Abstract
After uptake into target cells, many nonenveloped viruses undergo conformational changes in the low-pH environment of the endocytic compartment. This results in exposure of amphipathic viral peptides and/or hydrophobic protein domains that are inserted into and either disrupt or perforate the vesicular membranes. The viral nucleic acids thereby gain access to the cytosol and initiate replication. We here demonstrate the in vitro transfer of the single-stranded positive-sense RNA genome of human rhinovirus 2 into liposomes decorated with recombinant very-low-density lipoprotein receptor fragments. Membrane-attached virions were exposed to pH 5.4, mimicking the in vivo pH environment of late endosomes. This triggered the release of the RNA whose arrival in the liposomal lumen was detected via in situ cDNA synthesis by encapsulated reverse transcriptase. Subsequently, cDNA was PCR amplified. At a low ratio between virions and lipids, RNA transfer was positively correlated with virus concentration. However, membranes became leaky at higher virus concentrations, which resulted in decreased cDNA synthesis. In accordance with earlier in vivo data, the RNA passes through the lipid membrane without causing gross damage to vesicles at physiologically relevant virus concentrations.
INTRODUCTION
One of the crucial steps in virus infection is the transfer of the viral genome from within the protective capsid through a hostile environment in endocytic vesicles into the cytosol of the host cell. This process is aided by amphipathic peptides that become exposed during viral uncoating (26). In the case of enveloped viruses, such sequences are inserted into the lipid bilayer, causing lipid mixing and fusion of viral and cellular membranes (13, 15); as a result, the viral core enters the cytoplasm without ever being exposed to the outer world. In naked viruses, exposure to low pH and/or contact with a receptor similarly leads to exposure of amphipathic protein domains. However, as these viruses lack a membrane, fusion cannot occur and polypeptides are rather believed to form a channel through the lipid bilayer, thus connecting the viral interior with the cytosol. Alternatively, polypeptides might disrupt the membrane altogether (38). In the first case, it is assumed that viral nucleic acids travel through this pore into the cytosol whereas empty capsids remain in endosomes and are shuttled to lysosomes for degradation. In the case of membrane disruption, entire subviral particles arrive in the cytoplasm where they are further dismantled into nucleic acids and proteins. It is not clear whether pore formation and disruption are mutually exclusive or just reflect two extremes of the same membrane-destabilizing mechanism.
Isolated amphipathic viral proteins and derived synthetic peptides destabilize and disrupt lipid membranes in vitro (12, 41, 43). However, in the context of a virion, proteins act as oligomers with a defined stoichiometry dictated by the icosahedral symmetry of the viral shell. This necessarily affects their mode of action on membranes; therefore, results from experiments with isolated viral peptides alone must be interpreted with caution. Although suggestive, it has not yet been explicitly proven that peptides and/or hydrophobic stretches of capsid proteins could line a channel through the lipid bilayer, thus shielding the charged nucleic acids from the apolar lipids.
Aiming at analyzing genome transfer from naked virions through lipid membranes in vitro, we used human rhinovirus type 2 (HRV2) as a model. HRVs, the major cause of the common cold (24), belong to the genus Enterovirus within the large family of Picornaviridae. They possess a single-stranded (+)RNA genome of approximately 7.1 kb in length enclosed in a 30-nm capsid made from 60 copies of each of the four proteins VP1, VP2, VP3, and VP4. Minor receptor group viruses, such as HRV2, bind low-density lipoprotein receptor (LDLR), very-LDLR, and LDLR-related protein (LRP) for infection (17, 25); they enter the cell by clathrin-dependent receptor-mediated endocytosis (36). On arrival in early endosomes, the acidic pH triggers conformational changes, leading to dissociation from the receptor, release of the innermost capsid protein VP4, and exposure of the amino-terminal amphipathic region of VP1. The now-exposed hydrophobic domains of the resulting subviral A particle (22) are presumably inserted into the lipid bilayer (19). Similarly to a synthetic peptide derived from the N terminus of VP1 (41, 43), isolated VP4 has membrane-permeabilizing activity (12). As shown for the major receptor group virus HRV14 that binds intercellular adhesion molecule 1 (ICAM-1) for cell entry, release of VP4 is necessary for infection, as virus production is halted when maturation cleavage of the capsid protein precursor VP0 to VP2 and VP4 is prevented by mutations (21).
The genomic HRV2 RNA exits the virion and is delivered to the cytoplasm by an unknown mechanism. The empty capsid, also called B particle (22), remains in the endosome (9). Cointernalization of HRV2 and fluorescein isothiocyanate (FITC)-dextrans of 10 and 70 kDa into HeLa cells revealed that the 10-kDa dextran was released from endosomes, whereas the larger dextran was retained. This was taken to indicate the formation of pores of limited size in the endosomal membrane (33, 35). Furthermore, similar to observations for the related poliovirus (37), patch-clamp techniques indicated the opening of ion-permeable pores on triggering conformational changes in membrane-bound HRV2 (14). Rapid degradation of the HRV2 capsid proteins in lysosomes indicates that they do not arrive in the cytosol but rather travel to lysosomes. All these facts support the hypothesis that RNA of HRV2 is shuttled through a pore rather than accessing the cytosol as a consequence of membrane disruption.
As a first step toward a detailed analysis of the early processes in viral infection, we set up an in vitro model system to allow for experiments under well defined conditions; HRV2 was attached to receptor-decorated liposomes filled with reverse transcriptase and reagents required for cDNA synthesis. Upon triggering viral uncoating by lowering the pH, genomic viral RNA entered the protective liposomal lumen, where it was reverse transcribed into cDNA that was subsequently revealed via PCR. Such a nanocontainer system is extremely versatile, as receptors, lipid composition, triggers of viral uncoating, and physicochemical parameters can be varied more easily than in cell culture or under in vivo conditions (6, 7, 40); the influence of these parameters on genome transfer can now be investigated in detail.
MATERIALS AND METHODS
Chemicals.
The lyophilized lipids cholesterol (Ch), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), phosphatidylethanolamine (PE), sphingomyelin (SM), 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid)succinyl] nickel salt (DOGS-NTA), and 1-oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]lauroyl]-sn-glycero-3-phosphocholine (NBD-PC) were from Avanti Polar Lipids (Alabaster, AL) and purchased via Instruchemie (Delfzyl, the Netherlands). Lipids were dissolved in chloroform (pro analysi; Merck) at 10 mM except for DOGS-NTA and NBD-PC, which were at 9.3 and 11.3 mM, respectively. Primers were the same as those of Lu et al (23). The forward primer (CPXGCCZGCGTGGC) was from Exiqon (Vedbaek, Denmark), and the reverse primer (GAAACACGGACACCCAAAGTA) was from VBC Biotech (Vienna, Austria). The deoxynucleoside triphosphate (dNTP) mix and recombinant RNasin were from Promega (Madison, WI). Fivefold reverse transcription (RT) first-strand buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2) and SuperScript III reverse transcriptase were from Invitrogen (Carlsbad, CA). Kapa Sybr Fast quantitative PCR (qPCR) Universal was from Peqlab (Erlangen, Germany). CaCl2, NaCl, Na-acetate (all p.a.), and NaOH (ACS grade) were from E. Merck (Darmstadt, Germany), and Sephadex G50 (medium) was from GE HealthCare (Uppsala, Sweden). Tris (ultrapure) was from AppliChem (Darmstadt, Germany). Nuclease-free water employed for the preparation of RT liposomes was from Qiagen (Hilden, Germany); other water was of Millipore grade. All other chemicals were from Sigma Aldrich (Steinheim, Germany).
Buffers.
RT buffer was prepared by mixing 30 μl reverse primer (10 μM), 15 μl dNTP mix (all dNTPs at 10 mM), 60 μl 5-fold RT first strand buffer, 15 μl recombinant RNasin, 15 μl SuperScript III reverse transcriptase, and 165 μl nuclease-free water. Reaction (RE) buffer was 50 mM Tris-HCl (pH 8.3), 80 mM NaCl. TBSC was 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 10 mM CaCl2.
Virus and receptor.
HRV2 was prepared and its concentration and purity were assessed as described previously (16, 31). MBP-V33333, the recombinant concatemer of five copies of repeat number 3 of human very-low-density lipoprotein receptor (VLDLR) fused to MBP at its N terminus and to a His6 tag at its C terminus, was produced, purified, and folded as described previously (27, 34). Briefly, Escherichia coli DH5α1 carrying the expression plasmid for His6-tagged MBP-V33333 was grown to an A600 of 0.7 in LB medium (Roth, Karlsruhe, Germany), and protein expression was induced with 0.3 mM IPTG (isopropyl-β-D-thiogalactopyranoside; Peqlab) at 30°C. Bacteria were harvested by centrifugation and broken by ultrasonication on ice. Cell debris was removed by centrifugation, and MBP-V33333 was batch purified over Ni2+-NTA beads (Qiagen). The slurry was washed with 10 mM imidazole in TBSC, followed by elution with 250 mM imidazole in TBSC. The sample was extensively dialyzed against TBSC. Cystamin and cysteamin were added to final concentrations of 1 mM and 10 mM, respectively, and folding was allowed to proceed at 4°C for 2 days. The sample was concentrated in a Centricon (Centriprep, Millipore) tube to 2.4 mg/ml, as determined with a Nanodrop (Peqlab) instrument.
Instrumentation.
Spin size exclusion chromatography (spin SEC) was carried out in a table top centrifuge (5415D; Eppendorf, Hamburg, Germany) as described below. Reverse transcription (RT) and PCR were performed using an Eppendorf Mastercycler gradient 5331, and quantitative real-time PCR (qPCR) was performed using an Eppendorf Mastercycler ep realplex. Ultracentrifugation was done in a Beckmann Optima TLX Ultracentrifuge (Brea, CA) for 4 h at 4°C and 4.5 × 104 rpm, employing a TLS55 swing-out rotor and 11- by 34-mm centrifuge tubes. Fluorescence was measured using a Wallac 1420 Victor2 V plate reader (Perkin Elmer, Waltham, MA).
Preparation of nanocontainers.
Stock solutions of lipids were mixed at a molar POPC/PE/SM/Ch/DOGS-NTA/NBD-PC ratio of 1:1:1:1.5:0.5:0.05; NBD-PC was solely included in nanocontainers used for flotation experiments to render the liposomes fluorescent. A lipid film was prepared from 504 μl of this lipid mix (5 μmol total lipids) under a constant stream of nitrogen gas via rotation for 3 h. RT reagents were encapsulated by vortexing the dried lipid film with 300 μl RT buffer several times shortly over a total time period of 2 h. Unilamellar vesicles of uniform size were generated by extrusion of the raw liposomal material (41 times, with 400-nm polycarbonate filters; mini extruder from Avanti Polar Lipids). Dithiothreitol as usually included in RT reaction mixtures was omitted because it appeared to interact with Ni2+ ions on the liposome surface, resulting in brown color. The total phosphate content of extruded nanocontainer stocks was determined via a standard protocol from Avanti Polar Lipids (http://www.avantilipids.com/index.php?option=com_content&view=article&id=1686&Itemid=405 and reference 44); virtually no loss of lipids during liposome preparation was observed.
DOGS-NTA at the outer liposomal membrane was saturated with Ni2+ by incubation with 1.3 mM NiCl2. Subsequently, nonencapsulated RT buffer and excess Ni2+ were removed by spin SEC (2 successive steps in Corning X centrifuge tubes; cellulose acetate membrane; pore size, 0.45 μm; obtained from Szabo-Scandic, Vienna, Austria) using 10-μl aliquots of the liposome suspension on 0.9-ml Sephadex columns in RE buffer as described previously (7, 40, 41). Liposomes collected from the exclusion volume (40 μl) corresponded to 170 nmol total lipid of purified nanocontainers.
Assay for RNA transfer into nanocontainers.
Fifteen microliters of spin SEC-purified suspension of nanocontainers (64 nmol lipid; see above) was incubated for 30 min with 2.4 μl (35 pmol) MBP-V33333, followed by 30 min of incubation with virus suspended in RE buffer at concentrations indicated in the figures. RNA release from bound virus was triggered by pH adjustment to 5.4 for 15 min via addition of a suitable volume (tested in large scale) of 1 M Na-acetate, pH 5.0. Subsequently, the solution was reneutralized with 1 M NaOH. All reaction steps were carried out at room temperature. Negative controls included permeabilization of the nanocontainers by incubation with Triton X-100. RT was performed at 37°C for 1 h, and reverse transcriptase was then inactivated at 70°C for 15 min. Addition of 1% Triton X-100 to the RT and/or PCR had no influence on the signal.
cDNA synthesized in the lumens of the nanocontainers was released by incubation with Triton X-100 (final concentration, 1%) and amplified via PCR (23) as follows: 2 min at 95°C, followed by 60 cycles of 45 s at 95°C, 45 s at 60°C, and 2 min at 72°C, with a final elongation step of 10 min at 72°C. PCR products were analyzed by electrophoresis on a 1% agarose gel (0.5× Tris-acetate-EDTA [TAE] buffer). Gels were scanned and bands quantified with ImageJ (http://rsb.info.nih.gov/ij/). Where flotation was used prior to qPCR, the fraction recovered from the step gradient (about 1 ml) was incubated for 1 h at 37°C as described above to allow for cDNA synthesis. The resulting cDNA was recovered by ethanol precipitation, using glycogen (40 μg) as a carrier. The qPCR was performed by activation at 95°C for 2 min followed by 45 cycles of amplification at 95°C for 15 s and 60°C for 1 min. Standard curves were generated from in vitro-transcribed viral RNA, using 4 serially diluted RNA standards per curve. These showed good linear correlation (R2 = 0.998).
Flotation.
Where indicated, flotation was used to remove either unbound receptor or unbound virus from nanocontainers. A suspension of purified nanocontainers (30 μl) (containing NBD-PC lipid as a fluorescent label) was incubated with 72 pmol MBP-V33333 and subsequently with HRV2 at concentrations indicated in the figure legends under the conditions described above. Samples were brought to 50 μl with RE buffer, mixed with 150 μl 67% (wt/vol) sucrose (in RE buffer), and transferred into centrifuge tubes. This cushion was sequentially overlaid with 900 μl 25% sucrose-RE buffer and 900 μl RE buffer, followed by ultracentrifugation. Twelve fractions of 167 μl each were collected from the top of the gradient and nanocontainer-containing fractions localized by fluorescence measurement. Furthermore, the virus concentration of corresponding fractions was determined as 50% tissue culture infective dose (TCID50) (8). The 4 fractions exhibiting highest fluorescence values were combined (including >90% of the total fluorescence) and assayed for RNA transfer into nanocontainers. Additionally, the material was inspected by negative-stain transmission electron microscopy (TEM). As an additional control, nanocontainers (with or without receptor decoration) were incubated with virus and acidified at room temperature. After reneutralization, they were separated from excess unbound virus by flotation at 4°C, recovered, and incubated at 37°C to allow for cDNA synthesis as described above.
Leakage determination.
Leakage of Atto 637 from liposomes was assessed as described previously (41). Briefly, the dye was encapsulated at 11 μM in 50 mM Na-borate, pH 8.3, and nonencapsulated dye was removed by spin SEC. The purified liposomes were incubated with MBP-V33333 and subsequently with HRV2. The complexes were acidified for 15 min and subjected to chip electrophoresis. Leakage is expressed as the ratio of the peak area of encapsulated (i.e., liposome-associated) and free fluorophore. This value was related to the average ratio determined in the absence of virus (no leakage) and expressed as percent intact liposomes.
Transmission electron microscopy.
Carbon-coated copper grids (Agar Scientific; purchased via Groepl, Tulln, Austria) were glow discharged at 20 mA for 1 min in a modified Bal-Tec SCD005 sputter coater with the Au target removed. Four microliters of the sample was adsorbed for 1 min, and the grid was washed and stained with 2% sodium phosphotungstate (pH 7.2) for 1 min. TEM imaging was done on a FEI Morgagni instrument (FEI Tecnai, Eindhoven, the Netherlands) employing an 11-Mpixel Morada charge-coupled-device (CCD) camera at 5.6 × 104-fold magnification.
RESULTS
In vivo, uncoating of HRV2 occurs in late endosomes (32). Nanocontainers were thus prepared from a lipid mixture similar in composition to endosomal membranes (18, 39) and to liposomes previously used in virus-membrane interaction studies (7, 10, 42). To allow for attachment of His6-tagged recombinant receptors, DOGS-NTA was incorporated into membranes. We found that 10 mol% of this lipid was necessary for His6-tagged receptors to remain firmly attached during subsequent purification and incubation steps. Replacement with the trivalent DOGS-tris-NTA (2) reduced receptor loss at lower lipid concentrations but also led to substantial aggregation (data not shown); therefore, only DOGS-NTA was used in all following experiments. Liposomes were prepared in a buffer containing all components for RT. This mixture became enclosed in the liposomal lumen, giving rise to nanocontainers. With the aid of such nanocontainers, we intended to follow the RNA transfer of the HRV2 genome through the liposomal membrane.
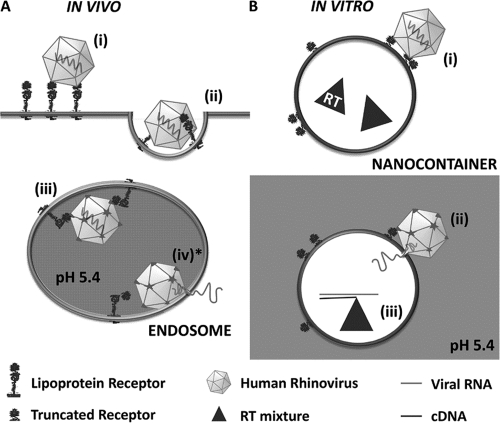
In Fig. 1, the different (in part hypothetic) steps in viral RNA transfer through lipid membranes in vivo (Fig. 1A) and in vitro (Fig. 1B) are schematized. Whereas viral uncoating in endosomes occurs on the inner face of the membrane, it must take place from the outside in the case of nanocontainers. However, since acidification of HRV2 prebound to the plasma membrane of HeLa cells results in productive uncoating (from outside) (4, 5), small differences in lipid composition (such as those between endosomes and the plasma membrane), membrane curvature, and sidedness seemingly do not play a significant role in rhinoviral RNA transfer.
Fig. 1.
Scheme of in vivo and in vitro HRV2 uncoating. (A) In vivo uncoating in the cell. Virus binds to the receptor on the host cell (i) and is taken up via clathrin-dependent endocytosis (ii). Upon arrival in endosomes (iii), the low pH triggers virus release from its receptor. VP4 molecules and N-terminal sequences of VP1 are externalized, and low-pH-induced conformational changes of the receptor cause handing over of the hydrophobic particle to the lipid membrane. The hydrophobic domains of VP1 are inserted into the membrane and, most probably assisted by VP4, a pore is formed allowing for transfer of the RNA into the cytosol (iv*). (B) In vitro uncoating on nanocontainers. Virus is bound to truncated recombinant receptors attached via His6 tags to Ni2+-loaded NTA lipids on the surfaces of nanocontainers (i). Upon triggering the structural changes by exposure to acidic buffer, the RNA is transferred through the liposomal membrane into the aqueous lumen (ii), where it is transcribed into cDNA (iii) that is subsequently detected via PCR. Note that in vivo uncoating occurs from within whereas in the in vitro system it is from without. The step modeled experimentally in the present communication is marked with an asterisk.
Conversion of liposome-bound HRV2 from native virions to empty capsids at low pH is time dependent and involves intermediate structures.
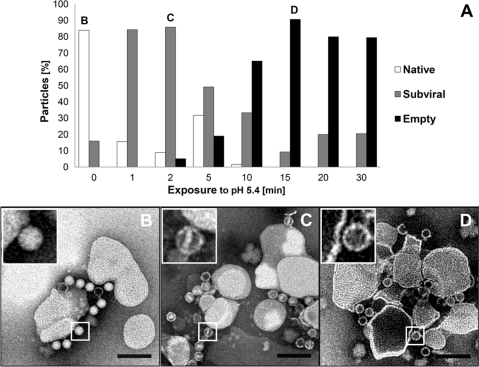
Nanocontainers purified by spin SEC were decorated with His6-tagged MBP-V33333 via the DOGS-NTA incorporated into the lipid membrane to allow for subsequent HRV2 binding (nanocontainer/receptor/virus [NC+R+V] complexes). Since the multivalent receptor impedes movements of the capsid proteins with respect to each other (19, 29), HRV2-receptor complexes require a lower pH for conversion into subviral particles than free virus. Therefore, in order to keep the concentration of free receptor low (to avoid formation of virus-receptor complexes in solution), it was used at a molar ratio of about 0.01 with respect to Ni2+-NTA groups (assuming that 50% of the NTA groups are found on the inner leaflet). Additionally, NC+R samples were subjected to flotation. After ultracentrifugation, HRV2 was added to the pooled fractions with the highest fluorescence at a virus-to-lipid ratio of 1.85 × 10−6 and incubated for 1 h to allow for viral attachment. The pH was brought to 5.4 to trigger viral uncoating (see scheme in Fig.1Bii), and aliquots of the acidified complexes were removed after different incubation times, directly adsorbed to carbon-coated copper grids, and stained for TEM. Imaging clearly showed a time-dependent conversion of native virions to empty capsids (Fig. 2A). Within 5 min of exposure to pH 5.4, native virions (Fig. 2B and inset) converted into intermediate particles whose interior was accessible to negative stain. Part of the internal density was lost, suggesting partial RNA release and stain penetration through openings in the capsid. Most particles contained residual rod-shaped internal density (Fig. 2C and inset), suggesting an intermediate stage of the viral uncoating process. Extension of exposure to pH 5.4 for 15 min led to a significant loss of this density inside the virions, giving rise to empty capsids (B particles) that remained membrane associated (Fig. 2D and inset). It is noteworthy that empty capsids generated in solution by exposure to a temperature above 50°C are hydrophilic and fail to attach to membranes (30). Whether acidification and the nonphysiologic exposure to elevated temperature give rise to (slightly) different subviral particles is a matter of debate and still under investigation.
Fig. 2.
Negative-stain transmission electron microscopy demonstrates HRV2 uncoating at the surfaces of receptor-decorated liposomes. Liposomes containing DOGS-NTA lipid were decorated with His6-tagged receptors and nonbound receptor was removed via flotation. Virus was attached at a virus-to-lipid ratio of 1.85 × 10−6, exposed to pH 5.4 for the times indicated and adsorbed to carbon-coated copper grids. Samples were stained with 2% phosphotungstate, pH 7.2, and viewed in a transmission electron microscope at 5.6 × 104-fold magnification. Particles were classified as native, subviral, and empty according to core density in negative stain and quantified from TEM images. (A) Distribution of the particle class as a function of incubation time. About 200 particles were examined per time point. Images taken at 0 min (B), 2 min (C), and 15 min (D) are shown. The typical appearance of the particles is depicted in the respective insets. Note that the sequence of formation of subviral and empty particles was reproducible but the absolute times varied for different virus preparations. Size bar = 200 nm.
Viral RNA arrives in nanocontainers.
Having visualized viral uncoating intermediates (early after acidification) and empty capsids (later after acidification) on the surfaces of liposomes by negative-stain TEM, we asked whether the RNA lost from the virions was transferred into the lumens of the nanocontainers. To exclude that RT occurs in the outside medium, removal of nonencapsulated reverse transcriptase and RT reaction components via spin SEC was important. Thereby, only RNA transferred through the lipid bilayer into the lumens of the nanocontainers would be detected via cDNA synthesis by the encapsulated reverse transcriptase and subsequently be amplified by PCR (see scheme in Fig.1Biii). Flotation of NC+R+V samples could be omitted, as only those virions uncoating in close vicinity of the nanocontainers would be able to transfer their RNA into the nanocontainers and result in amplified DNA. Nevertheless, as an additional control, we also subjected nanocontainers to flotation after triggering RNA transfer. The cDNA synthesis was then allowed to proceed in these purified nanocontainers by incubation at 37°C (see below).
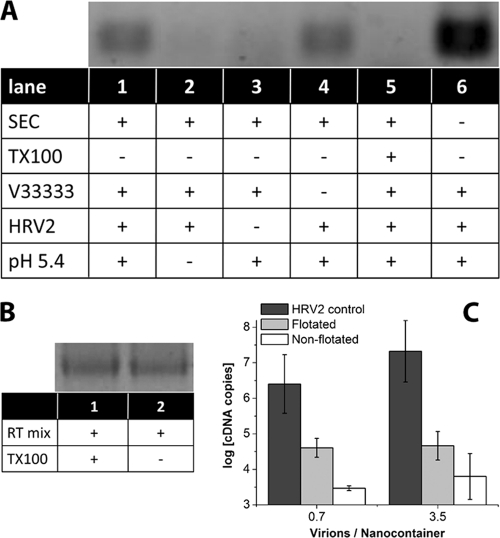
Receptor-decorated nanocontainers were incubated with virus, and uncoating was triggered by acidification for 15 min, the time point where most of the virus had been converted into empty capsids (Fig. 2). Analysis for the presence of cDNA demonstrated that on acidification, viral RNA indeed arrived in the nanocontainers (Fig. 3A, lane 1). No signal was apparent when incubation with the acidic buffer (Fig. 3A, lane 2), or HRV2 (Fig. 3A, lane 3) was omitted, excluding amplification of contaminating nucleic acids. Furthermore, no signal was seen when the nanocontainers were permeabilized with detergent prior to receptor decoration (Fig. 3A, lane 5), demonstrating that RT reagents leaking out of nanocontainers are too dilute for cDNA synthesis to occur outside the vesicles. To exclude an influence of the detergent on the enzymatic activities, an RT-PCR on in vitro-transcribed viral RNA was run in the presence (Fig. 3B, lane 1) and in the absence (Fig. 3B, lane 2) of 1% Triton X-100, giving rise to bands with essentially the same intensity. This excludes that addition of Triton X-100 reduces the signal and definitely shows that the nanocontainers must remain intact for the RT reaction to occur.
Fig. 3.
Viral RNA is transferred into nanocontainers upon incubation at low pH. (A) HRV2 (0.36 pmol) was bound to 17.4 μl of receptor-decorated nanocontainers (at a virus-to-lipid ratio of 5.6 × 10−6). The mixture was adjusted to 20 μl, brought to pH 5.4 via addition of 1.2 μl 1 M sodium acetate, pH 5.0, and incubated for 15 min at room temperature prior to reneutralization by addition of 0.6 μl 1 M sodium hydroxide. RNA transferred into the liposomal lumen was reverse transcribed by the encapsulated reverse transcriptase at 37°C for 1 h (see scheme in Fig. 1B). Synthesized cDNA was made accessible for subsequent PCR by addition of Triton X-100. The amplified 206 bp fragment was detected by agarose gel electrophoresis. Components present or absent and treatments of the nanocontainers are indicated by (+) and (−), respectively. SEC, not encapsulated RT buffer was removed from the nanocontainers via spin SEC; TX-100, nanocontainers were destroyed with detergent Triton X-100 prior to decoration with receptor; MBP-V33333, the receptor was present on the surfaces of nanocontainers; HRV2, virus was added; pH 5.4, the outside medium was brought to acidic pH. (B) In order to exclude inhibition of the RT-PCR by TX-100, reverse transcription and amplification were carried out by using in vitro-transcribed viral RNA in the presence (lane 1) and in the absence (lane 2) of the detergent. Note that the signal strength is identical. (C) RNA transfer into nanocontainers (without receptor decoration) was triggered as described for panel A, 1 aliquot was directly subjected to qRT-PCR, and 1 was first subjected to flotation. For comparison, the number of RNA copies released from virus into the medium on incubation at pH 5.4 in the absence of nanocontainers is also depicted (HRV2 control).
The signal in Fig. 3A, lane 6, emphasizes the importance of spin SEC in removal of excess reagents. Apparently, upon acidification, substantial quantities of viral RNA are shed into the medium and only part of it arrives in the lumens of the nanocontainers. In the presence of undiluted RT mixture outside the nanocontainers, cDNA synthesis occurs.
Unexpectedly, cDNA synthesis also occurred upon incubation at pH 5.4 when no receptor was present (Fig. 3A, lane 4). Apparently, structural changes of the virion close to the bare lipid membrane result in viral attachment via externalized amphipathic sequences followed by RNA transfer. This is in agreement with low but consistent infection of receptor-negative cells by A particles of the closely related poliovirus (11). However, as shown below, when free virus was removed by flotation of the nanocontainers prior to triggering of RNA release with low-pH buffer, the assay was completely dependent on the presence of the receptor. As an additional control, nanocontainers were incubated with virus and RNA transfer into the lumen was triggered via incubation at pH 5.4 as described above. However, the lipid vesicles were then separated from free virus and eventually released subviral particles by flotation at 4°C. The nanocontainers recovered from the sucrose step gradient were subsequently incubated at 37°C to allow for reverse transcription. Synthesized cDNA was quantified by qPCR. As seen in Fig. 3C, viral RNA had arrived in the nanocontainers, where it remained during the separation step. Assuming 100% recovery from the step gradient, in the case of on average of 3.5 virions per nanocontainer (see also below), roughly 1 RNA molecule out of 400 virions made it into the lipid vesicles. This is based on the difference (about 2.6 logs) (Fig. 3C) between RNA released by acidification of the same amount of virus (i.e., control incubation in the absence of nanocontainers) and that recovered from the nanocontainers. For an average of 0.7 virions per nanocontainer, this value was even around 60. This is in the same order of magnitude as the ratio between noninfectious physical particles and infectious virions observed in vivo (between 24 and 240, according to reference 1, and 2210 and 6500, according to reference 20). It is noteworthy that the signal was consistently higher for the sample subjected to flotation. A control experiment excluded that cDNA synthesis was modified by the sucrose present in the gradient fraction (data not shown). The lower signal observed without flotation could be due to leakage when excess virus and/or subviral particles remained in the sample during the incubation at 37°C (see below). Taken together, all these controls confirm that cDNA is synthesized only when viral RNA is transferred through the lipid membrane into the aqueous lumen without disruption of the nanocontainers.
Coflotation of virions and nanocontainers depends on the receptor.
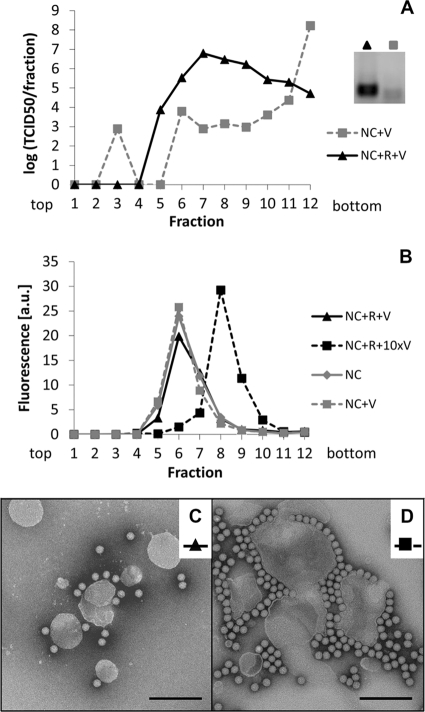
Nanocontainers with or without receptors were incubated with virus and subjected to flotation. Fractions were taken from the top of the sucrose step gradient. Nanocontainer-containing fractions were determined via fluorescence measurements, and the concentration of infectious virus was determined as the TCID50 by an endpoint dilution assay (8). Additionally, the material was examined by negative-stain TEM (Fig. 4C and D). In the presence of the receptor, most of the virus attached to nanocontainers (Fig. 4A). On the sucrose step gradient, the complex was marginally shifted toward lower density than that of nanocontainers without a receptor. At a ratio between virus and nanocontainers 10 times higher, this peak shift was substantial (Fig. 4B), indicating a sensible increase in the density of the nanocontainers when carrying virus. Examination of the peak fractions by TEM showed about 3 virions attached per nanocontainer (Fig. 4C) in the case of the low virus concentration (calculated virus/lipid ratio of 1.4 × 10−6). At a 10-fold-higher virus/lipid ratio, substantial aggregation was observed (Fig. 4D). When nanocontainers without receptor were incubated with virus and subjected to flotation, most of the infectivity remained in the high-density fraction at the bottom of the step gradient (Fig. 4A). No cDNA synthesis was seen after acidification of the sample lacking receptor (pool of fractions 5 to 9; squares), whereas a clear signal was observed in the sample of the nanocontainers carrying receptors (pool of fractions 5 to 9; triangles) and, consequently, virus (Fig. 4A, inset).
Fig. 4.
About 3 virions per liposome are sufficient to detect RNA transfer. Nanocontainers (NC) containing a fluorescent tracer lipid were decorated with receptor (R) and virus (V) was attached. Unbound particles were removed by flotation on a sucrose step gradient. (A) Infectious virus in the recovered fractions was detected via its infectivity as TCID50. (B) Nanocontainer-containing fractions were identified by fluorescence measurement. Binding of about 3 virus particles per liposome (i.e., resulting in a virus-to-lipid ratio of about 1.4 × 10−6) (see corresponding TEM image in panel C) led to slight broadening of the liposome peak. Higher virus concentrations (about 30 particles per liposome, i.e., at a virus-to-lipid ratio of about 1.4 × 10−5; see panel D) caused a shift of the liposome peak to higher density fractions. Coflotation of HRV2 occurred only with receptor-decorated vesicles; in the absence of the receptor, virtually all virus remained in the bottom fraction of the gradient (note the logarithmic scale) and no signal was seen for the RNA transfer reaction (A, inset). (C and D) Material in the pooled peak fractions in panel B was adsorbed to glow discharged, carbon-coated copper grids and stained with 2% phosphotungstate, pH 7.2. Images were taken under a transmission electron microscope at 5.4 × 104-fold magnification. Size bar = 200 nm. Note that the faint band in the sample without receptor (inset) is irrelevant, as it was also present in samples without a template.
Liposomal nanocontainers become leaky at high virus concentrations.
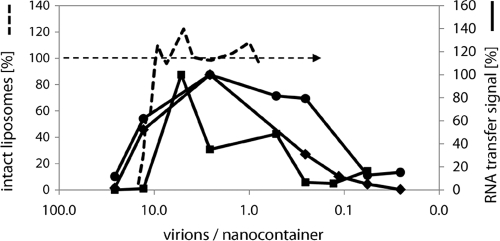
First experiments repeatedly showed a peak in synthesized cDNA at a virus-to-lipid ratio of 1.4 × 10−6, roughly corresponding to a mean of 3 virus particles per 400 nm liposome, as also detected by TEM (see above) (Fig. 4C). We thus assessed the dependence of cDNA synthesis on virus concentration. A positive correlation was found below a virus/lipid ratio of about 10−6; at higher virus concentrations, the cDNA signal diminished again (Fig. 5).
Fig. 5.
Luminal cDNA synthesis and liposomal leakage depend on virus concentration. Increasing virus concentration results in increased cDNA synthesis up to a virus/lipid ratio of about 10−6 (<10 virions/nanocontainer); at higher ratios the cDNA signal decreases again. RNA transfer was measured in three independent assays as described for Fig. 3, but using HRV2 at different concentrations as indicated. Bands corresponding to the amplified cDNA were quantified by using ImageJ (solid black lines). Data on leakage of a fluorophore from receptor-decorated liposomes measured with chip electrophoresis as in (41) are included for comparison (dashed line). Note that the decrease of the cDNA signal with higher virus concentrations correlates with the decrease of intact (tight) liposomes.
Weiss and colleagues previously showed that HRV2 induces leakage of low-molecular-weight dye from the liposomal lumen in a concentration-dependent manner (41). Therefore, substantial leakage, starting around a virus-to-lipid ratio of 10−5 (corresponding to roughly 10 virions per liposome), counteracts cDNA synthesis by releasing the components of the reverse transcriptase reaction into the outside medium where they are being diluted to a degree that does not allow the RT reaction to proceed (data obtained with receptor-decorated liposomes as described in reference 41 are also shown in Fig. 5 for comparison). Under the same conditions, but using liposomes without receptor, similar leakage effects were observed (data not shown). As stated above, the lack of a stimulating effect of the receptor might be explained by the multivalent nature of the binding impeding movements of viral capsid proteins with respect to each other (29), thus inhibiting the structural changes necessary for exposure of amphipathic domains. The stabilization of the virus might thus counteract the virus-concentrating effect of the receptor and explain the slightly increased RNA transfer signal observed in the absence of the receptor (Fig. 3A, lane 4).
DISCUSSION
Transfer of the nucleocapsids of enveloped viruses via membrane fusion is relatively well understood (13, 15). Much less is known on the release of genomic nucleic acids of nonenveloped animal viruses and, in particular, on viruses containing a single-stranded RNA molecule (3, 14, 38). Not only is the question as to what unwinds and drives the highly structured RNA through a pore opened in the viral capsid ignored, but also its transit through cellular membranes is enigmatic. For the latter step, essentially two mechanisms have been discussed; the RNA enters the cytosol either by disrupting the (endosomal) membrane or via a pore in the lipid bilayer, presumably lined by viral proteins. Invariably, the hydrophobic/amphiphilic peptides and/or stretches of viral capsid proteins present in these viruses and becoming accessible upon exposure to low pH or contact with receptors are involved in destabilization of the membrane. In the case of HRV2, it is unequivocally established that the low endosomal pH initiates the structural changes of the virion (28) that lead to irreversible exposure of N-terminal sequences of VP1 and release of VP4, which both play a role in RNA transfer into the cytosol (12, 41, 43).
In this report, we established an assay for the detection of the passage of the viral RNA through a lipid membrane that closely mimics the process occurring in vivo. This was possible by incorporating reverse transcriptase, together with all components required for cDNA synthesis, in the lumens of liposomes. Only RNA arriving in these nanocontainers can be transcribed into cDNA, providing the template for the following PCR used for its detection. This experimental setup allowed us to demonstrate a strict dependence of RNA transfer on exposure of HRV2 to low pH.
On the one hand, we observed that the signal reflecting the amount of RNA arriving in the nanocontainers was dependent on the viral concentration. However, upon exceeding a threshold that marked the detectable leakage of a fluorescent dye from similar liposomes, as demonstrated previously (41), the signal diminished again. We interpret this behavior as an indication of the loss of the components of the reverse transcriptase mixture from the nanocontainers and their dissipation in the outside medium. They thereby become too dilute to sustain RT of the viral RNA. In conclusion, at low virus concentrations and exposure to acidic pH, as occurring in vivo inside endosomes, the viral RNA accesses the cytoplasm via a pore. Conversely, at high concentrations, the viruses disturb the lipid membrane to an extent that results in leakage. This strongly suggests that in vivo, the RNA, at least in the case of the minor receptor group virus HRV2, is transferred through a pore.
As seen after flotation of nanocontainers in a sucrose density gradient, either bare or decorated with receptors, RNA transfer was dependent on the presence of the receptor. When absent, the virus failed to attach and did not comigrate upon ultracentrifugation; consequently, acidification did not result in cDNA production. However, when bare nanocontainers were not separated from the virus by centrifugation, we observed that the receptor was dispensable for RNA transfer itself. This is in accordance with previous results that showed that HRV2 dissociates from its receptor at the low pH in endosomes and that a receptor without the β-propeller domain, as used in our present experiments, even stabilizes the virus (19, 29). When nanocontainers were mixed with virus, acidified, and then separated from excess virus and/or residual subviral particles by flotation at 4°C, their incubation at 37°C led to synthesis of cDNA. This clearly shows that the viral RNA had been transferred into the nanocontainers, where it remained during the flotation and was available for subsequent reverse transcription.
Ongoing studies will show whether RNA transfer is increased when virus dissociation occurs more easily in the presence of the β-propeller domain of the LDL receptor. It is noteworthy that subviral 135S particles of poliovirus are infectious, although to a much reduced degree (11). Therefore, the hydrophobic nature of the subviral particles might be the main driving force for direct membrane interaction and the ensuing infection.
We shall also investigate whether major group rhinoviruses, such as HRV14, that bind ICAM-1 instead of members of the LDL receptor family also transfer their RNA through a pore in the endosomal membrane or rather access the cytosol via disruption of the membrane, as suggested by cointernalization and dye release experiments (35). We believe that the system presented in this report will be of general utility for the study of viral genome transfer through membranes during infection with nonenveloped viruses and allow distinguishing between pore formation and disruption.
ACKNOWLEDGMENTS
We thank Irene Gösler for virus preparation, Günter Resch, Marlene Brandstetter, and Nicole Fellner for help with the electron microscope, and Agilent Technologies, GmbH, Waldbronn, Germany, for support with the Bioanalyzer. Robert Tampé, Goethe-University Frankfurt, Germany, kindly provided us with DOGS-tris-NTA.
This project was supported by Austrian Science Foundation (FWF) grants P18693-B09, P19365, and P20915-B13. X.S. thanks the Government of Catalonia for financial support (grant 2008BPA00029).
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Abraham G., Colonno R. J. 1984. Many rhinovirus serotypes share the same cellular receptor. J. Virol. 51:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andre T., et al. 2009. Selectivity of competitive multivalent interactions at interfaces. Chembiochem 10:1878–1887 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee M., Johnson J. E. 2008. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr. Protein Pept. Sci. 9:16–27 [DOI] [PubMed] [Google Scholar]
- 4. Baravalle G., Brabec M., Snyers L., Blaas D., Fuchs R. 2004. Human rhinovirus type 2-antibody complexes enter and infect cells via Fc-gamma receptor IIB1. J. Virol. 78:2729–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berka U., Khan A., Blaas D., Fuchs R. 2009. Human rhinovirus type 2 uncoating at the plasma membrane is not affected by a pH gradient but is affected by the membrane potential. J. Virol. 83:3778–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bilek G., Kremser L., Wruss J., Blaas D., Kenndler E. 2007. Mimicking early events of virus infection: capillary electrophoretic analysis of virus attachment to receptor-decorated liposomes. Anal. Chem. 79:1620–1625 [DOI] [PubMed] [Google Scholar]
- 7. Bilek G., Weiss V. U., Pickl-Herk A., Blaas D., Kenndler E. 2009. Chip electrophoretic characterization of liposomes with biological lipid composition: coming closer to a model for viral infection. Electrophoresis 30:4292–4299 [DOI] [PubMed] [Google Scholar]
- 8. Blake K., O'Connell S. 1993. Virus culture, p. 81–122. In D. R. Harper (ed.), Virology labfax. Blackwell Scientific Publications, London, United Kingdom [Google Scholar]
- 9. Brabec-Zaruba M., Pfanzagl B., Blaas D., Fuchs R. 2009. Site of human rhinovirus RNA uncoating revealed by fluorescent in situ hybridization. J. Virol. 83:3770–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bubeck D., Filman D. J., Hogle J. M. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 12:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curry S., Chow M., Hogle J. M. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis M. P., et al. 2008. Recombinant VP4 of human rhinovirus induces permeability in model membranes. J. Virol. 82:4169–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falanga A., Cantisani M., Pedone C., Galdiero S. 2009. Membrane fusion and fission: enveloped viruses. Protein Pept. Lett. 16:751–759 [DOI] [PubMed] [Google Scholar]
- 14. Fuchs R., Blaas D. 2010. Uncoating of human rhinoviruses. Rev. Med. Virol. 20:281–297 [DOI] [PubMed] [Google Scholar]
- 15. Harrison S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hewat E. A., et al. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hofer F., et al. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. U. S. A. 91:1839–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kobayashi T., et al. 2002. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 277:32157–32164 [DOI] [PubMed] [Google Scholar]
- 19. Konecsni T., et al. 2009. Low pH-triggered beta-propeller switch of the low-density lipoprotein receptor assists rhinovirus infection. J. Virol. 83:10922–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korant B. D., Lonberg Holm K., Noble J., Stasny J. T. 1972. Naturally occurring and artificially produced components of three rhinoviruses. Virology 48:71–86 [DOI] [PubMed] [Google Scholar]
- 21. Lee W. M., Monroe S. S., Rueckert R. R. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lonberg-Holm K., Gosser L. B., Shimshick E. J. 1976. Interaction of liposomes with subviral particles of poliovirus type 2 and rhinovirus type 2. J. Virol. 19:746–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu X., et al. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mackay I. M. 2008. Human rhinoviruses: the cold wars resume. J. Clin. Virol. 42:297–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marlovits T. C., Abrahamsberg C., Blaas D. 1998. Soluble LDL minireceptors—minimal structure requirements for recognition of minor group human rhinovirus. J. Biol. Chem. 273:33835–33840 [DOI] [PubMed] [Google Scholar]
- 26. Marsh M., Helenius A. 2006. Virus entry: open sesame. Cell 124:729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moser R., et al. 2005. Neutralization of a common cold virus by concatemers of the third ligand binding module of the VLDL-receptor strongly depends on the number of modules. Virology 338:259–269 [DOI] [PubMed] [Google Scholar]
- 28. Neubauer C., Frasel L., Kuechler E., Blaas D. 1987. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology 158:255–258 [DOI] [PubMed] [Google Scholar]
- 29. Nicodemou A., et al. 2005. Rhinovirus-stabilizing activity of artificial VLDL-receptor variants defines a new mechanism for virus neutralization by soluble receptors. FEBS Lett. 579:5507–5511 [DOI] [PubMed] [Google Scholar]
- 30. Noble J. N., Lonberg-Holm K. 1973. Interactions of components of human rhinovirus type 2 with HeLa cells. Virology 51:270–278 [DOI] [PubMed] [Google Scholar]
- 31. Okun V. M., Ronacher B., Blaas D., Kenndler E. 1999. Analysis of common cold virus (human rhinovirus serotype 2) by capillary zone electrophoresis: the problem of peak identification. Anal. Chem. 71:2028–2032 [DOI] [PubMed] [Google Scholar]
- 32. Prchla E., Kuechler E., Blaas D., Fuchs R. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prchla E., Plank C., Wagner E., Blaas D., Fuchs R. 1995. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J. Cell Biol. 131:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ronacher B., Marlovits T. C., Moser R., Blaas D. 2000. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology 278:541–550 [DOI] [PubMed] [Google Scholar]
- 35. Schober D., Kronenberger P., Prchla E., Blaas D., Fuchs R. 1998. Major and minor-receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol. 72:1354–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snyers L., Zwickl H., Blaas D. 2003. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J. Virol. 77:5360–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tosteson M. T., Chow M. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23–43 [DOI] [PubMed] [Google Scholar]
- 39. van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiss V. U., Bilek G., Pickl-Herk A., Blaas D., Kenndler E. 2009. Mimicking virus attachment to host cells employing liposomes: analysis by chip electrophoresis. Electrophoresis 30:2123–2128 [DOI] [PubMed] [Google Scholar]
- 41. Weiss V. U., et al. 2010. Liposomal leakage induced by virus-derived peptides, viral proteins, and entire virions: rapid analysis by chip electrophoresis. Anal. Chem. 82:8146–8152 [DOI] [PubMed] [Google Scholar]
- 42. White J., Kartenbeck J., Helenius A. 1980. Fusion of semliki forest virus with the plasma membrane can be induced by low pH. J. Cell Biol. 87:264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zauner W., Blaas D., Kuechler E., Wagner E. 1995. Rhinovirus-mediated endosomal release of transfection complexes. J. Virol. 69:1085–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zuidam N. J., de Vrueh R., Crommelin D. J. A. 2003. Characterization of liposomes, p. 31–34. In V. P. Torchilinand and V. Weissig (ed.), Liposomes, 2nd ed Oxford University Press, Oxford, United Kingdom [Google Scholar]