Abstract
Reports of a high frequency of the transmission of minority viral populations with drug-resistant mutations (DRM) are inconsistent with evidence that HIV-1 infections usually arise from mono- or oligoclonal transmission. We performed ultradeep sequencing (UDS) of partial HIV-1 gag, pol, and env genes from 32 recently infected individuals. We then evaluated overall and per-site diversity levels, selective pressure, sequence reproducibility, and presence of DRM and accessory mutations (AM). To differentiate biologically meaningful mutations from those caused by methodological errors, we obtained multinomial confidence intervals (CI) for the proportion of DRM at each site and fitted a binomial mixture model to determine background error rates for each sample. We then examined the association between detected minority DRM and the virologic failure of first-line antiretroviral therapy (ART). Similar to other studies, we observed increased detection of DRM at low frequencies (average, 0.56%; 95% CI, 0.43 to 0.69; expected UDS error, 0.21 ± 0.08% mutations/site). For 8 duplicate runs, there was variability in the proportions of minority DRM. There was no indication of increased diversity or selection at DRM sites compared to other sites and no association between minority DRM and AM. There was no correlation between detected minority DRM and clinical failure of first-line ART. It is unlikely that minority viral variants harboring DRM are transmitted and maintained in the recipient host. The majority of low-frequency DRM detected using UDS are likely errors inherent to UDS methodology or a consequence of error-prone HIV-1 replication.
INTRODUCTION
Using standard population-based genotypic assays of HIV from individuals not yet treated with antiretroviral drugs, several studies (43, 64, 69, 76) have estimated the rate of transmitted drug resistance to be between 8 and 27% in countries with the highest rates of antiretroviral therapy (ART) use. In resource-limited settings, in which the introduction of ART has been more recent, the estimated frequency of transmitted drug resistance mutations (DRM) is substantially lower (41). It appears, however, to be increasing in these settings as well (2, 14, 51). Using more-sensitive genotypic assays, different research groups (15, 30, 32, 37, 48, 58, 65) have reported higher proportions of transmitted DRM in ART-naive individuals. The clinical importance of these low-level DRM remains unclear, as they have been associated with clinical consequences in some (21, 24, 32, 36, 37, 40, 46, 54, 55, 63, 66, 71) but not all (30, 47, 58) studies.
Highly sensitive assays for detecting low-frequency DRM include point mutation assays and high-resolution sequencing techniques. Point mutation assays, such as allele-specific PCR, can detect DRM at frequencies as low as 0.01% of the sampled viral population (31, 45, 53, 54), but they do not provide information about the sequence context surrounding a given DRM and may be prone to false positives at the lower level of detection (22). High-resolution sequencing techniques, such as single genome sequencing (SGS) and ultradeep sequencing (UDS), permit analyses of DRM in the context of the surrounding genetic sequence and this allows the investigation of accessory mutations (AM) that are often associated with DRM during DRM selection (50). Recent studies have suggested that highly sensitive PCR-dependent sequencing techniques could also lead to spurious detection of DRM at low levels (29, 68, 72).
Recent analyses of genetic diversity in the HIV-1 env gene during acute and early infection indicated that productive infections are derived predominantly from a single, or less frequently several, founder strains (1, 19, 25, 33). Differences in routes of virus transmission, clinical stage, and viral load of the source partner, as well as the presence of coinfections, may influence the complexity of transmitted viral populations (7, 33, 39). The reported high prevalence of transmitted drug resistance when the DRM in pol are present at very low levels is not consistent with the descriptions of mono- or oligoclonal transmission in the env coding region by SGS and UDS (1, 19, 25, 33). These discordant observations could be explained in several ways: (i) DRM emerge de novo early after infection and replicate as a relatively substantial proportion of the viral population, (ii) highly sensitive assays generate high rates of false positives, and (iii) recombination occurs between env and pol during acute infection and is followed by selection (on env) to yield a phylogenetically homogeneous population in env but not in pol. To examine these hypotheses, we performed UDS of the baseline samples of 32 recently HIV-infected individuals and quantified sequence diversity, DRM prevalence and proportion, linkage to AM, selective pressure and diversity levels at each site, experimental reproducibility, and impact on first-line ART.
MATERIALS AND METHODS
Participants, sample collection, and clinical assays.
Blood and urine samples from 32 subjects from the San Diego Primary Infection Cohort (42) were analyzed. All samples were collected less than 4 months after each participant's estimated date of infection, as calculated using established algorithms (18, 27, 42). Upon collection, samples were aliquoted, frozen, and stored at −80°C. At all time points, CD4 cell counts (LabCorp) and blood plasma HIV-1 RNA levels (Amplicor HIV-1 Monitor Test; Roche Molecular Systems, Inc.) were quantified.
Population-based sequencing and subtyping.
Standard genotypic HIV-1 drug resistance tests were performed on baseline blood plasma samples and on the first available specimen after viral failure using the population-sequencing-based Viroseq platform (version 2.0; Celera Diagnostics, Foster City, CA). Baseline pol sequences were used for HIV-1 subtype assignment using SCUEAL (http://www.datamonkey.org/GASP) (35).
UDS.
HIV-1 RNA was isolated from blood plasma (QIAamp viral RNA minikit; Qiagen, Hilden, Germany), and cDNA was produced (RETROscript kit; Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's instructions. If HIV-1 RNA levels (i.e., viral load) exceeded 20,000 HIV-1 RNA copies/ml in the sample, then 500 μl of blood plasma was used; if the viral load was below 20,000 HIV RNA copies/ml, then 1 ml of blood plasma was used. Three coding regions—gag p24 (HXB2 coordinates 1366 to 1619), pol reverse transcriptase (RT) (HXB2 coordinates 2708 to 3242), and env C2-V3 (HXB2 coordinates 6928 to 7344)—were amplified by PCR with region-specific primers as previously described (8, 52). Rubber gaskets were used to physically separate 16 samples on a single 454 GS FLX titanium picoliter plate (454 Life Sciences/Roche, Branford, CT), as previously described (52). For each sample, the cDNA template input was calculated assuming 100% reverse transcription efficiency and was expressed as the number of templates (log10) present in the 10-μl reaction volume used for the first round of nested PCR (i.e., cDNA input before any PCR amplification procedure). To validate our cDNA input estimation, we quantified cDNA for 4 samples with real-time quantitative PCR (qPCR) (8). To evaluate reproducibility, UDS was performed twice on the baseline samples for eight participants, using the same cDNA products and the same experimental conditions.
SGS.
Using the same viral cDNA that was produced for UDS, SGS was performed on seven patients (C7, I4, S1, U1, U7, N1, and Q9), as previously described (8). The targeted regions for PCR amplification included env C2-V3 (HXB2 coordinates 6928 to 7344) and pol RT (HXB2 coordinates 2708 to 3242), identical to the RT and C2-V3 regions amplified for UDS.
Quality control.
To avoid cross-contamination, extraction procedures and PCR were performed in separate rooms and on separate days with different sets of micropipettes. Extraction was performed in a laminar flow cabinet. For every experiment, negative controls were included to monitor cross-contamination. Reads were checked for intersample and lab strain contamination by performing MEGABLAST homology searches against each other and against the online public Los Alamos HIV sequence database (http://www.hiv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html [accessed August 2010]). We also evaluated UDS data for linked AM and divergent branches in phylogenies as evidence for possible contamination.
UDS data analysis.
Our UDS data analysis was performed as described elsewhere (52; W. Delport, A. F. Y. Poon, and S. L. Kosakovsky Pond, submitted for publication). This HIV-1 454 bioinformatics pipeline is publically available as a part of the Datamonkey sequence analysis tool (http://www.datamonkey.org) (12, 60). Briefly, the analysis consisted of eight main steps.
(i) Quality filtering and extraction of gene-specific reads from multiplexed samples.
We utilized site-specific PHRED scores (16, 17) to filter the low-quality base. By default, reads at least 100 nucleotides long and containing consecutive PHRED scores greater than 20 (equivalent to ≤1% base calling error) were retained for successive analyses. Since UDS errors are typically localized to homopolymers (68), we allowed for reads to be broken into multiple fragments, removing only the regions for which PHRED scores were less than the predefined cutoff. These quality-controlled reads were subsequently filtered for each of the sequenced coding regions (gag, env, and pol) by the use of an iterative alignment procedure described elsewhere (Delport et al., submitted). Briefly, we identified reads that were “mappable” against an HXB2 reference sequence through the successive amino acid alignment of each read in each of six potential reading frames (three forward and three reverse complemented). The reading frame with the highest scoring read was retained if the per-site alignment score exceeded five times the expected alignment score of a random sequence with sample-specific base composition. The consensus of these mapped high protein alignment scoring (HPAS) reads was constructed and utilized as a sample-specific reference sequence for subsequent alignments. The remaining sequences were aligned at the nucleotide level against this HPAS reference sequence and retained if their alignment score exceeded the median score of the distribution of HPAS reads. This nucleotide alignment step permits the correction of out-of-frame indels or homopolymer length errors.
(ii) Estimate of sequence diversity.
Overall sequence diversity was estimated for each coding region as the maximum likelihood divergence using the HKY85 substitution model (26) in sliding windows of 125 nucleotides in width, with 25-nucleotide shifts in window placement, and a minimum site coverage of 500 reads.
(iii) Identification of DRM and AM.
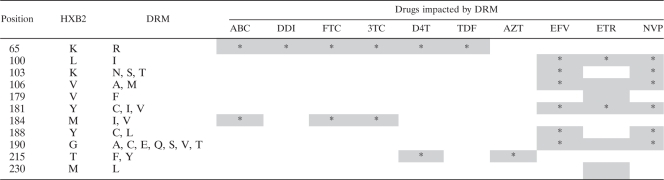
Using the Stanford Drug-Resistance Database (http://hivdb.stanford.edu), we first identified 11 sites with associated nucleoside reverse transcriptase inhibitor (NRTI) and nonnucleoside reverse transcriptase inhibitor (NNRTI) DRM with scores greater than 35, indicating that they confer moderate- to high-level resistance to ART (Table 1). Next, we prepared a list of AM (see Table S1 in the supplemental material) that have been reported to compensate for the loss of viral fitness incurred by acquiring DRM (5, 6, 11, 23, 28, 49, 50, 56, 57, 62, 70, 75). UDS reads containing at least one site of interest, either DRM or AM, were then analyzed. For each sample, reads were categorized into four classes containing (i) both DRM and AM, (ii) DRM but no AM, (iii) AM but no DRM, and (iv) neither DRM nor AM. We tested for the enrichment of linked AM in each sample (i.e., reads with both DRM and AM) using Fisher's exact test (20), which was Bonferroni corrected to account for multiple testing.
Table 1.
Drug resistance mutation sites with Stanford scores of >35a
Position based on HXB2 numbering. DRM indicates mutations associated with drug resistance with a Stanford score of >35. Letters represent the standard one-letter amino acid code. ABC, abacavir; DDI, didanosine; FTC, emtricitabine; 3TC, lamivudine; D4T, stavudine; TDF, tenofovir; AZT, zidovudine; EFV, efavirenz; ETR, etravirine; NVP, nevirapine. Shading indicates drugs affected by listed mutation, with the asterisk indicating a higher impact.
(iv) Estimation of site-specific diversity levels and diversifying/purifying selection.
To determine whether DRM sites tended to have higher diversity levels or unique patterns of selection, we inferred nucleotide profiles at all sites with sufficient UDS coverage (≥50 reads). We estimated the per-site diversity level as the proportion of non-HXB2 amino acids, assuming that each amino acid residue is independent. Although this is probably an overestimate of the amount of diversity, it is sufficient for the purposes of our analysis in the absence of a phylogeny (4, 13). For each DRM site, we ranked the diversity level against all other codons and assessed a median mutation rank for all DRM sites combined. We determined whether this median mutation rank was significantly different from random samples of non-DRM sites using a permutation test (n = 1,000).
Thereafter, we determined diversifying (or positive) and purifying (or negative) selection at each codon by inferring whether the ratio of observed nonsynonymous to synonymous substitutions was significantly greater (diversifying selection) or less (purifying selection) than expected, given the genetic code and observed codon frequencies (34; Delport et al., submitted). The ratio of synonymous to nonsynonymous substitutions was approximated by averaging over all possible single-nucleotide substitution pathways between all pairs of observed codons at a site, while assuming neutral evolution, as previously described (34; Delport et al., submitted). We finally tested for the enrichment of diversifying and purifying selection at DRM sites using Fisher's exact test (20).
(v) Interrogation for methodological error.
First, we determined multinomial confidence intervals (CI) of percent DRM at any given codon to estimate the reliability of our point estimate. Those DRM sites whose CI included zero were considered to be a UDS error. Furthermore, in order to estimate a sample-specific threshold for the identification of biologically meaningful minority polymorphisms, we fitted a binomial mixture model to site-specific diversity levels, as previously described (10; Delport et al., submitted). This binomial mixture model starts by evaluating the maximum likelihood fit of the data to a model that assumes that single nucleotide polymorphisms (SNP) have equal probabilities of being observed across all sites. Briefly, given the depth of sequence (c) and the number of observed mutations (m), a binomial estimate of the likelihood of the data at a site (i), given the probability of observing an SNP (r), can be obtained as , and assuming independent sites, the mean probability of observing an SNP across all sites can be estimated using the product of these binomials, i.e., . Next, we iteratively added additional SNP probability classes, each time estimating the model fit using a binomial mixture model such that , where pj is the mixing proportion for each of K classes, each with its own probability of observing an SNP (rj). All parameters were optimized using standard maximum likelihood optimization techniques. The procedure of adding classes and optimizing the assignment of sites to classes was repeated until the model fit, as evaluated using Akaike information criterion (AIC) (3), was no longer improved. The result of this binomial mixture model fitting procedure was (i) a statistical estimate of the number of classes supported by the data, (ii) the estimation of a class-specific probability of observing an SNP, and (iii) the assignment of sites to each of those classes. We then assumed that the class with the lowest level of diversity for each sample was equivalent to the proportion of the SNP expected to arise from a UDS error, after filtering for context-dependent homopolymer errors. This lowest level of diversity was then used as a sample-specific threshold for the identification of minority variants. Given the estimated diversity level (i.e., SNP probability), we calculated the posterior probability that a site belonged to each class using a naïve empirical Bayes procedure (74) and subsequently the posterior probability that a site does not belong to the background diversity-level class, which was calculated as the sum of the posterior probabilities for the nonbackground diversity-level classes.
(vi) Reproducibility of UDS data.
We assessed the reproducibility of UDS across eight samples by comparing the proportions of detected DRM for each of the 11 analyzed codons between duplicate UDS runs. We also compared the nucleotide composition of the entire sequence by analyzing every position (not only DRM sites) with coverage of >200 reads in both duplicates. The primary minority mutation was defined as the mutation with the second-highest frequency at a site in the first run. Linear correlation was computed using the square root-transformed frequencies.
(vii) Correlation analysis between sequence diversity, template input, and EDI.
To evaluate whether the occurrence of sites with detected minority variants is the result of continued viral evolution, we evaluated for correlations between sequence diversity, estimated duration of infection (EDI), and the number of codons with detected minority variants in each sample for each coding region. Minority variant sites were defined as those with a frequency of less than 10% in the UDS reads and which were unlikely to have arisen from instrument error. The latter was evaluated using three different approaches: (i) residue frequency of >1%, (ii) residue frequency that is greater than the background error rate estimated by the binomial mixture model, and (iii) residue sites that were not probabilistically assigned to the background diversity-level class based on an empirical Bayesian procedure (74). We also evaluated for correlations between (i) sequence diversity for the three coding regions, (ii) input of cDNA templates (and HIV-1 RNA viral loads) and sequence diversity, and (iii) sequence diversity and number of codons with detected minority variants. Correlation analyses were performed using both parametric (Pearson) and nonparametric (Spearman) tests. The level of significance for all analyses was a P value of ≤0.05. Lastly, we also compared mean sequence diversities for each coding region within participants according to the Fiebig classification (18) using the Wilcoxon signed-rank test.
(viii) Treatment response.
Using available longitudinal clinical data, we evaluated the impact that low-level (<20%) DRM detected by UDS may have had on the observed rate of clinical failure of first-line ART. To investigate a possible connection between virologic failure and observed low-level DRM at baseline, we sequenced (by population-based sequencing) the HIV-1 RNA from the earliest available sample after therapy failure for the six patients who experienced virologic failure.
Virologic failure was defined as the observation of two or more viral loads of greater than 500 HIV RNA copies/ml after initial suppression of the virus to undetectable levels, i.e., <50 HIV RNA copies/ml.
RESULTS
Study cohort.
Study participants were, predominantly, white men with a mean age of 31 years who reported sex with other men as their HIV risk factor. All but one were infected with HIV-1 subtype B virus, the exception being an HIV-1 subtype B/D recombinant. The mean EDI at sample collection was 2.8 months (range, 1.1 to 3.9 months; Fiebig stage IV or V). At the time of sampling, the mean CD4 count was 602 cells/ml (range, 228 to 937 cells/ml) and the mean blood plasma viral load was 6.1 log10 HIV-1 RNA copies/ml (range, 3.5 to 7.5 log10). The mean calculated cDNA input in the first-round nested PCR was 4.8 log10 copies/10 μl (range, 2.5 to 6.2 log10). For 4 samples, the calculated and the measured cDNA values correlated at an R2 value of 0.92 (P = 0.04). Baseline characteristics are summarized in Table 2.
Table 2.
Participant characteristicsa
| Characteristics | Values |
|---|---|
| No. of participants in study (%) | 32 (100) |
| No. male (%) | 31 (97) |
| Mean age, yr (range) | 31 (20-58) |
| No. of MSM (%) | 31 (97) |
| No. Caucasian (%) | 26 (81.2) |
| No. HIV subtype B (%) | 31 (97) |
| No. of DRM by bulk sequencing (%) | 4 (12.5) |
| No. of DRM by UDS (%) | 29 (91) |
| Mean EDI, mo (range) | 2.8 (1.1-3.9) |
| Mean CD4, cells/ml (range) | 602 (228-937) |
| Mean HIV-1 VL, log10 copies/ml (range) | 6.1 (3.5-7.5) |
| Mean cDNA input, log10 copies/10 μl (range) | 4.8 (2.5-6.2) |
MSM, men who have sex with men; DRM, drug resistance mutations with Stanford score of >35; VL, viral load; UDS, ultradeep sequencing; EDI, estimated duration of infection. Average cDNA input in log10 cDNA copies/10-ul reaction (range).
UDS and SGS coverage.
The UDS methodology yielded an average of 17,152 high-quality reads across all gene regions (range, 2,216 to 35,160), with a mean read length of 152 nucleotides (range, 98 to 195) (see Table S2 in the supplemental material). In comparison, SGS produced on average 25 reads (range, 20 to 30) with a mean read length of 400 nucleotides for RT and 300 nucleotides for C2-V3. On average, 76.5% of the quality-filtered UDS reads were successfully aligned to the HXB2 reference sequences. Of these aligned reads, 38.4% on average were C2-V3 (mean read length, 227 nucleotides), 33.7% p24 (mean read length, 192 nucleotides), and 27.9% RT (mean read length, 166 nucleotides). No evidence for laboratory contamination of any sample was seen.
Reproducibility of UDS.
The differences in estimated DRM proportions between duplicate UDS runs were significant. When at least one of the runs reported a DRM at any level <20%, there was little agreement with the other run (Table 3). Specifically, only in 3 of the 23 evaluated cases that had at least one detected DRM did the second UDS run confirm a minority DRM greater than the calculated background. We further compared all amino acid positions among those UDS runs with coverage of >200 reads between the duplicate runs (not only DRM sites). The consensus agreement was mostly conserved within the two duplicate runs (range, 95 to 100%), and the frequencies of primary minority residues (i.e., mutations with the second-highest frequency) between duplicated runs were strongly correlated (R2 range, 0.68 to 0.98). However, there was much variability in the primary minority mutations identified in the replicated runs (range, 25 to 59% agreement between runs) (see Table S3 in the supplemental material).
Table 3.
Percentages of detected DRM for 8 duplicate UDS runsa
| PID | UDS run | BG error rate | % DRM for indicated HXB2 position in reverse transcriptase |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 65 | 100 | 103 | 106 | 179 | 181 | 184 | 188 | 190 | 215 | 230 | |||
| I4 | 1 | 0.18 | 0.23 | 0.33 | 99.77 | 0.05 | 0.00 | 0.00 | 0.20 | 0.15 | 0.05 | 0.00 | 0.00 |
| I4 | 2 | 0.18 | 0.18 | 0.00 | 99.32 | 0.10 | 0.00 | 0.06 | 0.25 | 0.06 | 0.00 | 0.08 | 0.00 |
| J6 | 1 | 0.27 | 0.33 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.30 | 0.40 | 0.20 | 0.00 | 0.00 |
| J6 | 2 | 0.18 | 0.60 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 1.90 | 0.00 | 0.38 | 0.00 | 0.00 |
| L3 | 1 | 0.17 | 0.12 | 0.00 | IR | 0.00 | 0.00 | 0.00 | 0.14 | 0.57 | 0.00 | 0.00 | 0.00 |
| L3 | 2 | 0.22 | 0.57 | 0.13 | 4.55 | 0.04 | 0.00 | 0.10 | 0.24 | 0.14 | 0.00 | 0.10 | 0.00 |
| R2 | 1 | 0.38 | 0.00 | 0.00 | 17.74 | 0.00 | 0.00 | 0.00 | 0.00 | 0.81 | 0.00 | 0.00 | IR |
| R2 | 2 | 0.27 | 2.27 | 0.00 | IR | 0.00 | 0.23 | 0.00 | 0.89 | 0.22 | 0.00 | 0.00 | IR |
| R6 | 1 | 0.22 | 0.15 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.14 | 0.07 | 0.07 | 0.00 | 0.00 |
| R6 | 2 | 2.36 | 0.17 | 0.09 | 1.19 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.11 | 0.00 | 0.00 |
| U1 | 1 | 0.27 | 0.20 | 0.00 | IR | 0.00 | 0.00 | 0.17 | 0.17 | 0.00 | 0.00 | 0.00 | 0.64 |
| U1 | 2 | 0.34 | 0.00 | 0.00 | IR | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 | 0.00 | 0.00 | 0.00 |
| U6 | 1 | 0.25 | 0.00 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.25 | 0.08 | 0.00 | 0.00 | 0.00 |
| U6 | 2 | 0.11 | 0.84 | 0.00 | 0.15 | 0.34 | 0.00 | 0.09 | 0.36 | 0.00 | 0.27 | 0.00 | 0.00 |
| U7 | 1 | 0.35 | 0.00 | 0.00 | 100 | 0.00 | 0.00 | 0.25 | 0.25 | 0.00 | 0.00 | 0.00 | 0.63 |
| U7 | 2 | 0.14 | 0.61 | 0.00 | 100 | 0.00 | 0.00 | 0.00 | 0.24 | 0.00 | 0.00 | 0.00 | 0.00 |
PID, patient identification number; UDS, ultradeep sequencing; BG error rate, background UDS error (%) estimated using a binomial mixture model for each UDS sample; IR, insufficient reads at site/sample. Bold type indicates percentage of drug resistance mutations (% DRM) exceeding the background error rate. Underlining indicates those DRM that were detected by both population-based sequencing and SGS.
UDS error estimation.
We used two approaches to estimate UDS errors in our study: (i) multinomial CI for mutations and (ii) a binomial mixture model. For approximately 60% of the screened DRM sites, the multinomial CI for the DRM proportions included the value zero and were therefore considered instrumental errors (see Table S4 in the supplemental material). The mean frequency for the true DRM was calculated to be 0.56% (95% CI, 0.43 to 0.69). In our binomial mixture model approach, we estimated diversity-level classes across all sites in the UDS alignment. We assumed that the smallest diversity level represented the background diversity level and conservatively reflected the UDS background error rate. Using this approach, we detected between two and eight diversity-level classes, with a mean minimum diversity of 0.0021 ± 0.0008 per site (Table 4). This corresponds to an expected error percentage of 0.21 ± 0.08 per site and is similar to a previously published study (29). For most samples, a large proportion of sites were assigned to this lower diversity class (Table 4) and variants at these sites were therefore considered to be likely technical errors. We then evaluated all UDS results in relation to the specific diversity-level class and inferred the background error rate in the sample identified by the binomial mixture model.
Table 4.
Background error rate as determined by the binomial mixture modela
| PID | No. of diversity levels | BG error rate (%) | BG proportion (%) |
|---|---|---|---|
| A7 | 8 | 0.02 | 8.56 |
| R8 | 5 | 0.07 | 10.78 |
| I9 | 5 | 0.11 | 33.20 |
| U6 | 5 | 0.11 | 35.50 |
| J7 | 5 | 0.12 | 54.47 |
| L1 | 4 | 0.13 | 45.84 |
| R4 | 5 | 0.13 | 60.53 |
| U7 | 3 | 0.14 | 63.02 |
| J8 | 5 | 0.17 | 27.51 |
| I4 | 5 | 0.18 | 44.30 |
| C4 | 4 | 0.19 | 71.34 |
| N3 | 4 | 0.19 | 77.86 |
| F8 | 5 | 0.20 | 73.97 |
| N1 | 4 | 0.20 | 68.14 |
| Q9 | 4 | 0.20 | 58.35 |
| L2 | 6 | 0.22 | 69.16 |
| L3 | 7 | 0.22 | 54.15 |
| R6 | 4 | 0.22 | 82.12 |
| S1 | 3 | 0.22 | 66.70 |
| L6 | 3 | 0.23 | 91.27 |
| R3 | 4 | 0.23 | 45.94 |
| N6 | 5 | 0.24 | 60.04 |
| Q1 | 4 | 0.24 | 77.00 |
| C7 | 4 | 0.25 | 47.63 |
| L5 | 2 | 0.25 | 86.65 |
| M4 | 2 | 0.25 | 81.02 |
| J6 | 5 | 0.27 | 58.84 |
| N0 | 5 | 0.27 | 56.86 |
| U1 | 7 | 0.27 | 64.86 |
| R2 | 5 | 0.38 | 72.71 |
| S3 | 2 | 0.39 | 83.89 |
| J5 | 5 | 0.46 | 84.18 |
PID, patient identification number. The number of diversity levels is the number found in each sample. The background (BG) error rate is the lowest inferred diversity-level class, and BG proportion is the proportion of sites assigned to this lowest diversity-level class per sample. Those sites are conservatively assumed to have no mutations other than sequencing errors. Rows are sorted according to the BG rate, from lowest to highest.
Identification of DRM and AM and estimation of site-wise diversity level and diversifying/purifying selection.
We evaluated the detection of DRM by (i) comparing inferred DRM from UDS, SGS, and a population-based sequencing method, (ii) estimating the frequency of DRM by each method, (iii) comparing per-site diversity and selection patterns at DRM and non-DRM sites, and (iv) evaluating AM linked to detected DRM.
For 11 well-described DRM sites (Table 1), we compared amino acid compositions identified by population-based sequencing, SGS, and UDS. Out of the 32 samples, population-based sequencing and SGS (if available) detected DRM at 3 of the 11 sites in four individuals (12.5%) (see Table S5 in the supplemental material). In addition to the detection of this same DRM, UDS also detected at least one additional DRM at a frequency greater than the estimated UDS error rate (median, 2 positions; range, 0 to 6) in 29 of the 32 individuals (91%). The frequencies of detected DRM by UDS were between 0.1 and 17.74% (see Table S5 in the supplemental material). Other non-HXB2 amino acid substitutions not associated with DRM were identified in five samples (15.6%) by population-based sequencing and SGS. UDS also detected other non-HXB2 and non-DRM variants (at a frequency of 0.1 to 70%) at 3 or more positions (median, 8; range, 3 to 11) in every subject (100%).
We found no evidence for increased (or decreased) diversity at DRM sites compared to all other sites. Indeed, for all individuals the rank of the median diversity level for DRM sites was not significantly greater than a random selection of an equivalent number of non-DRM sites from the same alignment (see Table S6 in the supplemental material). Similarly, we found evidence for diversifying or purifying selection at DRM sites in only 2 of the 32 screened patients, despite detection of DRM by UDS in 91% of the participants (P < 0.05). Of the two subjects with evidence of selection at DRM sites, one (subject R8) had evidence for diversifying selection, whereas the other (subject U1) showed purifying selection. Interestingly, a repeated UDS run for patient U1 did not confirm purifying selection at DRM sites.
Finally, we screened for AM since their co-occurrence with observed DRM would support the notion that a detected DRM was less likely to arise as a random mutation. We evaluated whether UDS reads with identified high-level DRM were more likely to have linked compensatory AM than expected (Bonferroni-corrected Fisher's exact test). Only in one sample (I4) did we observe the enrichment of reads containing DRM (with a Stanford score of >35) and linked AM.
Comparison of sequence diversity between coding regions.
The transmission of minority drug-resistant viral variants, as detected in pol, would require polyclonal transmission, which contradicts the current evidence supporting mono- or oligoclonal transmission in env (1, 19, 25, 33). This disparity may be the result of recombination between strains from multiple transmission events in which pol diversity is maintained, while env diversity is not. To investigate this possibility, we compared sliding windows of sequence diversity in three gene regions for each sample (gag p24, pol RT, env C2-V3). Mean sequence diversity was 0.012 for pol (range, 0 to 0.074), 0.016 for gag (range, 0 to 0.086), and 0.068 for env (range, 0 to 0.217), and diversity measures were highly correlated within coding regions (P < 0.01).
Sequence diversity may also increase over time as a consequence of viral evolution and is expected to be lower during the earliest phases of HIV-1 infection, especially assuming a mono- or oligoclonal transmission event (1, 19, 25, 33). Although our sampling was within 4 months of the estimated time of infection, we did not find any significant correlation between viral population diversity and EDI for any of the three analyzed coding regions (env, P = 0.67; gag, P = 0.99; pol, P = 0.93). Repeating the analysis using the Fiebig classification, we did not find a significant difference between the mean diversities in patients with stage IV compared to stage V infection for any of the three coding regions (env, P = 1.00; gag, P = 0.9441; pol, P = 0.3828). However, we found a significant correlation between higher cDNA levels (and HIV-1 RNA viral load) and lower sequence diversity in all three regions (Spearman correlation, P = 0.017 for pol and P < 0.01 for gag and env) (see Fig. S1 in the supplemental material).
There was no correlation between overall sequence diversity and number of codons with detected minority variants in pol, gag, and env. This was true for each of the three described approaches to filtering for methodological errors (i.e., [i] minority mutations at a frequency of >1%, [ii] minority mutations at a frequency greater than the background diversity estimated using a binomial mixture model, and [iii] frequency of minority mutations probabilistically not assigned to the background diversity class using an empirical Bayesian procedure). Moreover, the number of codons with detected minority mutations did not correlate with EDI or cDNA input (or HIV-1 RNA viral load) for any of the three coding regions.
Treatment response.
Since HIV-1 DRM can abrogate the efficacy of ART (43, 59), we investigated whether the presence of DRM identified by UDS at positions 65, 103, 181, 184, and 215 adversely impacted response to first-line ART. Virologic failure was defined as two or more viral loads of >500 HIV-1 RNA copies/ml after initial HIV-1 suppression of the virus to <50 copies/ml during first-line ART. Before the DRM results from UDS were known, 17 of the 32 subjects started ART during follow-up (Table 5). The ART regimen for seven of these patients included the NNRTI efavirenz (EFV) and two or three NRTI medications. Using UDS, two of these patients appeared to be infected by a virus possessing a low-level DRM at position 103 or 181, which can confer resistance to NNRTI. None of the seven patients receiving an NNRTI experienced virologic failure after a mean follow-up of 608 days (range, 28 to 1,743 days), including those two patients with an apparent DRM that would influence NNRTI sensitivity. Additionally, 10 patients with a detected low-level DRM conferring resistance to NRTI (range, 1 to 3) at position 65, 184, 188, or 215 started ART, including NRTI. Four of these patients (40%) experienced virologic failure after an average of 329 days of treatment (range, 247 to 579). A review of clinical data revealed clear nonadherence to ART in two of these four patients. Also, two more patients without any detected DRM at baseline experienced virologic failure. Only two of the six patients with virologic failure had an M184V DRM at the time of virologic failure, and one of these had an apparent low-level DRM at RT position 184 at baseline. By clinical records, he was also not compliant with the prescribed ART (Table 5).
Table 5.
Outcome of first-line ART in 17 participants
| PID | % DRM for HXB2 position: |
ART | VF | Duration of ART (days) | UDS to ART (days) | DRM at VF | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 65 | 103 | 181 | 184 | 188 | 215 | ||||||
| A7 | 0.1 | 0.94 | 0 | 0.39 | 0 | 0 | 3TC/AZT/IND | Yes** | 148 | 1 | M184MV |
| C4 | 0 | 0 | 0 | 0 | 0 | 0.51 | 3TC/AZT/NFV | Yes | 579 | 1 | M184V, D30N, N88D |
| I4 | *0.23 | 99.77 | *0.06 | 0.2 | 0.15 | *0.08 | 3TC/TDF/ATV/RTV | No | 1040 | 1223 | |
| I9 | 0.3 | 0.66 | 0 | 0.82 | 0 | 0 | 3TC/D4T/TDF/EFV | No | 1743 | 10 | |
| J5 | 0 | IR | 0 | 0 | 0 | 0 | 3TC/AZT/SQV/RTV | Yes** | 180 | 3 | A71V |
| J6 | 0.6 | 0 | 0 | 1.9 | *0.38 | 0 | 3TC/AZT/EFV | No | 1965 | 102 | |
| J8 | 1.36 | IR | 0 | 1 | 0.17 | 0 | 3TC/ABC/D4T/APV/RTV | Yes | 314 | 1 | NONE |
| L1 | 0 | IR | 0 | 0.56 | 0.56 | 0 | 3TC/AZT/LPV/RTV | Yes** | 277 | 20 | NONE |
| L6 | 0.27 | 2.99 | 0.12 | 0.12 | 0 | 0 | FTC/TDF/ATZ/RTV | No | 115 | 613 | |
| M4 | 0.09 | 0.5 | 0 | 0.13 | 99.35 | 0.17 | 3TC/AZT/ABC/LPV/RTV | No | 1500 | 19 | |
| N0 | 0.23 | IR | 0.13 | 0.26 | 0.13 | 0 | FTC/TDF/EFV | No | 840 | 574 | |
| R2 | *2.27 | 17.74 | 0 | *0.89 | *0.81 | 0 | FTC/TDF/EFV | No | 86 | 857 | |
| R3 | 0 | 0 | 0 | 0.17 | 0 | 0 | FTC/TDF/EFV | No | 28 | 983 | |
| R4 | 0 | IR | 0 | 0 | 0 | IR | ABC/3TC/EFV | No | 168 | 676 | |
| S3 | IR | 0 | 0 | 0 | 0 | IR | TDV/FTC/ATZ/RTV | Yes | 378 | 10 | L10I |
| U1 | *0.2 | IR | *0.17 | *0.17 | *0.19 | 0 | FTC/TDF/EFV | No | 34 | 490 | |
| U7 | *0.61 | 100 | *0.25 | *0.25 | 0 | 0 | ABC/3TC/FOS/RTV | No | 227 | 233 | |
PID, patient identification number; ART, antiretroviral therapy; VF, virologic failure of ART; IR, insufficient reads at site/sample;*, DRM that were found in only one of repeated runs; **,likely nonadherence to ART. Bold type indicates percentages of drug resistance mutations (% DRM) exceeding the background error rate. Underlining indicates those DRM that were detected by both population-based sequencing and SGS. Duration of ART, follow-up in days from therapy start to viral failure or to last available time point; UDS to ART, follow-up time in days from UDS to ART start; 3TC, lamivudine; AZT, zidovudine; IND, indinavir; NFV, nelfinavir; TDF, tenofovir; ATV, atazanavir; RTV, ritonavir; D4T, stavudine; EFV, efavirenz; SQV, sequinavir; ABC, abacavir; APV, amprenavir; LPV, lopinavir; FTC, emtricitabine; FOS, fosamprenavir.
DISCUSSION
Using ultrasensitive genotypic assays, many research groups have reported high proportions of transmitted DRM among ART-naïve individuals (15, 30, 32, 37, 48, 58, 65), but these findings are not consistent with the descriptions of mono- or oligoclonal transmission in the env coding region by SGS and UDS (1, 19, 25, 33). The primary goal of this analysis was to investigate whether detected minority viral variants in the earliest part of HIV-1 infection were (i) truly transmitted, (ii) a consequence of viral evolution and selection early after transmission, (iii) technical errors in highly sensitive detection methods, or (iii) de novo mutations resulting as a consequence of the high error rate of HIV-1 replication. To address these issues, we performed bulk sequencing of HIV-1 pol and UDS of three HIV-1 coding regions (partial gag, pol, and env) sampled from 32 recently infected individuals. These data were then used to evaluate overall and per-site diversity levels, selective pressure, sequence reproducibility, DRM, and AM. A series of statistical and computational analyses were applied to help differentiate biologically meaningful mutations from those caused by methodological errors. We also examined the association between detected minority DRM and virologic failure of first-line ART. UDS of viral populations confirmed all DRM detected by bulk sequencing, but it also identified multiple low-level variants in every individual evaluated, even after removing likely sequencing errors through model-based statistical filtering. Similar to previous studies, the use of UDS was very likely to detect low-frequency variants harboring DRM during recent infection, especially at frequencies of <1%.
Early recombination between env and pol could possibly explain the conundrum of the frequent detection of low-level DRM when evaluating pol but of oligo- or monoclonal transmission when evaluating env using SGS (22). This study found, however, that the overall sequence diversities for the three coding regions were highly correlated (P < 0.01). This observation speaks against early recombination, since it would theoretically decrease diversity in some coding regions (e.g., env) while maintaining diversity in others (e.g., pol). Additionally, if multiclonal transmission had occurred (7, 39), then we would expect that detected DRM would be associated with AM in the same sequence, but we did not find that this was the case. Alternatively, DRM could be positively selected for in a minority of transmitted variants, or transmitted DRM at a higher frequency could be replaced by wild-type virus (negative selection) in the initial stages of infection (42). This study found, however, that DRM sites had the same levels of residue diversity as all other sites in pol from the same subject, and all detected DRM were not significantly enriched for positive or negative selection (except for one DRM site in one individual). Taken together, these data do not support frequent transmission and early (negative or positive) selection of viral variants with DRM. This is consistent with recent observations by Wang et al. (73) and Fischer et al. (19), who investigated viral diversity during acute HCV and HIV-1 infection by the use of UDS. These studies also showed that one or a few viral variants were present during transmission and that the detected low-frequency variants were only one or two mutations distant from the inferred transmitted variants.
As an alternative explanation, the detected DRM could emerge de novo early after infection and replicate as a minor but measurable proportion of the viral population. In contrast to previous studies (33, 38), we did not find a significant correlation between viral population diversity and EDI or Fiebig stage of infection, as might be expected with viral evolution following mono- or oligoclonal transmission. Our analysis did find, however, that lower viral diversity correlated with higher cDNA input into UDS and higher HIV-1 viral load and that higher viral load correlated with more recent EDI (P = 0.02). Overall, these data suggest that viral diversity is lower in the earliest stage of infection when viral load is higher and that over time, as viral load decreases, viral diversity increases. Interestingly, there was no correlation between the number of sites presenting minority mutations and overall sequence diversity for any of the three coding regions. This likely indicates that the frequency of codons carrying minority mutations does not increase over time in parallel with overall sequence diversity, and these detected mutations are the likely consequence of the daily appearance and disappearance of mutations across the HIV-1 genome secondary to the error-prone reverse transcriptase of HIV-1.
The detection of DRM at very low levels during primary HIV-1 infection is likely either a consequence of variants emerging during viral replication or the result of technical artifacts, especially near the lower limit of detection. Distinguishing true DRM present at low levels from technical error is a challenge when using highly sensitive genotypic methods such as UDS. To investigate this, we used a binomial mixture model to identify different classes of diversity levels in each UDS run and then used these diversity levels to evaluate possible error rates (10; Delport et al., submitted). This model-based approach has the advantage of examining all of the data to deduce an appropriate cutoff for each residue site by accounting for (i) per-site coverage by UDS, (ii) the overall distribution of mutations, and (iii) the diversity of the underlying viral population. In addition to instrument error and baseline HIV-1 mutation rate, the number of diversity-level classes would be influenced by overall sequence diversity (i.e., how many different viral sequences are circulating), natural selection (i.e., some sites appearing more or less variable than others), and population structure. Therefore, we conservatively assumed that the lowest estimated diversity level corresponds to the technical background error rate. In this analysis, the estimated mean background error rate was 0.21 ± 0.08% per site, which is consistent with previous studies that reported UDS error rates ranging from 0.05 to 1.0% (29, 44, 67, 68, 72, 77). This variation is likely due to multiple factors, including subtle differences in the methods, gene-specific mutation patterns, and UDS read coverages. Similarly, a previous study also found that by using a filtering procedure similar to that applied in this study, observed error rates were reduced from 0.5% to 0.25% (29). Taken together, these computed background error rates allow for the most conservative estimation of detectable DRM by UDS in our sample.
If technical errors were the reason for the frequent detection of low-level DRM, then, we hypothesized, duplicated UDS runs would demonstrate different results. We assessed the reproducibility of UDS in eight samples by comparing the proportions of detected DRM and the nucleotide composition of the entire sequence when UDS coverage was >200 reads in each duplicated run. Despite excellent agreement between runs in recovering the majority residue at each position (see Table S3 in the supplemental material), significant differences in the inferred prevalence and frequencies of low-level DRM in the eight replicated UDS assays were observed (Table 3). The agreement was generally poorer for runs with lower numbers of reads. It is difficult to determine whether these inconsistencies are the result of technical errors or simply the failure to amplify, sequence, and detect consistently low-frequency variants. Variability between replicated UDS runs on the 454 platform has previously been reported (61, 68). For example, Poon et al. (61) found that many minority variants detected at levels between 1 and 5% in one replicate were undetectable in another. Therefore, we believe that our filtering and background-calling procedure is unlikely to be the sole source of incongruent results between duplicate UDS runs and that other sources of noise could play a role, including PCR and sampling biases and various UDS instrument errors. Moreover, in our analysis we still observed >60% of replicated UDS runs to have some levels of discordance in detected/nondetected amino acid residues. Thus, it remains a challenge to distinguish low-level “true” variants from assay artifacts. The confirmation of whether detection represents a “true” viral variant or a false-positive result may be assisted by the use of clonal analysis, as exemplified by Varghese and colleagues (68), who found that when the K65R DRM was detected at a frequency around 1% in an HIV-1 subtype C population by UDS, these variants could not be confirmed by subsequent clonal analysis.
We also hypothesized that if low-level DRM represent transmitted variants, then they may negatively impact an individual's response to ART. We therefore investigated if the presence of DRM identified by UDS at positions 65, 103, 181, 184, and 215 adversely impacted response to first-line ART in our study cohort. There was no apparent correlation between detected minority DRM and clinical outcome or detected DRM in the first available blood sample after virologic failure of ART. Of course, these observations are limited by the small cohort sample size, the short duration of follow-up for most of the participants, and other factors, like differences in medication adherence. These limitations could lead us to underestimate the negative impact of detected minority DRM during primary infection on first-line ART.
Other limitations include a large number of low-quality UDS reads that had to be excluded from the analysis. Relatively short lengths and uneven coverage of UDS reads represent a challenge for linked mutation analysis and the estimation of underlying viral diversity. Additionally, low template input into the UDS reaction could cause resampling of the original viral population and negatively impact our ability to estimate sequence diversity for these samples; however, this study found a negative correlation between input cDNA and observed viral population diversity, and this provides some evidence against oversampling by UDS. Lastly, EDI determination could be biased by various factors, including inaccurate reporting of recent sexual risk contacts and variations in assays used to estimate timing of infection. These biases could explain the observed lack of correlation between EDI and diversity in the study.
The sensitivity of detection methods depends on different factors, namely, the number of templates that are interrogated, the error rate and amplification bias of the PCR, and sequencing procedures. For UDS, the number of reads obtained for each position and the efficiency of filtering procedures applied during the data analysis play an important role. Relying on UDS for the detection of low-level DRM will require the development of more realistic statistical and computational models, experimental replication, and confirmation using other sequencing techniques. Low-level DRM are more likely to be clinically relevant if present at frequencies higher than a specific model-based background threshold and in the right sequence context (e.g., with AM). However, no filtering can fully account for errors due to sampling or experimental biases, and new sequencing technologies that promise to deliver longer reads with lower per-base error rates will undoubtedly prove to be useful for future investigations of transmitted DRM and minority populations. In conclusion, this study used one of the largest available sets of UDS data and found a diverse population of viruses rich in low-frequency mutants at nearly every sequenced residue position in pol, even early during infection. However, there was no evidence to support the hypothesis that minor populations of DRM are frequently transmitted or selected during the first months after infection. In conclusion, we believe that these DRM detected at low levels during primary HIV-1 infection likely represent the consequence of the high error rate of HIV-1 replication (9) or experimental artifacts.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the participants in the San Diego Primary Infection Cohort, to Parris Jordan, Pok Man Cheng, Caroline Ignacio, and Stephen Espitia for excellent technical support, and to Nadir Weibel, Christopher Woelk, and Josué Perez for helpful discussions. We also thank and commemorate our dear friend and outstanding research colleague Marek Fischer for all his contributions and support to our research over many years.
This work was supported by grants from the U.S. National Institutes of Health, AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, GM093939, and AI080353, the James Pendleton Trust, Swiss National Science Foundation grant PBZHP3-125533, and the Ettore Balli Foundation (Switzerland).
S.G. participated in the study design and data analyses and wrote the core of the manuscript. M.E.P. participated in the generation of sequencing data and data analysis and revised the manuscript. J.A.Y., W.D., and S.L.K.P. developed the bioinformatics platform for UDS data analysis, devised statistical analyses, performed data analyses, and revised the manuscript. J.Y.C. participated in data analysis and revised the manuscript. S.J.L. enrolled patients and revised the manuscript. D.D.R. and D.M.S. designed the study, participated in data analysis, and revised the manuscript. All authors read and approved the final manuscript.
S.G. does not have any commercial or other associations that might pose a conflict of interest. D.D.R. has served as a consultant for Biota, Bristol-Myers Squibb, Chimerix, Gen-Probe, Gilead Sciences, J & J, Merck & Co., Monogram Biosciences, Tobira Therapeutics, and Vertex. D.M.S. has received research support from ViiV Pharmaceuticals. The remaining authors do not report any conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aghokeng A. F., et al. 2010. Frequency of antiretroviral resistance mutations among ART-naive HIV-1-infected populations in rural areas from Cameroon, 2010. Antivir. Ther. 15(Suppl. 2):A151 [Google Scholar]
- 3. Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19:716–723 [Google Scholar]
- 4. Anisimova M., Kosiol C. 2009. Investigating protein-coding sequence evolution with probabilistic codon substitution models. Mol. Biol. Evol. 26:255–271 [DOI] [PubMed] [Google Scholar]
- 5. Bacheler L., et al. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balzarini J., Pelemans H., Esnouf R., De Clercq E. 1998. A novel mutation (F227L) arises in the reverse transcriptase of human immunodeficiency virus type 1 on dose-escalating treatment of HIV type 1-infected cell cultures with the nonnucleoside reverse transcriptase inhibitor thiocarboxanilide UC-781. AIDS Res. Hum. Retroviruses 14:255–260 [DOI] [PubMed] [Google Scholar]
- 7. Bar K. J., et al. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241–6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler D. M., Pacold M. E., Jordan P. S., Richman D. D., Smith D. M. 2009. The efficiency of single genome amplification and sequencing is improved by quantitation and use of a bioinformatics tool. J. Virol. Methods 162:280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coffin J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489 [DOI] [PubMed] [Google Scholar]
- 10. Cummings S. M., McMullan M., Joyce D. A., van Oosterhout C. 2010. Solutions for PCR, cloning and sequencing errors in population genetic analysis. Conserv. Genet. 11:1095–1097 [Google Scholar]
- 11. Delaugerre C., et al. 2001. Prevalence and conditions of selection of E44D/A and V118I human immunodeficiency virus type 1 reverse transcriptase mutations in clinical practice. Antimicrob. Agents Chemother. 45:946–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delport W., Poon A. F., Frost S. D., Kosakovsky Pond S. L. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delport W., Scheffler K., Seoighe C. 2009. Models of coding sequence evolution. Brief. Bioinform. 10:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diakite M., et al. 2010. High prevalence of transmitted drug resistance in HIV-1-infected antiretroviral-naive patients from Conakry, Guinea-Conakry. Antivir. Ther. 15(Suppl. 2):A149 [Google Scholar]
- 15. Ellis G. M., Page L. C., Burman B. E., Buskin S., Frenkel L. M. 2009. Increased detection of HIV-1 drug resistance at time of diagnosis by testing viral DNA with a sensitive assay. J. Acquir. Immune Defic. Syndr. 51:283–289 [DOI] [PubMed] [Google Scholar]
- 16. Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 17. Ewing B., Hillier L., Wendl M. C., Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 18. Fiebig E. W., et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 19. Fischer W., et al. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fisher A. 1922. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 85:87–94 [Google Scholar]
- 21. Geretti A. M., et al. 2009. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 52:569–573 [DOI] [PubMed] [Google Scholar]
- 22. Gianella S., Richman D. D. 2010. Minority variants of drug-resistant HIV. J. Infect. Dis. 202:657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzales M. J., et al. 2003. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS 17:791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goodman D. D., et al. 2011. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS 25:325–333 [DOI] [PubMed] [Google Scholar]
- 25. Haaland R. E., et al. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hasegawa M., Kishino H., Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 27. Hecht F. M., et al. 2006. A multicenter observational study of the potential benefits of initiating combination antiretroviral therapy during acute HIV infection. J. Infect. Dis. 194:725–733 [DOI] [PubMed] [Google Scholar]
- 28. Hertogs K., et al. 2000. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob. Agents Chemother. 44:568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huse S. M., Huber J. A., Morrison H. G., Sogin M. L., Welch D. M. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jakobsen M. R., et al. 2010. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin. Infect. Dis. 50:566–573 [DOI] [PubMed] [Google Scholar]
- 31. Johnson J. A., et al. 2007. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS One 2:e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson J. A., et al. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kosakovsky Pond S. L., Frost S. D. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 35. Kosakovsky Pond S. L., et al. 2009. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput. Biol. 5:e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuritzkes D. R., et al. 2008. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J. Infect. Dis. 197:867–870 [DOI] [PubMed] [Google Scholar]
- 37. Lataillade M., et al. 2010. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One 5:e10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee H. Y., et al. 2009. Modeling sequence evolution in acute HIV-1 infection. J. Theor. Biol. 261:341–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H., et al. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J. Z., et al. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 305:1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao L., et al. 2010. The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J. Acquir. Immune Defic. Syndr. 53(Suppl. 1):S10–S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Little S. J., et al. 2008. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J. Virol. 82:5510–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Little S. J., et al. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385–394 [DOI] [PubMed] [Google Scholar]
- 44. Margulies M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Metzner K. J., et al. 2003. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J. Infect. Dis. 188:1433–1443 [DOI] [PubMed] [Google Scholar]
- 46. Metzner K. J., et al. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239–247 [DOI] [PubMed] [Google Scholar]
- 47. Metzner K. J., et al. 2010. Efficient suppression of minority quasispecies of drug-resistant viruses present at primary HIV-1 infection by RTV-boostered protease inhibitor containing ART. J. Infect. Dis. 201:1063–1071 [DOI] [PubMed] [Google Scholar]
- 48. Metzner K. J., et al. 2005. Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS 19:1819–1825 [DOI] [PubMed] [Google Scholar]
- 49. Montes B., Segondy M. 2002. Prevalence of the mutational pattern E44D/A and/or V118I in the reverse transcriptase (RT) gene of HIV-1 in relation to treatment with nucleoside analogue RT inhibitors. J. Med. Virol. 66:299–303 [DOI] [PubMed] [Google Scholar]
- 50. Nijhuis M., Deeks S., Boucher C. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23–28 [DOI] [PubMed] [Google Scholar]
- 51. Nzeyimana S. D., et al. 2010. Monitoring of HIV-1 drug resistance and associated programmatic factors in patients initiating antiretroviral therapy at two ART sites in Bujumbura, Burundi. Antivir. Ther. 15(Suppl. 2):A153 [Google Scholar]
- 52. Pacold M., et al. 2010. Comparison of methods to detect HIV dual infection. AIDS Res. Hum. Retroviruses 26:1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Palmer S., et al. 2006. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 20:701–710 [DOI] [PubMed] [Google Scholar]
- 54. Paredes R., Marconi V. C., Campbell T. B., Kuritzkes D. R. 2007. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J. Virol. Methods 146:136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paredes R., et al. 2010. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral failure. J. Infect. Dis. 201:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parkin N. T., Gupta S., Chappey C., Petropoulos C. J. 2006. The K101P and K103R/V179D mutations in human immunodeficiency virus type 1 reverse transcriptase confer resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 50:351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pelemans H., et al. 1997. Characteristics of the Pro225His mutation in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase that appears under selective pressure of dose-escalating quinoxaline treatment of HIV-1. J. Virol. 71:8195–8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peuchant O., et al. 2008. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS 22:1417–1423 [DOI] [PubMed] [Google Scholar]
- 59. Pillay D., et al. 2006. The impact of transmitted drug resistance on the natural history of HIV infection and response to first-line therapy. AIDS 20:21–28 [DOI] [PubMed] [Google Scholar]
- 60. Pond S. L., Frost S. D. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 61. Poon A. F., et al. 2010. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol. Biol. Evol. 27:819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rhee S. Y., et al. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 103:17355–17360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Simen B. B., et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701 [DOI] [PubMed] [Google Scholar]
- 64. Simon V., et al. 2002. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS 16:1511–1519 [DOI] [PubMed] [Google Scholar]
- 65. Toni T. A., et al. 2009. Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob. Agents Chemother. 53:1670–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Laethem K., et al. 2007. No response to first-line tenofovir+ lamivudine+efavirenz despite optimization according to baseline resistance testing: impact of resistant minority variants on efficacy of low genetic barrier drugs. J. Clin. Virol. 39:43–47 [DOI] [PubMed] [Google Scholar]
- 67. Varghese V., et al. 2009. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J. Acquir. Immune Defic. Syndr. 52:309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Varghese V., et al. 2010. Nucleic acid template and the risk of a PCR-induced HIV-1 drug resistance mutation. PLoS One 5:e10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vercauteren J., et al. 2009. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J. Infect. Dis. 200:1503–1508 [DOI] [PubMed] [Google Scholar]
- 70. Vingerhoets J., et al. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Violin M., et al. 2004. Risk of failure in patients with 215 HIV-1 revertants starting their first thymidine analog-containing highly active antiretroviral therapy. AIDS 18:227–235 [DOI] [PubMed] [Google Scholar]
- 72. Wang C., Mitsuya Y., Gharizadeh B., Ronaghi M., Shafer R. W. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 17:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang G. P., Sherrill-Mix S. A., Chang K. M., Quince C., Bushman F. D. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J. Virol. 84:6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Z., Wong W. S., Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107–1118 [DOI] [PubMed] [Google Scholar]
- 75. Yap S. H., et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yerly S., et al. 2007. Transmission of HIV-1 drug resistance in Switzerland: a 10-year molecular epidemiology survey. AIDS 21:2223–2229 [DOI] [PubMed] [Google Scholar]
- 77. Zagordi O., Geyrhofer L., Roth V., Beerenwinkel N. 2010. Deep sequencing of a genetically heterogeneous sample: local haplotype reconstruction and read error correction. J. Comput. Biol. 17:417–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.