Abstract
Herpesvirus nucleocapsids assemble in the nucleus but mature to infectious virions in the cytoplasm. To gain access to this cellular compartment, nucleocapsids are translocated to the cytoplasm by primary envelopment at the inner nuclear membrane and subsequent fusion of the primary envelope with the outer nuclear membrane. The conserved viral pUL34 and pUL31 proteins play a crucial role in this process. In their absence, viral replication is strongly impaired but not totally abolished. We used the residual infectivity of a pUL34-deleted mutant of the alphaherpesvirus pseudorabies virus (PrV) for reversion analysis. To this end, PrV-ΔUL34 was serially passaged in rabbit kidney cells until final titers of the mutant virus PrV-ΔUL34Pass were comparable to those of wild-type PrV. PrV-ΔUL34Pass produced infectious progeny independently of the pUL34/pUL31 nuclear egress complex and the pUS3 protein kinase. Ultrastructural analyses demonstrated that this effect was due to virus-induced disintegration of the nuclear envelope, thereby releasing immature and mature capsids into the cytosol for secondary envelopment. Our data indicate that nuclear egress primarily serves to transfer capsids through the intact nuclear envelope. Immature and mature intranuclear capsids are competent for further virion maturation once they reach the cytoplasm. However, nuclear egress exhibits a strong bias for nucleocapsids, thereby also functioning as a quality control checkpoint which is abolished by herpesvirus-induced nuclear envelope breakdown.
INTRODUCTION
Morphogenesis of herpesvirus particles proceeds in two different cellular compartments (reviewed in references 17a, 18, and 19). Autocatalytic assembly of capsids as well as cleavage and encapsidation of newly replicated viral genomic DNA takes place in the nuclei of infected cells, whereas subsequent virion morphogenesis, including the acquisition of major portions of the tegument and final envelopment, occurs in the cytoplasm. To leave the nucleus and gain access to the cytosol, nucleocapsids have to cross the nuclear envelope in a process designated nuclear egress. This comprises the acquisition of a primary envelope by budding at and fission from the inner nuclear membrane (INM), resulting in the formation of an enveloped virus particle residing between the two leaflets of the nuclear envelope. The primary envelope then fuses with the outer nuclear membrane (ONM), releasing the nucleocapsid into the cytoplasm for further maturation. Although the molecular details of nuclear egress are not yet clear, it is conserved within the herpesviruses. Homologs of the herpes simplex virus 1 (HSV-1) proteins pUL34 and pUL31 are crucial for nuclear egress in all herpesviruses analyzed in this respect (1, 2, 4, 5, 9, 15, 20, 21, 22, 23). pUL34, a tail-anchored membrane protein, which is targeted to the nuclear envelope, forms a heterodimeric complex with the nucleoplasmic pUL31. Formation of this nuclear egress complex (NEC) is a prerequisite for proper positioning of both complex partners at the INM, recruitment of viral and/or cellular kinases for local dissolution of the nuclear lamina, modification of host cell chromatin, and efficient nuclear egress of nucleocapsids (reviewed in reference 19). Coexpression of pUL34 and pUL31 from the alphaherpesvirus pseudorabies virus (PrV) resulted in the formation of vesicles from the INM resembling primary envelopes, which indicates that these two viral proteins are required and sufficient for the budding process (11). Whereas these vesicles may also fuse with the ONM without the involvement of other viral proteins, fusogenic glycoproteins gB and gH might enhance the efficiency of this process in HSV-1 (3) but are apparently not involved in nuclear egress of PrV (8). Besides these conserved herpesviral proteins, the pUS3 protein kinase, which is present only in the alphaherpesviruses, has been shown to be involved in nuclear egress. In its absence, primary enveloped virions accumulate in the perinuclear cleft, indicating an impairment of fusion of the primary envelope with the ONM (10, 22, 25).
Besides the widely accepted nuclear egress pathway via primary envelopment-deenvelopment-reenvelopment, exit of intranuclear capsids through grossly dilated nuclear pores has been proposed for HSV-1 and bovine herpesvirus 1 (16, 26), as has transport of (primary) enveloped HSV-1 virions via the endoplasmic reticulum (ER) and Golgi apparatus within the secretory pathway (6).
In PrV, which is relevant in agriculture as the causative agent of Aujeszky's disease in swine (17), productive replication is severely impaired in the absence of the NEC but not completely abolished. Deletion mutants lacking pUL34 and/or pUL31 produce infectious progeny, though at 3- to 4-log-reduced titers compared to wild-type PrV (4, 9, 11). This residual infectivity allowed us to use reversion analysis by repeated passages in cell culture cells, i.e., evolution in vitro, to check whether the requirement for the NEC can be overcome and whether nuclear egress would be possible via alternative routes.
MATERIALS AND METHODS
Cells and viruses.
Wild-type PrV strain Kaplan (PrV-Ka) (7) was used throughout these studies. Mutants PrV-ΔUL34 (10) and PrV-ΔUL31 (4) have been described elsewhere. To facilitate further manipulation, mutant PrV-ΔUL34Pass-Bsa was isolated by selection of nonfluorescent progeny after cotransfection of DNA from PrV-ΔUL34Pass, which carries a green fluorescent protein (GFP) expression cassette at the UL34 locus (Fig. 1A), with a recombination plasmid containing a 5.5-kb BstEII fragment lacking an 825-bp SalI/BsaBI fragment from the UL34 gene region (Fig. 1A). Transgenic RK13 cells constitutively expressing pUL34 (RK13-UL34) or containing a genomic fragment comprising genes UL31 to UL35 (RK13-DrdI) have been described elsewhere (4, 9, 11). PrV-ΔUL34Pass-ΔUL31 was isolated after cotransfection of PrV-ΔUL34Pass-Bsa DNA with recombination plasmid pUC-ΔUL31gfp carrying a GFP expression cassette at the UL31 locus and subsequent selection for autofluorescent plaques. To this end, flanking regions of the UL31 open reading frame were amplified by PCR using primers del31_1 (5′-CACAGGATCCAACTGCGCCTTCATCAACG-3′; nucleotides [nt] 28038 to 28057; accession no. BK001744) (14) and del31_2 (5′-CACAGGATCCGGTGGTGCGAAAGAAGGGAG-3′; nt 28819 to 28838) comprising the 3′ part of the UL30 open reading frame and del31_3 (5′-CACACTGCAGCGCTCAAACGTAGCTGCCG-3′; nt 29478 to 29496) and del31_4 (CACAAAGCTTCTCGTGGACGGCAGCGG-3′; nt 30480 to 30497) covering the 3′ part of the UL32 gene (Fig. 1B). Primer del31_3 was designed with a mismatch (shown in bold), resulting in mutation of the UL31 start codon from ATG to ACG without affecting the overlapping UL32 open reading frame. PCR fragments were eluted, cleaved with the corresponding restriction enzymes for which sites were provided with the primers (restriction sites underlined; see above), and successively cloned into pUC19. The GFP cassette under the control of the immediate-early promoter/enhancer complex from human cytomegalovirus was inserted as a BamHI/PstI fragment, giving rise to plasmid pUC-ΔUL31gfp. PrV-ΔUL34Pass-ΔUS3 (Fig. 1C) was isolated after cotransfection of PrV-ΔUL34Pass-Bsa DNA with pUC-ΔUS3gfp (10) and selection for autofluorescent plaques.
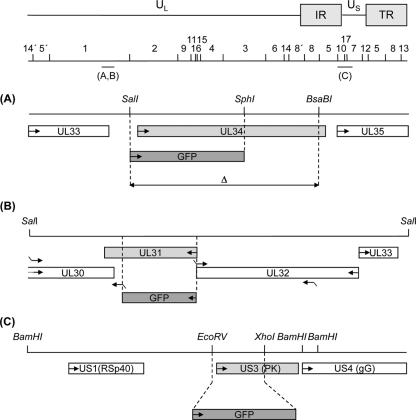
Fig. 1.
Diagram of mutant viruses. The PrV genome consisting of long (UL) and short (US) unique regions and inverted repeat sequences (IR and TR), as well as location and numbering of BamHI restriction fragments, is shown. Locations of the genomic regions shown enlarged are indicated. (A) UL34 gene region with relevant restriction sites. Open reading frames are drawn as rectangles. The regions which had been deleted for generation of PrV-ΔUL34 (SalI/SphI) and PrV-ΔUL34Pass-Bsa (SalI/BsaBI) are delineated by dashed lines. Insertion of the reporter gene in PrV-ΔUL34 is indicated. (B) UL31 gene region and relevant restriction sites. Locations of primers used for amplification of UL31 flanking regions are indicated by bent arrows. Deletion of UL31 coding sequences is shown by dashed lines, and insertion of the GFP expression cassette is indicated. (C) Substitution of US3 coding sequences between the dashed lines with a GFP reporter cassette is indicated (10).
One-step growth analysis.
Confluent monolayers of RK13 or RK13-DrdI cells were infected at a multiplicity of infection (MOI) of 5 with PrV-Ka or the indicated mutants and incubated on ice for 1 h. Thereafter, the inoculum was removed, prewarmed medium was added, and cells were incubated for one additional hour at 37°C. Extracellular virus was inactivated by low-pH treatment, and cells and supernatant were harvested either immediately (0 h) or after 4, 8, 12, 24, and 36 h. Progeny virus titers were determined by plaque assays on RK13 or RK13-DrdI cells (11). Mean values of two independent experiments and the corresponding minimum and maximum values were plotted.
Western blot analysis.
Cells were lysed approximately 18 h after infection with the indicated viruses at an MOI of 5. Lysates were harvested as described previously (4), separated in sodium dodecyl sulfate-10% or 12% polyacrylamide gels, blotted, and incubated with monospecific anti-pUL34 (dilution, 1:50,000 [9]), anti-pUL31 (dilution 1:50,000 [4]), anti-US3 (1:50,000 [10]), anti-UL25 (1:50,000 [12]), or anti-UL37 (1:100,000 [13]) rabbit sera. Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (Super Signal; Pierce, Bonn, Germany) recorded on X-ray films.
Electron microscopy.
RK13 or RK13-UL34 cells were infected with PrV-ΔUL34 which had been propagated on pUL34-expressing cells, PrV-ΔUL31 which had been propagated on pUL31-expressing cells, PrV-ΔUL34Pass, PrV-ΔUL34Pass-ΔUL31, PrV-ΔUS3, or PrV-ΔUL34Pass-ΔUS3 at an MOI of 1 and processed for electron microscopy 14 h after infection as described previously (9). Counterstained ultrathin sections were analyzed with a transmission electron microscope (Philips Tecnai 12; Eindhoven, the Netherlands).
RESULTS
Isolation and characterization of PrV-ΔUL34Pass.
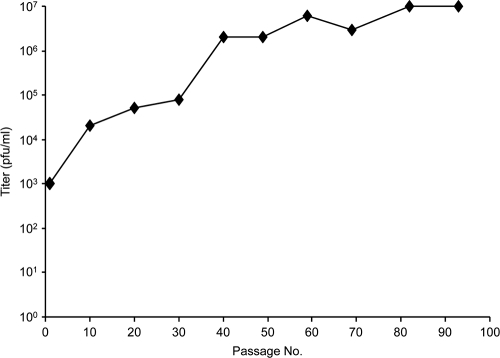
We used the residual infectivity of PrV-ΔUL34 for serial passaging in rabbit kidney (RK13) cells by initial repeated trypsinization and reseeding of infected cells followed by passaging of supernatants after a complete cytopathic effect had developed. The titer of infectious progeny rose until it remained stable at approximately wild-type levels after ca. 90 passages (Fig. 2). From this virus population, six single plaques were picked and analyzed by Southern blotting, Western blotting, and one-step replication kinetics (data not shown). Since they were identical in these assays, one isolate was randomly chosen, designated PrV-ΔUL34Pass, and used for further studies.
Fig. 2.
Selection of PrV-ΔUL34Pass. Rabbit kidney (RK13) cells were infected with PrV-ΔUL34 and passaged by initial repeated trypsinization and reseeding of infected cells followed by passaging of supernatants after a complete cytopathic effect had developed. Extracellular infectivity was determined on RK13 cells. After ca. 90 passages, virus progeny was plaque purified and further analyzed.
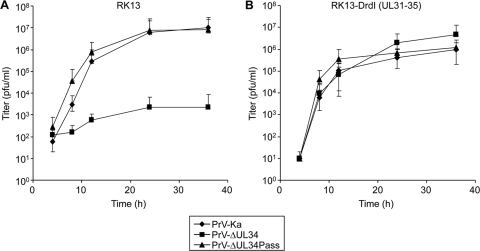
Compared to the parental virus PrV-ΔUL34 which, in noncomplementing cells, reached titers of only 103 PFU/ml (Fig. 3A), PrV-ΔUL34Pass showed one-step replication kinetics similar to those of PrV-Ka with final titers of approximately 107 PFU/ml (Fig. 3A). Titers of PrV-ΔUL34 were elevated to wild-type levels on complementing RK13-DrdI cells, whereas kinetics and final titers of PrV-ΔUL34Pass were unchanged (Fig. 3B). Thus, PrV-ΔUL34Pass replication occurs independently of pUL34.
Fig. 3.
One-step growth kinetics of wild-type and mutant viruses. RK13 (A) or RK13-DrdI (B) cells were infected with PrV-Ka, PrV-ΔUL34, or PrV-ΔUL34Pass at an MOI of 5 and analyzed at the indicated times after low-pH treatment. The cells were lysed by freezing and thawing, and progeny virus titers were determined on RK13-DrdI cells by plaque assays. Mean values of at least two independent experiments were calculated, and corresponding minimum and maximum values were plotted.
To assess whether PrV-ΔUL34Pass still lacked pUL34, infected cell lysates were immunoblotted with a monospecific anti-pUL34 rabbit serum (9). As shown in Fig. 4, in PrV-Ka-infected cells, pUL34 was readily detectable as a ca. 28-kDa protein. No pUL34-specific immunoreactivity was evident in cells infected by PrV-ΔUL34 or PrV-ΔUL34Pass. In contrast, the pUL34 complex partner pUL31 was present in both lysates. Demonstration of pUL25 and pUL37 served as loading controls.
Fig. 4.
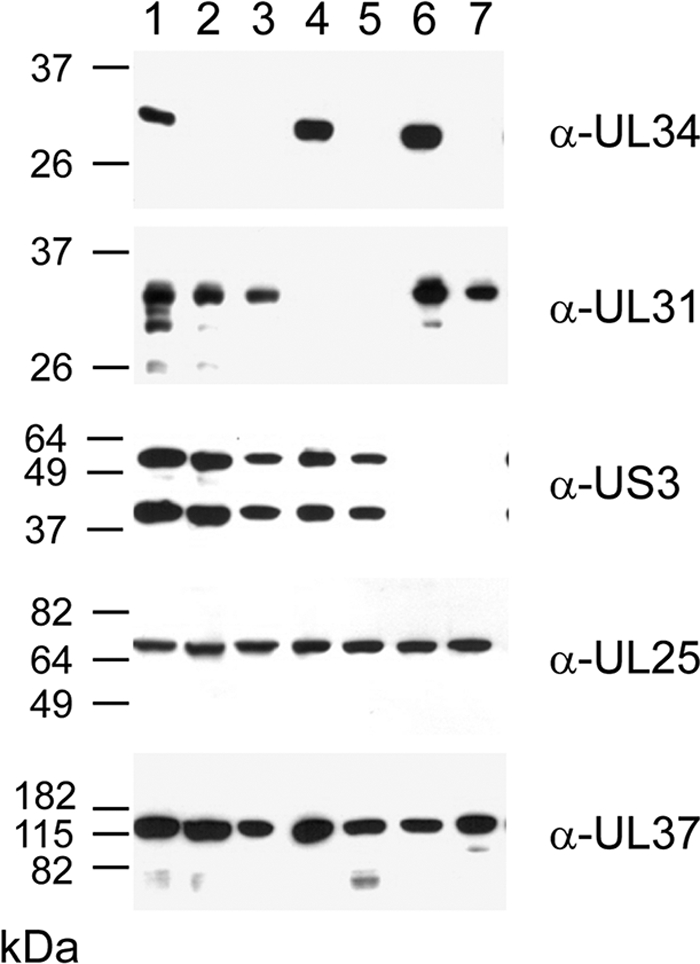
Immunoblots of infected cells. RK13 cells infected at an MOI of 5 with PrV-Ka (lane 1), PrV-ΔUL34 (lane 2), PrV-ΔUL34Pass (lane 3), PrV-ΔUL31 (lane 4), PrV-ΔUL34Pass-ΔUL31 (lane 5), PrV-ΔUS3 (lane 6), or PrV-ΔUL34Pass-ΔUS3 (lane 7) were lysed 18 h postinfection, and proteins were separated on SDS-10% or 12% polyacrylamide gels. Parallel blots were incubated with rabbit sera raised against pUL34, pUL31, pUS3, pUL25, or pUL37. The locations of molecular mass markers are indicated on the left.
Replication of PrV-ΔUL34Pass does not require the NEC.
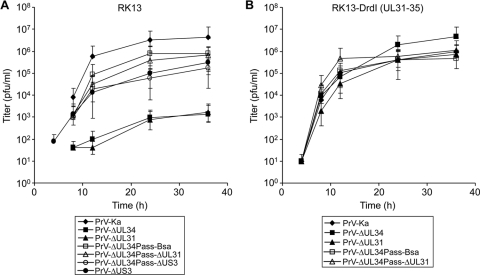
To assay whether the other component of the NEC, pUL31, was also dispensable for replication of PrV-ΔUL34Pass, we first isolated a deletion mutant of PrV-ΔUL34Pass devoid of GFP expression by cotransfection of PrV-ΔUL34Pass DNA with a recombination plasmid lacking a SalI/BsaBI fragment comprising codons 1 to 256 of the 262-amino-acid pUL34 open reading frame, yielding PrV-ΔUL34Pass-Bsa (Fig. 1A). Besides facilitating subsequent deletion of other viral genes using GFP as a marker, this also removed most of the UL34 sequences retained in PrV-ΔUL34 (Fig. 1A). Subsequently, the UL31 gene was removed from PrV-ΔUL34Pass-Bsa and replaced by a GFP expression cassette (Fig. 1B). Absence of pUL31 and pUL34 from PrV-ΔUL34Pass-ΔUL31 was verified by immunoblotting (Fig. 4, lane 5). As shown in Fig. 5A, one-step replication kinetics of PrV-ΔUL34Pass-Bsa and PrV-ΔUL34Pass-ΔUL31 on noncomplementing cells were nearly identical, with final titers slightly below 106 PFU/ml (Fig. 5A). Replication of either virus was not altered on pUL34- and pUL31-expressing cells (Fig. 5B), demonstrating that replication occurred independently of the NEC. In contrast, PrV-ΔUL34 and PrV-ΔUL31 reached wild-type-like titers only on complementing cells (Fig. 5B), whereas in noncomplementing cells titers were ca. 1,000-fold lower (Fig. 5A).
Fig. 5.
Growth kinetics of PrV-ΔUL34Pass-Bsa mutants lacking UL31 or US3. RK13 (A) or RK13-DrdI (B) cells were infected with PrV-Ka, PrV-ΔUL34, PrV-ΔUL31, PrV-ΔUL34Pass-Bsa, PrV-ΔUL34Pass-ΔUL31, PrV-ΔUS3, or PrV-ΔUL34Pass-ΔUS3 and processed as described in the legend to Fig. 3.
To test whether the pUS3 kinase is involved in PrV-ΔUL34Pass replication, a corresponding deletion mutant, PrV-ΔUL34Pass-ΔUS3 (Fig. 1C), was isolated and absence of the protein was verified by immunoblotting (Fig. 4). PrV expresses two isoforms of pUS3, resulting in two protein products of different sizes (24). Both isoforms are absent in cells infected with the pUS3 deletion mutants. In one-step replication kinetics, deletion of pUS3 decreased replication of PrV-Ka and PrV-ΔUL34Pass-Bsa by roughly 10- to 50-fold (Fig. 5A), indicating similar involvements of pUS3 in replication of either virus.
NEC-independent nuclear egress of PrV-ΔUL34Pass occurs by fragmentation of the nuclear envelope.
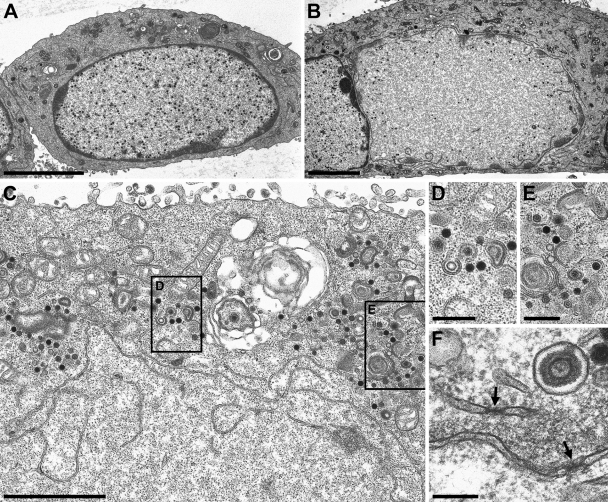
The data so far demonstrated that the formation of infectious virus progeny at wild-type levels occurred independently of the NEC in PrV-ΔUL34Pass-infected cells. To analyze in detail how PrV-ΔUL34Pass matures, ultrastructural analyses were performed by transmission electron microscopy of RK13 cells 14 h after infection with PrV-ΔUL34 or PrV-ΔUL34Pass. As observed before (9), in PrV-ΔUL34-infected cells nucleocapsids are unable to leave the nucleus, which is bounded by an intact nuclear envelope even late in infection (Fig. 6A). In contrast, in cells infected with PrV-ΔUL34Pass, approximately half of infected cell nuclei showed disruptions of the nuclear envelope (Fig. 6B and C). Mature genome-containing C capsids as well as genome-less immature empty A and scaffold-containing B capsids were detectable in the cytoplasm undergoing secondary envelopment (Fig. 6C to E). Thus, PrV-ΔUL34Pass was able to replicate independently of the NEC since capsids gained access to the cytoplasm after disintegration of the nuclear membrane. No intracytoplasmic capsids were observed in cells with intact nuclei (see Fig. 8). Disruption of the nuclear envelope was also observed on pUL34-expressing RK13-UL34 cells (Fig. 7A), demonstrating that even in the presence of pUL34, PrV-ΔUL34Pass maturation entails this novel pathway. In the remnants of the nuclear envelope present after nuclear envelope breakdown (NEBD), morphologically intact nuclear pores could still be found (Fig. 6F).
Fig. 6.
Ultrastructural analysis of PrV-ΔUL34Pass-infected cells. RK13 cells infected with PrV-ΔUL34 (A) or PrV-ΔUL34Pass (B to F) at an MOI of 1 were analyzed 14 h after infection by transmission electron microscopy. (A) An intact nucleus with trapped nucleocapsids is shown. (B) A partially dissolved nucleus. (C to E) Remnants of the nuclear envelope are visible together with capsids in the cytoplasm (C), several of them undergoing envelopment, magnified in panels D and E. (F) Morphologically intact nuclear pores in nuclear envelope fragments are labeled by arrows. Bars, 5 μm (A and B), 2 μm (C), 500 nm (D and E), and 200 nm (F).
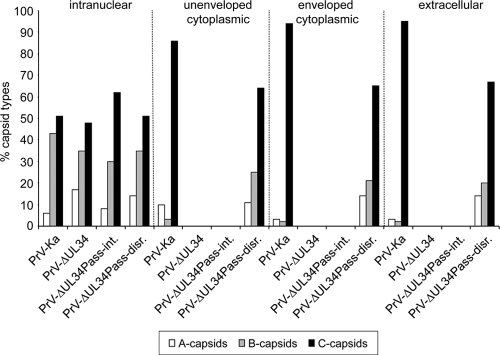
Fig. 8.
Quantitation of different capsid forms. Electron microscopic images of RK13 cells infected with PrV-Ka, PrV-ΔUL34, and PrV-ΔUL34Pass were used to count different capsid forms (empty A capsids, scaffold-containing B capsids, and DNA-containing C capsids) in the nucleus and nonenveloped and enveloped particles in the cytoplasm as well as in the extracellular space. Capsids in 10 cells per assay were counted. The total numbers of capsids counted were 666 for cells infected with PrV-Ka, 954 for PrV-ΔUL34-infected cells, 580 for PrV-ΔUL34Pass-infected cells with intact nuclei (PrV-ΔUL34Pass-int.; all intranuclear), and 1,188 for PrV-ΔUL34Pass-infected cells with disrupted nuclei (PrV-ΔUL34Pass-disr.).
Fig. 7.
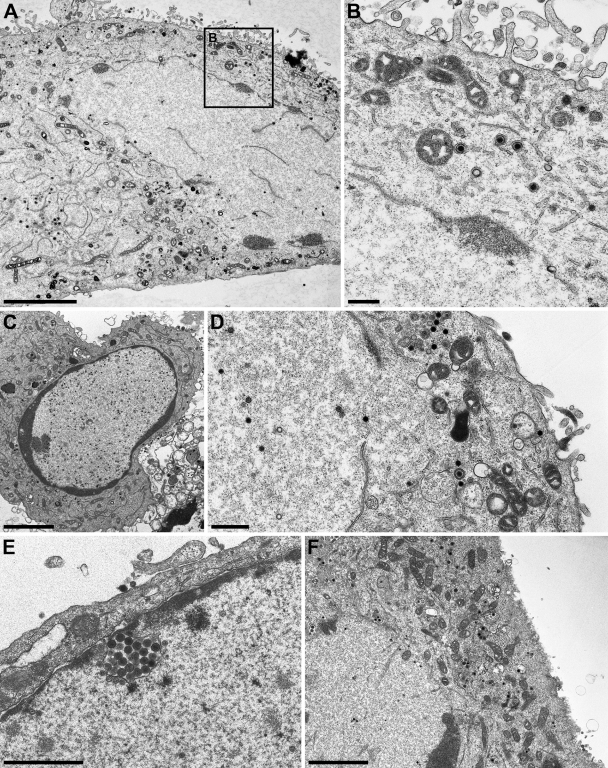
Ultrastructural analysis of cells infected with PrV mutants. RK13-UL34 cells infected with PrV-ΔUL34Pass (A and B), as well as RK13 cells infected with PrV-ΔUL31 (C), PrV-ΔUL34Pass-ΔUL31 (D), PrV-ΔUS3 (E), or PrV-ΔUL34Pass-ΔUS3 (F), at an MOI of 1 were analyzed 14 h after infection by transmission electron microscopy. Bars, 5 μm (A), 500 nm (B and D), 3.7 μm (C), 1.5 μm (E), and 3 μm (F).
In the absence of pUL31 from wild-type PrV-Ka, nucleocapsids are retained in intact nuclei (Fig. 7C) (4), similar to the situation in the absence of pUL34 (Fig. 6A) (9). Additional deletion of pUL31 from PrV-ΔUL34Pass did not alter the phenotype in infected cells. Disruption of the nuclear envelope occurred, and secondary envelopment proceeded in the cytosol (Fig. 7D). Whereas deletion of pUS3 from PrV-Ka resulted in the expected accumulation of primary enveloped virions in large invaginations of the INM (Fig. 7E), additional deletion of pUS3 from PrV-ΔUL34Pass-Bsa also did not alter the observed phenotype (Fig. 7F). These data corroborate the idea that PrV-ΔUL34Pass replicates by a mechanism which is independent of the NEC and drastically differs from nuclear egress as observed in wild-type virus infection.
Nuclear egress via nuclear envelope breakdown abolishes selection of nucleocapsids for virion formation.
In wild-type PrV-infected cells, genome-less capsids are only rarely observed in the cytoplasm, indicating a quality control at the stage of nuclear egress (12). In contrast, in PrV-ΔUL34Pass-infected cells with NEBD numerous capsid forms lacking DNA were present in the cytoplasm with or without an envelope. Quantitation of the different capsid forms present in the nucleus, in the cytoplasm, and in extracellular virions (Fig. 8) revealed that in PrV-Ka-infected cells, ca. 90% of intracytoplasmic and extracellular capsids contain DNA. In PrV-ΔUL34Pass-infected cells with NEBD, ca. 30% of these capsids lack DNA, indicating a significant impairment of selection of mature capsids for final envelopment in the cytoplasm. Cells infected with PrV-ΔUL34Pass showing intact nuclei do not contain cytoplasmic capsids equaling cells infected with PrV-ΔUL34. Thus, regulated primary envelope-mediated nuclear egress serves to discriminate mature from immature capsids for translocation into the cytoplasm to continue virus maturation.
DISCUSSION
The salient findings reported here are as follows. (i) Serial passaging of a UL34 deletion mutant of PrV in RK13 cells resulted in a mutant virus, PrV-ΔUL34Pass, which was able to produce infectious progeny similar to wild-type PrV in the absence of pUL34. (ii) Neither pUL34 nor pUL31 is required for replication of PrV-ΔUL34Pass in RK13 cells. Thus, the conserved herpesviral NEC is not required for PrV-ΔUL34Pass replication. (iii) Nuclear egress of PrV-ΔUL34Pass occurs via disintegration of the nuclear envelope and direct access of capsids to the cytoplasm. (iv) Nuclear escape via disintegration of the nuclear envelope impairs selection of mature nucleocapsids for final envelopment.
The exit pathway of herpesvirus nucleocapsids, which assemble in the nucleus of infected cells, to the cytoplasmic maturation compartment has long been discussed. An initial proposal which included budding of nucleocapsids at the INM followed by lumenal transport through the ER and Golgi apparatus along the secretory pathway (6) was shown to be inconsistent with the apparent lack of integrity between enveloped virus particles in the perinuclear cleft, which contain the NEC components pUL34 and pUL31, and mature extracellular virions, which do not (reviewed in references 17a, 18, and 19). To consider the fact that primary and mature herpesvirus virions differ in their composition and to accommodate ultrastructural and biochemical data (reviewed in reference 19), a model has been proposed which also entails budding at the INM leading to (primary) enveloped virions in the perinuclear cleft. This, however, is followed by fusion of the primary envelope with the ONM and translocation of the nucleocapsid to the cytosol. Egress of nucleocapsids via grossly dilated nuclear pores has been observed by one group (16, 26). Moreover, it is unclear whether nuclear egress serves to transport nucleocapsids from one maturation compartment to another or whether it entails maturational steps of the nucleocapsid by addition, e.g., of tegument proteins, or modification by viral and/or cellular enzymes during nuclear egress.
Our data show a mechanism by which capsids of a mutant herpesvirus gain access to the cytoplasm by disintegration of the nuclear envelope. This nuclear escape does not require the NEC, which mediates regulated nuclear egress. The fact that disintegration of the nuclear envelope and, thus, breaking the barrier between the nuclear and cytoplasmic compartments appears sufficient to rescue the nuclear egress defect otherwise associated with pUL34 and/or pUL31 deletion mutants indicates that intranuclear capsids are maturation competent once they reach the cytoplasm. Thus, the observed disintegration of the nuclear envelope apparently compensates for the controlled, primary envelope-mediated transfer of nucleocapsids to the cytosol which occurs in wild-type herpesvirus-infected cells. However, we cannot exclude the possibility that during passaging another compensatory mutation(s) with relevance for capsid maturation may also have been acquired.
Formation at and fission from the INM of membranous vesicles resembling primary envelopes require only pUL34 and pUL31 (11). However, the NEC is also involved in recruiting viral and cellular proteins for altering nuclear architecture, including local dissolution by phosphorylation of lamins A and B and chromatin displacement to allow access of intranuclear nucleocapsids to the primary envelopment site (reviewed in references 18 and 19). Since the NEC is neither required nor beneficial for nuclear egress of PrV-ΔUL34Pass, these functions are either not needed or are compensated for by other viral or cellular proteins. Apparently, the requirements for nuclear escape via disintegration of the nuclear envelope and for regulated primary envelope-mediated nuclear egress are strikingly different. This is also demonstrated by the lack of influence of a pUS3 deletion on nuclear escape, whereas it impairs deenvelopment of primary virions following the “classical” pathway. However, absence of pUS3 from PrV-Ka or PrV-ΔUL34Pass impaired viral replication, leading to decreased viral titers, indicating that this defect is obviously not correlated with exit of capsids from the nucleus.
If nuclear egress can occur efficiently by disruption of the nuclear envelope, why did herpesviruses invent an elaborate mechanism for translocation through the intact nuclear envelope? There are several possible explanations for this conundrum. First, as is true for all viruses, herpesvirus replication is dependent on the function of the cellular metabolism. Early dissolution of the nuclear envelope resulting in breakdown of cellular compartmentalization could, under natural conditions, strongly impair the formation of virus progeny. This could correlate with the inability of our passaged mutant virus to induce NEBD in cells other than those it was selected in. Second, herpesviruses evolved together with their host to maintain themselves in an intricate form of parasitism without doing excess harm to the host. Latency shows this to perfection. This is also realized by regulated nuclear egress. Third, the ancestors of herpesviruses may have left their prokaryotic hosts by budding at the cell membrane, just as many eukaryotic viruses do, and not by lysis. This might be reflected by the primary envelopment-deenvelopment pathway. Fourth, dramatic disruptions of cellular compartmentalization could trigger innate antiviral responses detrimental for virus replication.
However, we cannot exclude that under certain circumstances wild-type herpesviruses may use the mechanism that we uncovered in PrV-ΔUL34Pass. In our mouse model of intranasal alphaherpesvirus infection, PrV-ΔUL34Pass proved to be innocuous even when given at doses higher than those of wild-type PrV (data not shown). Thus, the phenotype that we selected for in RK13 cells appears not to compensate for pUL31/pUL34 function in vivo. Alternatively, we may have uncovered a mechanism which is cell type specific. First studies indeed show that in host cells other than RK13, which was used for the selection, PrV-ΔUL34Pass nucleocapsids are incapable of or at least significantly less efficient in reaching the cytoplasm, indicating a cell-type-specific effect. This also applies to explanted rat neurons, which do not support productive replication of PrV-ΔUL34Pass (data not shown). A cell-type-dependent requirement for the NEC has been described before (16a), although this was not related to NEBD.
Intriguingly, despite extensive plaque purifications, the phenotype observed after infection of RK13 cells with PrV-ΔUL34Pass is not uniform. Whereas in ca. 50% of infected cells the nuclear envelope is disrupted, the other half still retains an intact nucleus with nucleocapsids trapped inside, i.e., a PrV-ΔUL34 phenotype. It is currently unclear whether this is simply reflecting a temporal difference or whether some nuclei are indeed refractory to envelope disruption by PrV-ΔUL34Pass.
The nature of the compensatory mutation(s) that we selected for is unclear at present. Disintegration of the nuclear envelope occurs in PrV-ΔUL34Pass-infected cells only and not in cells infected with either wild-type PrV-Ka or UL31 or UL34 deletion mutants. Thus, it is clearly induced by infection with PrV-ΔUL34Pass. We are currently trying to pinpoint the mutation(s) in PrV-ΔUL34Pass which is responsible for this phenotype and identify potential cellular targets. These data should give further insight into the property of herpesviruses to modulate the host cell nucleus for their own benefit.
ACKNOWLEDGMENTS
We thank D. Werner, C. Meinke, P. Meyer, and A. Carnitz for expert technical assistance and M. Jörn for photographic help. We also thank an anonymous reviewer for suggesting avoidance of innate immune reactions.
This study was supported by the Deutsche Forschungsgemeinschaft Priority Program SPP 1175, grant Me 854/8-2.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Bubeck A., et al. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farina A., et al. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farnsworth A., et al. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuchs W., Klupp B. G., Granzow H., Osterrieder N., Mettenleiter T. C. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Granato M., et al. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82:4042–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson D. C., Spear P. G. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaplan A. S., Vatter A. E. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407 [DOI] [PubMed] [Google Scholar]
- 8. Klupp B. G., Altenschmidt J., Granzow H., Fuchs W., Mettenleiter T. C. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 82:6299–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klupp B. G., Granzow H., Mettenleiter T. C. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klupp B. G., Granzow H., Mettenleiter T. C. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363–2371 [DOI] [PubMed] [Google Scholar]
- 11. Klupp B. G., et al. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klupp B. G., Granzow H., Keil G. M., Mettenleiter T. C. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klupp B. G., Granzow H., Mundt E., Mettenleiter T. C. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klupp B. G., Hengartner C. J., Mettenleiter T. C., Enquist L. W. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lake C. M., Hutt-Fletcher L. M. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99–106 [DOI] [PubMed] [Google Scholar]
- 16. Leuzinger H., et al. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 79:13047–13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a. Liang L., Tanaka M., Kawaguchi Y., Baines J. D. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68–76 [DOI] [PubMed] [Google Scholar]
- 17. Mettenleiter T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99–115 [DOI] [PubMed] [Google Scholar]
- 17a. Mettenleiter T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mettenleiter T. C., Klupp B. G., Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423–429 [DOI] [PubMed] [Google Scholar]
- 19. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 20. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 21. Neubauer A., Rudolph J., Brandmüller C., Just F. T., Osterrieder N. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189–204 [DOI] [PubMed] [Google Scholar]
- 22. Reynolds A. E., et al. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roller R. J., Zhou Y., Schnetzer R., Ferguson J., DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Zijl M., van der Gulden H., de Wind N., Gielkens A., Berns A. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747–1755 [DOI] [PubMed] [Google Scholar]
- 25. Wagenaar F., et al. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851–1859 [DOI] [PubMed] [Google Scholar]
- 26. Wild P., et al. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]