN625MTWMQWDR | 1 | 1 |
| EINN637YTNTIYR | 1 | 1 |
PNGs are shown in boldface.
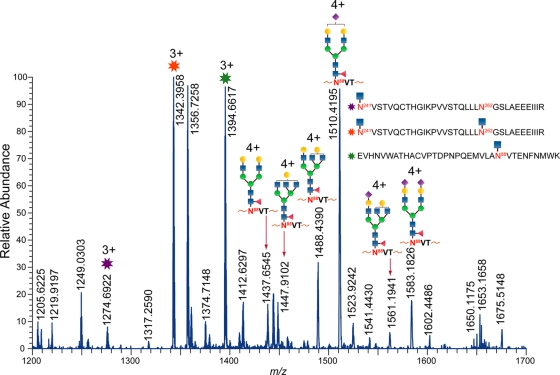
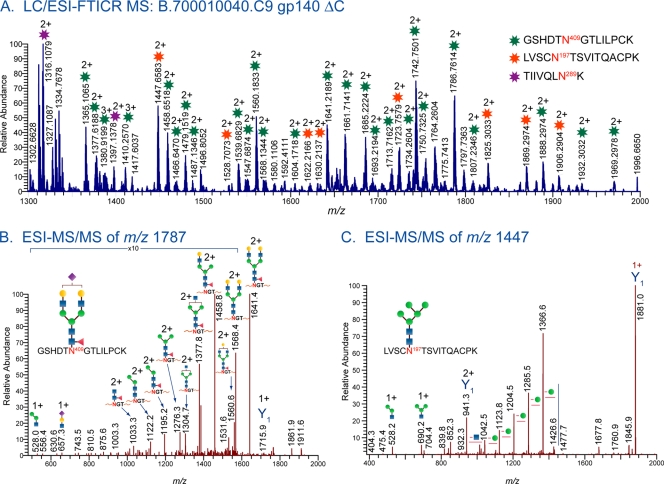
A representative piece of LC/ESI-FTICR MS data from the Endo H-treated clade C Env sample C.1086 gp140 depicts two peptides bearing HexNAc residues coeluting with several glycopeptides with the peptide portion EVHNVWATHACVPTDPNPQEMVLAN88VTENFNMWK (Fig. 3). The PNG is shown in boldface. Each identified peak in this high-resolution mass spectrum was validated from MS/MS data as described in Materials and Methods. This spectrum shows three partially deglycosylated peptides, EVHNVWATHACVPTDPNPQEMVLAN88VTENFNMWK, with one utilized glycosylation site and, N241VSTVQCTHGIKPVVSTQLLLN262GSLAEEEIIIR, with one and two utilized glycosylation sites. In addition to these deglycosylated peptides, the nonglycosylated form of the peptide, N241VSTVQCTHGIKPVVSTQLLLN262GSLAEEEIIIR, was also detected. These results are shown in Table 1, which summarizes the degree of glycosylation site occupancy for the two transmitted/founder Envs. The site occupancy analysis revealed that out of the 25 and 27 sites in B.700010040. C9 gp140 and C.1086 gp140, respectively, 13 sites in B.700010040.C9 gp140 and 20 sites in C.1086 gp140 are variably utilized. In addition to these variably utilized sites, the glycosylation sites N398 and N637 in B.700010040.C9 gp140 and the glycosylation sites N401 and N465 in C.1086 gp140 are not glycosylated at all. Not all of the variably utilized and unutilized glycosylation sites are located the same region of the two Envs when mapped onto the Env structure. Only 11 of the variably utilized glycosylation sites are common to both of the transmitted/founder Envs, and these sites are located in the V1-V2 (N156), C2 (N197, N276, and N289), V3 (N301), C3 (N334), V4 (N408/N409), C4 (N448), V5 (N462/N463), and gp41 (N611 and N616) regions. It should be noted that the env genes that expressed the transmitted Envs were codon optimized, as mentioned in Materials and Methods. At this point, it is unknown whether or not codon optimization has an impact on glycosylation site occupancy. Both the proteins in Fig. 1 and the proteins we characterized previously were codon optimized in the same fashion, so we would expect that in the unlikely event that codon optimization impacts glycosylation site occupancy, it would impact all of the proteins in Fig. 1 in the same manner—for example, all of the proteins would potentially have fewer unoccupied sites, if codon optimization were not done. Thus, codon optimization of recombinant env genes would not be expected to explain glycan differences observed between the transmitted/founder Envs and the chronic Envs.
Fig. 3.
LC/ESI-FTICR mass spectrum of the Env C.1086 gp140 ΔC, which was sequentially digested with Endo H and trypsin. Endo H cleaves high-mannose and hybrid glycans, leaving a single HexNAc on the glycosylated site while complex glycans are unaffected. Several peaks are present resulting from Endo H digestion on two tryptic peptides, EVHNVWATHACVPTDPNPQEMVLAN88VTENFNMWK in the C1 region and N241VSTVQCTHGIKPVVSTQLLLN262GSLAEEEIIIR in the C2 region. These peptides contain one and two HexNAcs, and these species coelute with glycopeptides with complex-type glycans and peptide portion EVHNVWATHACVPTDPNPQEMVLAN88VTENFNMWK. In addition, the nonglycosylated form of N241VSTVQCTHGIKPVVSTQLLLN262GSLAEEEIIIR was also observed. The peptide sequences were confirmed from tandem-MS data. This is a demonstration of using endoglycosidases to identify site occupancy for glycopeptides.
Partial deglycosylation using Endo H and Endo F3.
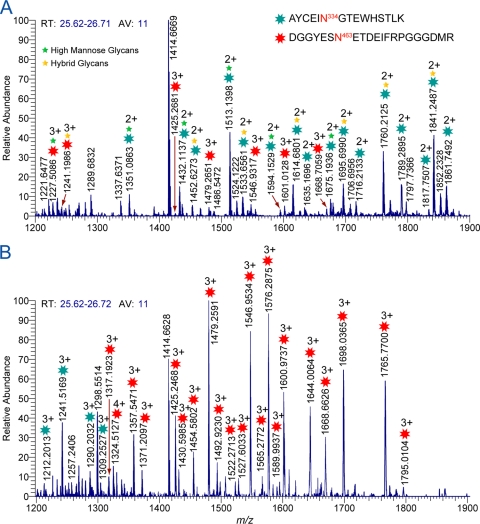
The use of Endo H and Endo F3 for partial release of N-glycans affords several benefits. First as described in the previous section and elsewhere (22-24, 44, 59, 60), these enzymes can be used to unambiguously identify glycosylation site occupancy since a clear distinction is made between partially deglycosylated and nonglycosylated forms of the same peptide. In addition to the above benefit, we show that these enzymes are useful for the reduction of glycan heterogeneity in the LC/MS data, which facilitates the characterization of glycopeptides that do not ionize efficiently or are not detected at all when all of the glycopeptides are present. An illustrative example is shown in Fig. 4. This figure shows high-resolution mass spectra of two clade B Env samples, one digested with trypsin (Fig. 4A) and the other sequentially digested with Endo H and trypsin (Fig. 4B). The data show a glycopeptide-rich fraction with two coeluting glycopeptides containing the peptide portions AYCEIN334GTEWHSTLK and DGGYESN463ETDEIFRPGGGDMR. Glycan compositions of the observed glycopeptide peaks in the mass spectra were verified from tandem-MS data (data not shown) and software analysis tools. Compositional analysis of the glycopeptide peaks observed in Fig. 4A reveals that glycopeptides containing the peptide portion AYCEIN334GTEWHSTLK were mostly high-mannose and hybrid-type glycopeptides coeluting with several complex type glycopeptides containing the peptide portion DGGYESN463ETDEIFRPGGGDMR. Comparison of the MS data obtained from the Endo H-treated B.700010040.C9 gp140 sample in the same elution window shows ions with higher abundance corresponding glycopeptides containing complex glycans and the peptide portion DGGYESN463ETDEIFRPGGGDMR and smaller ions corresponding to complex glycopeptides with peptide portion AYCEIN334GTEWHSTLK, with and without sialylation (Fig. 4B). Endo H cleaves high-mannose/hybrid glycans, while complex glycans are unaffected, thereby reducing glycan heterogeneity. As a result, improvements in both glycosylation coverage and signal/noise (S/N) ratio were observed for glycopeptides with peptide portion DGGYESN463ETDEIFRPGGGDMR, and the detection of both sialylated and nonsialylated species with peptide portion AYCEIN334GTEWHSTLK was also possible (Fig. 4B). Clearly, the sequential digestion with Endo H and trypsin helps minimize ionization suppression, enhances ionization of glycopeptides outside the glycan specificity of Endo H, and increases the dynamic range of the experiment and therefore increases glycopeptide coverage.
Fig. 4.
LC/ESI-FTICR mass spectra of a glycopeptide-rich fraction showing two coeluting glycopeptides found in the C3 region and V5 loop for the B.700010040.C9 gp140 ΔC Env sample after trypsin digestion (A) and sequential digestion (B) with Endo H and trypsin eluting between 25.6 and 26.7 min. The number of scans for each spectrum was 11. Asterisks in the MS data denote glycopeptide peaks. Green and yellow stars on top of asterisks in panel A denote glycopeptides with high-mannose and hybrid glycans, respectively. Glycan compositions of each observed glycopeptide peak in the mass spectra were verified from tandem-MS data. These data show that after Endo H treatment, strongly ionizing high-mannose and hybrid glycopeptides are depleted and low-abundance glycopeptides that were suppressed in panel A now are detectable in panel B.
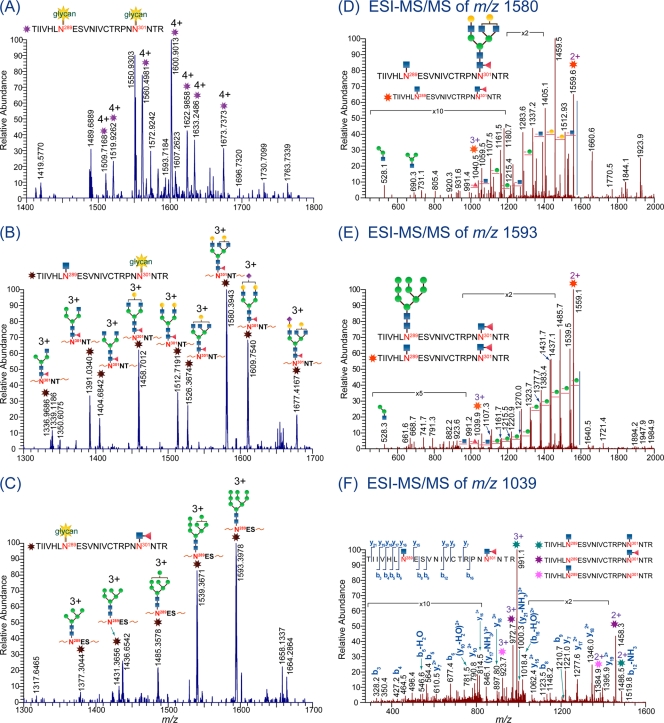
We also report another benefit from using the endoglycosidases. Due to the distinct glycan specificities of Endo H and Endo F3, the type of glycan attached at a particular glycosylation site of multiply glycosylated peptides can be deduced. More specifically, the use of these enzymes facilitates the determination of whether the site is populated with either high-mannose/hybrid-type glycans, complex-type glycans, or both. One example showing this benefit of using the glycosidases is in the analysis of glycopeptides from C.1086 gp140 with peptide portion TIIVHLN289ESVNIVCTRPNN301NTR, bearing two potential glycosylation sites at N289 and N301. Figure 5A shows the high-resolution mass spectrum of the glycopeptides that were not treated with Endo H or Endo F3. The data show the quadruply charged glycopeptide peaks with both of the glycosylation sites utilized. Compositional analysis of each glycopeptide peak in the spectrum is not straightforward due to ambiguity in the tandem-MS data arising from less intense peaks, missing diagnostic peaks, and peaks that exist in several charge states. Plausible compositions obtained from the compositional analysis of the quadruply charged peaks in Fig. 5A indicated that one of the sites is populated with high-mannose glycans and the other is populated with either bi- or triantennary complex glycans with core fucosylation (see Table S2 in the supplemental material). However, in the analysis of Env samples treated with Endo H or Endo F3, characterization of the glycan profile from these glycopeptides was straightforward (Fig. 5B and C). MS data of samples treated with Endo H (Fig. 5B) show glycopeptides containing one site occupied with complex glycans and the other with a single HexNAc. In contrast, when the same Env is treated with Endo F3 instead of Endo H (Fig. 5C), the glycopeptides for this part of the protein sequence have one occupied site containing high-mannose glycans and the other site containing fucosylated HexNAc. Taken together, the data in Fig. 5B and C clearly show that one of the two sites on this peptide contains high-mannose glycans, while the other site contains complex glycans. These assignments are supported by the MS/MS data shown in Fig. 5D and E. However, identification of which site is populated with high-mannose or complex glycans is not possible without the data in Fig. 5F. To accurately determine which site is populated with high-mannose or complex glycans, Env samples were treated with a combination of Endo H and Endo F3 to cleave all glycans except for the core HexNAc's with and without the attached core fucose. Tandem-MS analysis of the triply charged peak at m/z 1,039.5404 (Fig. 5F), corresponding to TIIVHLN289ESVNIVCTRPNN301NTR, with one site containing a single HexNAc and the other containing fucosylated HexNAc, revealed that N289 is populated with high-mannose glycans, while N301 is populated with complex glycans. These results show that Endo H and Endo F3 provide complementary information about the glycan profile of glycopeptides, and in some cases, they can be used to determine which glycans are present at which sites on multiply glycosylated peptides.
Fig. 5.
LC/ESI-FTICR MS data of the doubly glycosylated peptide TIIVHLN289ESVNIVCTRPNN301NTR in the C.1086 gp140 ΔC sample after trypsin digestion (A), sequential digestion with Endo H and trypsin (B), and sequential digestion with Endo F3 and trypsin (C). Glycan compositions in these high-resolution spectra (B and C) are verified from MS/MS data in shown in panels D and E. Note that panel D corresponds to the representative MS/MS data of a glycopeptide peak in panel B, while panel E corresponds to the representative MS/MS data of a glycopeptide peak in panel C. (F) MS/MS data of the peptide generated from sequential digestion of C.1086 gp140 ΔC Env sample with Endo F3, Endo H, and trypsin, confirming that N289 is populated with high-mannose glycans, while N301 is populated with complex glycans.
Glycan profile of transmitted/founder Envs.
Using the method described in Fig. 2A, glycan compositions were deduced from high-resolution and tandem mass spectra. The tandem-MS data were evaluated to elucidate the glycopeptide composition on the basis of the following criteria: (i) the presence of a fragment ion corresponding to the glycosidic cleavage ion, Y1, to determine the peptide portion of the glycopeptide, (ii) the presence of oxonium ions in the low-mass region (m/z <1,000), and (iii) the series of peaks that are separated by the mass of the monosaccharide units. Figure 6A shows a representative LC/ESI-FTICR mass spectrum from an elution window of glycopeptides located in the V2-C2, C2, and V4 regions for B.700010040.C9 gp140. The glycan composition of each peak in this high-resolution mass spectrum was determined using tandem-MS data as just described. Two examples of the MS/MS data are shown in Fig. 6B and C. The identified unique glycan compositions deduced from this analysis were verified with several experimental runs to check the reproducibility of the measurements in terms of glycan compositions and coverage. Overall, a high degree of glycosylation coverage was obtained, as demonstrated by the identification of ∼400 unique glycopeptide compositions per Env. Table 2 shows a partial list of the glycopeptides bearing high-mannose, hybrid, and complex glycans identified in this study (see Tables S1 and S2 in the supplemental material for the complete list). Akin to the glycan profile of the four Envs that we previously reported (19, 20), the glycosylation sites of the two transmitted/founder Envs, when utilized, are also populated with high-mannose, hybrid, and complex glycans with multiantennary structures with and without sialylation and core fucosylation. In addition, we also identified peptides with PNG sites modified with a single HexNAc with and without core fucosylation from Env samples that were not treated with glycosidases. Finally, in comparison to the Envs that we have previously analyzed, the transmitted/founder Envs have a higher level of sialylation.
Fig. 6.
(A) LC/ESI-FTICR mass spectrum of the glycopeptide-rich fraction showing three coeluting glycopeptides in the V2-C2, C2, and V4 regions for B.700010040.C9 gp140. (B and C) LC/ESI-MS/MS spectra showing the fragmentation pattern of the glycopeptides in the V4 loop (B) and the V2-C2 region (C). Note that peaks with asterisks in panel A correspond to glycopeptides.
Table 2.
Representative glycopeptide compositions for B.700010040.C9 gp140 ΔC and C.1086 gp140 ΔCa
| Env domain | Charge state |
m/z |
Mass error (ppm) | Peptide sequenceb | Glycan composition | |
|---|---|---|---|---|---|---|
| Experimental | Theoretical | |||||
| B.700010040.C9 gp140 ΔC | ||||||
| C1 | 4+ | 1,552.6714 | 1,552.6675 | 3 | NVTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | [Hex]5[HexNAc]6 |
| 4+ | 1,564.4305 | 1,564.4149 | 10 | NVTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | [Hex]6[HexNAc]4[NeuNAc]1 | |
| 4+ | 1,542.4202 | 1,542.4109 | 6 | NVTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | [Hex]6[HexNAc]5 | |
| 4+ | 1,615.1600 | 1,615.1847 | 15 | NVTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | [Hex]6[HexNAc]5[NeuNAc]1 | |
| 4+ | 1,593.2045 | 1,593.1807 | 15 | NVTENFNMWENNMVEQMHEDIISLWDQSLKPCVK | [Hex]6[HexNAc]6 | |
| V2 | 2+ | 1,455.6338 | 1,455.6340 | 0.1 | LDVVPINDTR | [Hex]5[HexNAc]4[Fuc]1 |
| 2+ | 1,601.1771 | 1,601.1817 | 3 | LDVVPINDTR | [Hex]5[HexNAc]4[Fuc]1[NeuNAc]1 | |
| 2+ | 1,746.7469 | 1,746.7294 | 10 | LDVVPINDTR | [Hex]5[HexNAc]4[Fuc]1[NeuNAc]2 | |
| 2+ | 1,557.1775 | 1,557.1737 | 2 | LDVVPINDTR | [Hex]5[HexNAc]5[Fuc]1 | |
| 2+ | 1,260.5483 | 1,260.5521 | 3 | LDVVPINDTR | [Hex]6[HexNAc]2 | |
| V4 | 2+ | 1,444.0971 | 1,444.0999 | 2 | LFNSTWPWNDTK | [Hex]6[HexNAc]2 |
| 2+ | 1,525.1272 | 1,525.1263 | 1 | LFNSTWPWNDTK | [Hex]7[HexNAc]2 | |
| 2+ | 1,606.1508 | 1,606.1527 | 1 | LFNSTWPWNDTK | [Hex]8[HexNAc]2 | |
| C4-V5 | 3+ | 1,227.4659 | 1,227.4860 | 16 | DGGYESNETDEIFRPGGGDMR | [Hex]6[HexNAc]2 |
| 3+ | 1,770.3555 | 1,770.3467 | 5 | DGGYESNETDEIFRPGGGDMR | [Hex]6[HexNAc]5[Fuc]1[NeuNAc]3 | |
| 3+ | 1,862.7033 | 1,862.7187 | 8 | DGGYESNETDEIFRPGGGDMR | [Hex]7[HexNAc]7[Fuc]1[NeuNAc]2 | |
| C.1086 gp140 ΔC | ||||||
| C2 | 2+ | 957.3689 | 957.3561 | 3 | CNNK | [Hex]6[HexNAc]2 |
| 2+ | 1,038.3882 | 1,038.3825 | 5 | CNNK | [Hex]7[HexNAc]2 | |
| 2+ | 1,119.4048 | 1,119.4089 | 4 | CNNK | [Hex]8[HexNAc]2 | |
| 2+ | 1,200.4365 | 1,200.4353 | 1 | CNNK | [Hex]9[HexNAc]2 | |
| 5+ | 1,373.2330 | 1,373.2182 | 11 | NVSTVQCTHGIKPVVSTQLLLNGSLAEEEIIIR | [Hex]15[HexNAc]4 | |
| 5+ | 1,405.6548 | 1,405.6288 | 18 | NVSTVQCTHGIKPVVSTQLLLNGSLAEEEIIIR | [Hex]16[HexNAc]4 | |
| 5+ | 1,438.0554 | 1,438.0394 | 11 | NVSTVQCTHGIKPVVSTQLLLNGSLAEEEIIIR | [Hex]17[HexNAc]4 | |
| 5+ | 1,470.3994 | 1,470.4499 | 34 | NVSTVQCTHGIKPVVSTQLLLNGSLAEEEIIIR | [Hex]18[HexNAc]4 | |
| C2-V3 | 4+ | 1,509.6629 | 1,509.6447 | 12 | TIIVHLNESVNIVCTRPNNNTR | [Hex]13[HexNAc]6[Fuc]1 |
| 4+ | 1,499.4001 | 1,499.3880 | 8 | TIIVHLNESVNIVCTRPNNNTR | [Hex]14[HexNAc]5[Fuc]1 | |
| 4+ | 1,622.9858 | 1,622.9611 | 15 | TIIVHLNESVNIVCTRPNNNTR | [Hex]14[HexNAc]6[Fuc]1[NeuNAc]1 | |
| V4 | 3+ | 1,167.7907 | 1,167.7895 | 1 | NGTYNHTGR | [Hex]6[HexNAc]6[NeuNAc]1 |
| 3+ | 1,270.5169 | 1,270.4931 | 19 | NGTYNHTGR | [Hex]7[HexNAc]6[Fuc]1[NeuNAc]1 | |
| 3+ | 1,367.5176 | 1,367.5249 | 5 | NGTYNHTGR | [Hex]7[HexNAc]6[Fuc]1[NeuNAc]2 | |
| 3+ | 1,464.5866 | 1,464.5567 | 20 | NGTYNHTGR | [Hex]7[HexNAc]6[Fuc]1[NeuNAc]3 | |
The complete list is found in Tables S1 and S2 in the supplemental material.
PNGs are shown in boldface.
While a very high degree of glycosylation diversity was detected among the characterized glycosylation sites, not all of the glycopeptides could be fully accounted for, even with the extreme measures that were undertaken to obtain full glycosylation coverage. Of the 20 and 21 tryptic peptides with single and multiple glycosylation sites that were identified for both transmitted/founder Envs, the glycan profile of the glycopeptide bearing three PNG sites in the V1-V2 loop with peptide portion LTPLCVTLN130CTDLGN136VTN139TTNSNGEMMEK from B.700010040.C9 gp140 was not identified. This particular glycopeptide has a high-mass peptide backbone (m/z 3,327) with two and three of the three PNG sites utilized, as determined from PNGase F-treated Env samples (Table 1). Due to the inherent low ionization efficiency of high-mass glycopeptide species, peaks corresponding to this glycopeptide have relatively low intensity and have high charge states that are not completely resolved in the mass spectrum. In addition to this peptide, we are unable to characterize the glycan profiles of two glycopeptides from B.700010040.C9 gp140. The first of these two peptides contains PNG sites at N611 and N616, while the second contains the site at N625. These peptides were detected with a single HexNAc and/or fucosylated HexNAc only from Env samples treated with Endo H and Endo F3. Results obtained from glycosidase-treated Env samples revealed that the PNG sites at N611 and N616 are variably utilized, while the PNG site at N625 is fully utilized.
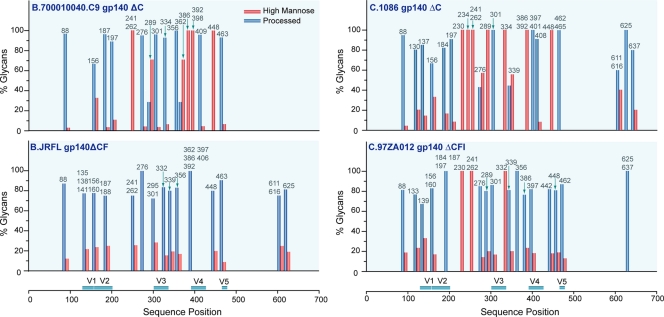
For the glycosylation sites that were fully characterized, the glycan distribution of the transmitted/founder Envs was deduced. Glycan compositions of each glycopeptide were sorted and broadly grouped according to the criteria used in our previous studies (19, 20). The glycan profile of each glycopeptide with either single or multiple glycosylation sites is represented by a pair of bars denoting the relative percentage of the type of glycan (high mannose or processed) arranged according to the Env sequence position, as shown in Fig. 7. The bar graph illustrates the differential glycan profiles between the two transmitted/founder Envs characterized herein, along with those of previously characterized Envs derived from chronic infections (19, 20). Comparison of the glycan profiles of the transmitted/founder Envs C.1086 gp140 and B.700010040.C9 gp140 reveals that C.1086 gp140 has more sites with exclusively high-mannose glycans (8 out of 27 PNG sites, or 30%) than B.700010040.C9 gp140 (5 out of 25 PNG sites, or 20%). Between the two transmitted/founder Envs, there are five conserved glycosylation sites bearing exclusively high-mannose glycans. These glycosylation sites are located in the outer domain, C2 (N241 and N262) and C4 (N448) regions, and the V4 loop (N386 and N392). The conserved high-mannose glycan profile in these regions of the Env indicates that both Envs have similar protein conformations proximal to these glycosylation sites.
Fig. 7.
Glycan profiles on each identified glycosylation site of Env immunogens derived from viruses isolated from acute (C.1086 gp140 ΔC and B.700010040.C9 gp140 ΔC) and chronic (B.JR-FL gp140 ΔCF and C.97ZA012 gp140 ΔCFI) HIV-1 infections. Glycan compositions (in percentages) were broadly categorized into two classes—high mannose and processed glycans—and arranged according to sequence position. Processed glycans include hybrid and complex glycans. Data presented for B.JR-FL and C.97ZA012 were obtained from our previous studies (19, 20).
DISCUSSION
Transmitted/founder viruses are those quasispecies of virus that establish productive infection in the first stages of acute HIV-1 infection (33). The founder viral quasispecies that establishes productive infection is initially homogeneous in 70 to 80% of subjects but diversifies, due to both random errors in reverse transcriptase and immune selective pressures (1, 32, 33, 38). The viral Env in acute HIV-1 infection has shorter variable loops and fewer potential glycosylation sites than chronic Envs (13, 36, 40, 42, 55). Information regarding the global glycosylation patterns of transmitted/founder Envs has been unavailable. As a first step toward characterizing transmitted/founder Env glycans, we have determined the degree of glycosylation site occupancy and characterized the founder/transmitted Env glycosylation patterns by elucidating the glycan motif at each potential glycosylation site on 293T cell-derived recombinant envelope proteins. While these analyses are not of Env derived from virus from CD4+ T cells, these studies report the technologies needed for this next study of CD4+ T-cell-derived virus and as well demonstrate that recombinant founder Envs have distinct glycosylation patterns compared to chronic Envs.
The data reported in this study, as summarized in Fig. 7 and fully reported in Tables S1 and S2 in the supplemental material, show that founder Env glycosylation sites are populated with high-mannose, hybrid, and complex glycans containing multianternnary structures with and without sialylation and core fucosylation; additionally, single HexNAcs with and without core fucosylation were also detected, even without the use of endoglycosidases. One distinctive glycan pattern of the two transmitted/founder Envs is that at least 20% of the sites were populated exclusively with high-mannose glycans, and five of these high-mannose-containing glycosylation sites (N241, N262, N386, N392, and N448) are conserved.
Having determined the glycan profiles of transmitted/founder Envs, an important question was whether the overall glycan profiles of founder Envs varied compared to those of chronic Envs. To identify the trends and highlight the differences in the glycosylation patterns, we compared the glycan profiles of chronic Envs, B.JR-FL gp140 and C.97ZA012 gp140, with those of the transmitted/founder Envs, C.1086 gp140 and B.700010040.C9 gp140. Data shown in Fig. 7 demonstrate several glycan features that differ with chronic versus founder Envs analyzed. (i) The transmitted/founder Envs had more high-mannose content compared to chronic Envs. The high level of high-mannose glycans observed suggested that transmitted/founder Envs may be adopting different structural features than chronic Envs since high-mannose glycans are known to promote protein folding and stabilize protein conformation (63, 64). (ii) Within clade C Envs, C.1086 gp140 and C.97ZA012 gp140, the glycosylation sites located in the C2 region (N230, N241, and N262) and the V3 loop, N334 for C.1086 and N332 for C.97ZA012, bear a similar glycosylation pattern consisting of predominantly high-mannose glycans. This finding is in agreement with our previous study (19), indicating that clade C Envs display a clade-specific pattern in these regions. (iii) The glycan profiles of the four Envs in the C1 region and the V1/V2 loops are also similar among all four proteins, wherein glycosylation sites bear predominantly processed glycans either with or without sialylation. This result suggested that these regions were accessible to glycotransferases and therefore were solvent exposed. Finally (iv) on the basis of the transmitted/founder and chronic Envs that we analyzed, the two transmitted/founder Envs have relatively similar glycan profiles regardless of clade, while chronic Envs showed more clade-specific glycan profiles.
Another key characteristic glycosylation feature inherent in HIV-1 Envs is the variation in the degree of glycosylation site occupancy. The degree to which a particular site will be modified with glycans in the ER largely depends on the protein sequence, availability of the glycosylation site, enzyme kinetics, and substrate concentration (30). We demonstrated that transmitted/founder Envs have 13 and 20 PNG sites that are variably utilized in B.700010040.C9 gp140 and C.1086 gp140, respectively, and both have two PNG sites that are not utilized at all. The chronic Env, B.JR-FL gp140, has seven sites that are not utilized at any time and one site that is variably utilized, while C.97ZA012 gp140 has two sites that are variably utilized (19, 20). Direct comparison of the glycosylation site occupancies among the Envs revealed that both transmitted/founder Envs have more sites that are variably utilized than B.JR-FL gp140 and C.97ZA012 gp140. To determine whether these glycosylation data can be used to group the Envs based on glycosylation site occupancy, we used one-dimensional hierarchical clustering to classify the glycosylation site occupancy profiles of the four Envs. A heat map was generated with a corresponding dendrogram to illustrate the clustering patterns of the four Envs (Fig. 8). The value used for site occupancy is between 0 and 1, depending on whether the glycosylation site is unutilized or missing, utilized, and variably utilized (see Materials and Methods). Interestingly, the data separate into three distinct clusters: two distinct clusters representing the characteristic glycosylation site occupancy profiles of the chronic Envs B.JR-FL gp140 and C.97ZA012 gp140 and one cluster for both C.1086 gp140 and B.700010040.C9 gp140. The results indicate that glycosylation site occupancy profiles for transmitted/founder Envs were similar to each other, while glycosylation site occupancy profiles for chronic Envs showed a higher degree of diversity, as they clustered separately from the transmitted/founder Envs, and as well clustered separately from each other. One important aspect of this work going forward is to resolve the issue of how closely the recombinant transmitted/founder Envs' glycosylation profiles mimic those on the virus, since it has been recently reported that glycan processing may be different with membrane-associated gp160 compared to soluble gp140 (15, 45).
Fig. 8.
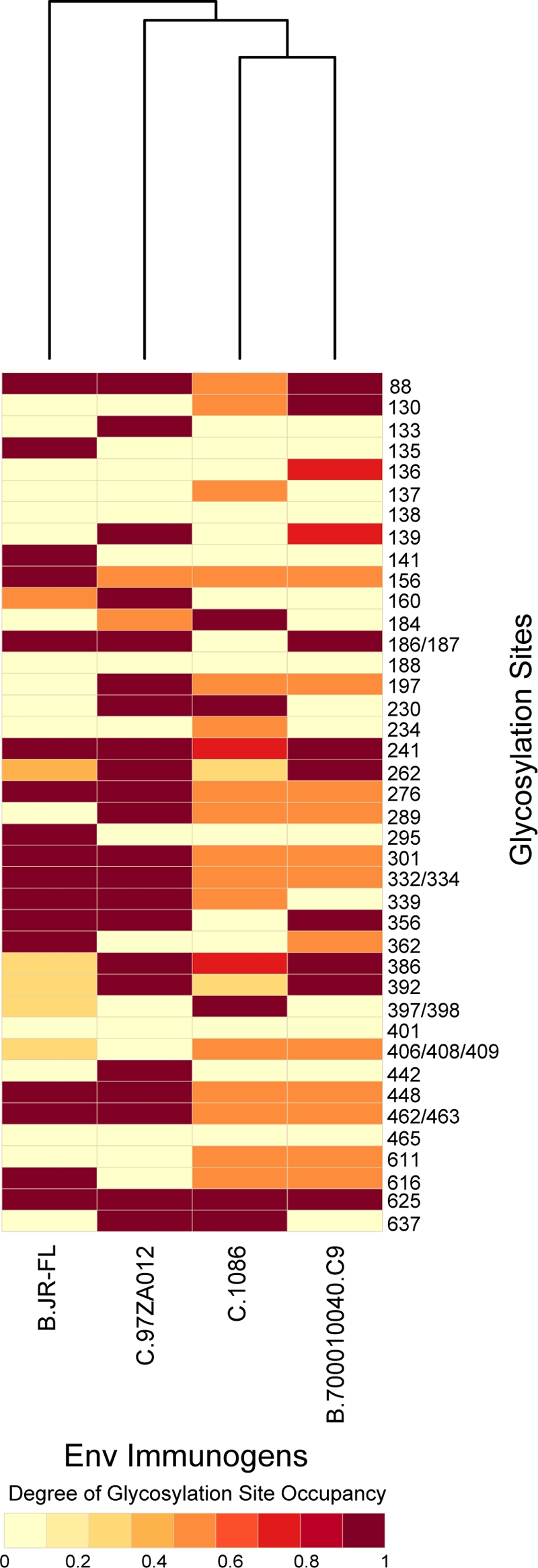
Heat map generated from the hierarchical clustering analysis of the glycosylation site occupancy of the early and late Env immunogens. Each column represents the glycosylation site occupancy of each Env immunogen. The dendrogram for glycosylation occupancy clustering is shown on the top.
In conclusion, we demonstrate the glycosylation profiles of transmitted/founder Envs using mass spectrometry in a glycosylation site-specific fashion. Our experimental workflow that included up-front complete and partial deglycosylation steps prior to trypsin digestion proved to be necessary to obtain a high degree of glycosylation coverage and to facilitate the characterization of the glycosylation patterns of two recombinant transmitted/founder Envs. Sequential digestion with glycosidases and trypsin prior to mass analysis allowed for the unambiguous identification of glycosylation site occupancy, identification of the type of glycan attached on a particular glycosylation site, and reduction in glycan heterogeneity. This approach has facilitated the characterization of glycopeptides that do not ionize efficiently or are not detected at all when all glycopeptides are present. Our results show that both B.700010040.C9 gp140 and C.1086 gp140 have similar degrees of glycosylation site occupancy and similar glycan profiles. This glycosylation profile is markedly different from those of chronic Envs B.JR-FL gp140 and C.97ZA012 gp140 in their degree of site occupancy and in the levels of complex glycans. Since glycosylation is known to affect Env immunogenicity, determination of glycan characteristics of different Envs should provide important correlates of antigenicity and immunogenicity of Env vaccine candidates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1RR026061 to H.D., grant PO1AI61734, the Center for HIV/AIDS Vaccine Immunology grant from the Division of AIDS, NIAID, NIH, and Collaboration for AIDS Vaccine Discovery grant from the Bill and Melinda Gates Foundation to B.F.H.
We also acknowledge the Analytical Proteomics Laboratory at KU for instrument time, technical assistance with the production of acute HIV-1 Env from Ashleigh Nagel, and helpful discussions with Ronald Swanstrom, Beatrice Hahn, and George Shaw.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An H. J., Froehlich J. W., Lebrilla C. B. 2009. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. An H. J., Kronewitter S. R., de Leoz M. L., Lebrilla C. B. 2009. Glycomics and disease markers. Curr. Opin. Chem. Biol. 13:601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andre S., et al. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binley J. M., et al. 2010. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 84:5637–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Budnik B. A., Lee R. S., Steen J. A. J. 2006. Global methods for protein glycosylation analysis by mass spectrometry. Biochim. Biophys. Acta 1764:1870–1880 [DOI] [PubMed] [Google Scholar]
- 7. Chakrabarti B. K., et al. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chohan B., et al. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cooper C. A., Gasteiger E., Packer N. H. 2001. GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1:340–349 [DOI] [PubMed] [Google Scholar]
- 10. Cutalo J. M., Deterding L. J., Tomer K. B. 2004. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J. Am. Soc. Mass Spectrom. 15:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dalpathado D. S., Desaire H. 2008. Glycopeptide analysis by mass spectrometry. Analyst 133:731–738 [DOI] [PubMed] [Google Scholar]
- 12. Dalpathado D. S., et al. 2006. Comparative glycomics of the glycoprotein follicle stimulating hormone: glycopeptide analysis of isolates from two mammalian species. Biochemistry 45:8665–8673 [DOI] [PubMed] [Google Scholar]
- 13. Derdeyn C. A., et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 14. Desaire H., Hua D. 2009. When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? Int. J. Mass Spectrom. 287:21–26 [Google Scholar]
- 15. Doores K. J., et al. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U. S. A. 107:13800–13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drake P. M., et al. 2010. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 56:223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geyer H., Holschbach C., Hunsmann G., Schneider J. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760–11767 [PubMed] [Google Scholar]
- 18. Geyer H., Geyer R. 2006. Strategies for analysis of glycoprotein glycosylation. Biochim. Biophys. Acta 1764:1853–1869 [DOI] [PubMed] [Google Scholar]
- 19. Go E. P., et al. 2009. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J. Proteome Res. 8:4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Go E. P., et al. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 7:1660–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Go E. P., et al. 2007. GlycoPep DB: a tool for glycopeptide analysis using a “Smart Search. ” Anal. Chem. 79:1708–1713 [DOI] [PubMed] [Google Scholar]
- 22. Go E. P., et al. Methods development for analysis of partially deglycosylated proteins and application to an HIV envelope protein vaccine candidate. Int. J. Mass Spectrom. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagglund P., Bunkenborg J., Elortza F., Jensen O. N., Roepstorff P. 2004. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3:556–566 [DOI] [PubMed] [Google Scholar]
- 24. Hagglund P., et al. 2007. An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J. Proteome Res. 6:3021–3031 [DOI] [PubMed] [Google Scholar]
- 25. Han J. C., Han G. Y. 1994. A procedure for quantitative determination of Tris(2-carboxyethyl)phosphine, an odorless reducing agent more stable and effective than dithiothreitol. Anal. Biochem. 220:5–10 [DOI] [PubMed] [Google Scholar]
- 26. Hirabayashi J. 2004. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconj. J. 21:35–40 [DOI] [PubMed] [Google Scholar]
- 27. Ihaka R., Gentleman R. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299–314 [Google Scholar]
- 28. Irungu J., Go E. P., Dalpathado D. S., Desaire H. 2007. Simplification of mass spectral analysis of acidic glycopeptides using GlycoPep ID. Anal. Chem. 79:3065–3074 [DOI] [PubMed] [Google Scholar]
- 29. Irungu J., et al. 2008. Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J. Am. Soc. Mass Spectrom. 19:1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones J., Krag S. S., Betenbaugh M. J. 2005. Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta 1726:121–137 [DOI] [PubMed] [Google Scholar]
- 31. Jung K. Y., Cho W. R., Regnier F. E. 2009. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J. Proteome Res. 8:643–650 [DOI] [PubMed] [Google Scholar]
- 32. Keele B. F., Derdeyn C. A. 2009. Genetic and antigenic features of the transmitted virus. Curr. Opin. HIV AIDS 4:352–357 [DOI] [PubMed] [Google Scholar]
- 33. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kong L., et al. 2010. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 403:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kornfeld R., Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631–664 [DOI] [PubMed] [Google Scholar]
- 36. Kraft Z., et al. 2008. Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses. J. Virol. 82:5912–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 38. Learn G. H., et al. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953–11959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leonard C. K., et al. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373–10382 [PubMed] [Google Scholar]
- 40. Li M., et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao H. X., et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y., et al. 2008. Env length and N-linked glycosylation following transmission of human immunodeficiency virus type 1 subtype B viruses. Virology 374:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195–204 [DOI] [PubMed] [Google Scholar]
- 44. Medzihradszky K. F. 2005. Characterization of protein N-glycosylation. Methods Enzymol. 405:116–138 [DOI] [PubMed] [Google Scholar]
- 45. Mizuochi T., et al. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J. Biol. Chem. 265:8519–8524 [PubMed] [Google Scholar]
- 46. Mizuochi T., et al. 1988. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem. J. 254:599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morelle W., Canis K., Chirat F., Faid V., Michalski J. C. 2006. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics 6:3993–4015 [DOI] [PubMed] [Google Scholar]
- 48. Morelle W., Michalski J. C. 2007. Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2:1585–1602 [DOI] [PubMed] [Google Scholar]
- 49. Nawaz F., et al. 2011. The genotype of early-transmitting HIV gp120s promotes alphabeta-reactivity, revealing alphabetaCD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 7:e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nicoli R., et al. 2010. Advances in LC platforms for drug discovery. Expert Opin. Drug Discov. 5:475–489 [DOI] [PubMed] [Google Scholar]
- 51. North S. J., Hitchen P. G., Haslam S. M., Dell A. 2009. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19:498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paulson J. C., Colley K. J. 1989. Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. J. Biol. Chem. 264:17615–17618 [PubMed] [Google Scholar]
- 53. Paulson J. C. 1989. Glycoproteins: what are the sugar chains for? Trends Biochem.Sci. 14:272–276 [DOI] [PubMed] [Google Scholar]
- 54. Qiu R. Q., Regnier F. E. 2005. Use of multidimensional lectin affinity chromatography in differential glycoproteomics. Anal. Chem. 77:2802–2809 [DOI] [PubMed] [Google Scholar]
- 55. Rademeyer C., et al. 2007. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368:172–181 [DOI] [PubMed] [Google Scholar]
- 56. Raska M., et al. 2010. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J. Biol. Chem. 285:20860–20869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rerks-Ngarm S., et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 58. Salazar-Gonzalez J. F., et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schulz B. L., Aebi M. 2009. Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol. Cell. Proteomics 8:357–364 [DOI] [PubMed] [Google Scholar]
- 60. Segu Z. M., Hussein A., Novotny M. V., Mechref Y. 2010. Assigning N-glycosylation sites of glycoproteins using LC/MSMS in conjunction with Endo-M/exoglycosidase mixture. J. Proteome Res. 9:3598–3607 [DOI] [PubMed] [Google Scholar]
- 61. Tarentino A. L., Plummer T. H., Jr 1994. Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230:44–57 [DOI] [PubMed] [Google Scholar]
- 62. Ungar D. 2009. Golgi linked protein glycosylation and associated diseases. Semin. Cell Dev. Biol. 20:762–769 [DOI] [PubMed] [Google Scholar]
- 63. Wyss D. F., et al. 1995. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science 269:1273–1278 [DOI] [PubMed] [Google Scholar]
- 64. Yamaguchi H. 2002. Chaperone-like functions of N-glycans in the formation and stabilization of protein conformation. Trends Glycosci. Glycotechnol. 14:139–151 [Google Scholar]
- 65. Yeh J. C., Seals J. R., Murphy C. I., van Halbeek H., Cummings R. D. 1993. Site-specific N-glycosylation and oligosaccharide structures of recombinant HIV-1 gp120 derived from a baculovirus expression system. Biochemistry 32:11087–11099 [DOI] [PubMed] [Google Scholar]
- 66. Zaia J. 2008. Mass spectrometry and the emerging field of glycomics. Chem. Biol. 15:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang M., et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 68. Zhang Y., Go E. P., Desaire H. 2008. Maximizing coverage of glycosylation heterogeneity in MALDI-MS analysis of glycoproteins with up to 27 glycosylation sites. Anal. Chem. 80:3144–3158 [DOI] [PubMed] [Google Scholar]
- 69. Zhu X., Borchers C., Bienstock R. J., Tomer K. B. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194–11204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.