Abstract
Interferons (IFNs) play a major role in orchestrating the innate immune response toward viruses in vertebrates, and their defining characteristic is their ability to induce an antiviral state in responsive cells. Interferons have been reported in a multitude of species, from bony fish to mammals. However, our current knowledge about the molecular function of fish IFNs as well as their evolutionary relationship to tetrapod IFNs is limited. Here we establish the three-dimensional (3D) structure of zebrafish IFNφ1 and IFNφ2 by crystallography. These high-resolution structures offer the first structural insight into fish cytokines. Tetrapods possess two types of IFNs that play an immediate antiviral role: type I IFNs (e.g., alpha interferon [IFN-α] and beta interferon [IFN-β]) and type III IFNs (lambda interferon [IFN-λ]), and each type is characterized by its specific receptor usage. Similarly, two groups of antiviral IFNs with distinct receptors exist in fish, including zebrafish. IFNφ1 and IFNφ2 represent group I and group II IFNs, respectively. Nevertheless, both structures reported here reveal a characteristic type I IFN architecture with a straight F helix, as opposed to the remaining class II cytokines, including IFN-λ, where helix F contains a characteristic bend. Phylogenetic trees derived from structure-guided multiple alignments confirmed that both groups of fish IFNs are evolutionarily closer to type I than to type III tetrapod IFNs. Thus, these fish IFNs belong to the type I IFN family. Our results also imply that a dual antiviral IFN system has arisen twice during vertebrate evolution.
INTRODUCTION
Interferons (IFNs) are small helical cytokines defined by their ability to inhibit viral replication in responsive cells (13). IFNs have been identified in vertebrates from bony fish to mammals (3, 5, 12, 32). They have been playing a key role in regulating host-pathogen interactions for more than 450 million years and are subjected to high evolutionary pressures.
In mammals, there exist three types of IFNs, defined by their receptor utilization. Type I IFNs are the archetypal IFNs; they consist of multiple subtypes with alpha interferon (IFN-α) and beta interferon (IFN-β) being the best described. Type I IFNs signal through a heterodimeric complex consisting of the IFN-α/β receptor 1 (IFNAR1) and IFNAR2 chains (37). Type II IFN has only one member designated gamma interferon (IFN-γ). IFN-γ does possess some antiviral activity but is mainly an immunomodulatory cytokine acting on leukocytes (42). Type III IFNs, also known as lambda interferon (IFN-λ), are structurally distinct from type I IFNs (10) and use a different receptor complex made up of the interleukin 10 receptor 2 (IL-10R2) and IFN-λ receptor 1 (IFNLR1) chains (15, 35). Despite the fact that the IL-10R2 chain is shared with several other class II cytokines (IL-10, IL-22, IL-24, and IL-26), type III IFN signaling in responsive cells is highly similar to that of type I IFNs but not to that of IL-10 (44). Type I and type III IFN signaling is mainly mediated via interaction of activated STAT1 and STAT2 (STAT stands for signal transducers and activators of transcription) with IRF9 (interferon regulatory factor 9), forming the transcription factor ISGF3 (interferon-stimulated gene factor 3). The primary biological difference between type I and III IFNs is receptor distribution, with the type I receptor found on essentially all nucleated cells, while only a limited subset of cells, primarily of epithelial origin, seems to respond to type III IFNs (27, 36).
The structures of all three types of mammalian IFNs together with those of other class II helical cytokines (IL-10, IL-19, and IL-22) are now available (4, 10, 24, 33, 41, 43). They share a structural basis, consisting of six secondary structure elements (A through F), which are connected by loops of different lengths. In general, elements A, C, D, E, and F form α-helices, whereas element B is less ordered and more variable. Helices A, C, D, and F form a four-helical bundle, which defines the structural core of class II helical cytokines. Helix F is straight in type I IFNs, whereas it contains a characteristic bend in the other cytokines. Due to the straight helix F, type I IFNs adopt a characteristic elliptical shape resembling that of an American football, clearly distinguishing their three-dimensional (3D) structure from the other class II helical cytokines. Thus, structural evidence indicates that cytokines of the IL-10 family, as well as type II and type III IFN, are structurally closer to one another than to type I IFN (11).
Bony fish have a clear homolog of IFN-γ (46), and a second relatively diverse family of IFNs, which is highly induced during viral infection and possesses characteristics of both mammalian type I and III IFNs. Since IFN-γ is not covered by the present study, we will from now on refer to the second set of IFNs simply as “fish IFNs.” These fish IFNs have been described in several fish species (3, 18, 20, 30) with most species possessing multiple IFN genes (39). Using a bioinformatics approach, Zou et al. identified two distinct groups of fish IFNs (45). Subsequent work demonstrated that both IFN groups possessed antiviral activity (19) and signaled through distinct but related receptor complexes (2). We deliberately use “group” referring to fish IFN and “type” referring to mammalian IFN as not to imply any direct relation, and thus, fish group I is not necessarily equivalent to mammalian type I. In zebrafish, group I includes two members, IFNφ1 and IFNφ4, and uses a receptor complex consisting of cytokine receptor family member b1 (CRFB1) and CRFB5. Group II also has two members, IFNφ2 and IFNφ3, that signal via a receptor complex consisting of CRFB2 and CRFB5 (2).
The evolution of IFNs, in particular type I and III IFNs, has been intensely debated (9, 16, 20). As described above, the cloning of IFNs from several fish species further fuelled this debate. On the basis of sequence similarities and predicted structural features, some authors have claimed fish IFNs to be related to type I IFNs (31, 45), but the similarity values are too low for traditional phylogenetic analysis to yield definite answers to this question (20, 23, 24). In contrast, using receptor structure, we proposed that fish IFN is related to type III IFNs (17). Another argument in this favor, namely, gene structure, has been discarded by the recent finding of intron-containing type I IFN genes in amphibians (28).
The three-dimensional structures of proteins carry substantial information on both function and possible phylogenetic relationships. We established the 3D structure of zebrafish IFNs from both groups and used this structure to produce structurally guided alignment of vertebrate class II cytokines. Our structural data firmly establish the two groups of zebrafish IFNs as homologs of the mammalian type I IFNs. We discuss the possible evolutionary scenarios of class II helical cytokines in light of these structural results.
MATERIALS AND METHODS
Protein expression and purification.
The IFNφ1 and IFNφ2 constructs were made as previously described (2). Expression and purification of the zebrafish IFNs were done as previously described for human IFN-λ3 (7). However, the buffer used for size exclusion chromatography was replaced with 150 mM NaCl and 20 mM morpholineethanesulfonic acid (MES) (pH 6.5), and the eluted protein was used immediately for crystallization.
Crystallization and structural determination. (i) IFNφ2.
The initial screening was done using the index Screen system (Hampton Research) at both 4°C and room temperature (RT). Optimal crystals were grown by mixing equal volumes of protein solution with reservoir solution containing 0.2 M MgCl2, 0.1 M HEPES (pH 7.5), and 25% polyethylene glycol 3350 (PEG 3350) at 4°C. Upon optimization of these conditions, it was established that 0.2 M MgCl2, 22% PEG 3350, and 0.1 M HEPES (pH 8) and 4°C gave the best crystals. The crystals appeared after 6 to 10 weeks. When the initial hits were used for seeding, crystals would appear in 2 or 3 days. They were flash frozen in a cryo buffer containing 0.2 M MgCl2, 25% PEG 3350, 0.1 M HEPES (pH 8), and 17.5% glycerol. Data were collected at the I911-3 beam line at the MAX laboratory (Lund, Sweden) at 0.8742 Å. The data were indexed using HKL2000 and scaled using scalepack to 1.49 Å. The crystals belonged to space group P61 with unit cell lengths a = 84.215 Å, b = 84.215 Å, and c = 45.209 Å and angles α = 9°, β = 9°, and γ = 12°.
The phases were obtained by quickly soaking the crystals in 0.2 M MgCl2, 25% PEG 3350, 0.1 M HEPES (pH 8), 15% glycerol, and 500 mM NaI. The data were collected at the I911-3 beam line at the MAX laboratory at a wavelength of 1.5 Å. The data were indexed using HKL2000 (26) and scaled using scalepack to 2.19 Å. A single iodine site was found by a single-wavelength, anomalous dispersion approach using SHELX (34). The site found using SHELX was transferred to SHARP (6), and a full single isomorphous replacement with anomalous scattering approach, using the iodide derivative and native data set, was adopted. The initial structure was built using Resolve (40), and successive cycles of building and refinement were done in coot (8) and Phenix (1). The final R and Rfree values were 0.1413 and 0.1874, respectively, at a resolution of 1.49 Å. See Table 1 for details.
Table 1.
Statistics of crystallographic data for the zebrafish interferonsa
| Parameterb | Value for IFNc |
||
|---|---|---|---|
| IFNφ1 (native) | IFNφ2 (native) | IFNφ2 (iodine) | |
| Data collection statistics (MAX laboratory) | |||
| Energy (keV) | 14,199 | 13,565 | 8,331 |
| Space group | P21 | P61 | P61 |
| Unit cell lengths | |||
| a, b, c (Å) | 48.8, 62.2, 51.3 | 84.2, 84.2, 45.2 | 84.1, 84.1, 45.2 |
| α, β, γ (°) | 90.00, 91.42, 90.00 | 90.00, 90.00, 120.00 | 90, 90, 120 |
| Mosaicity (°) | 0.83 | 0.21 | 0.24 |
| Resolution (Å) | 50–2.09 | 50–1.49 | 50–2.2 (2.3–2.2) |
| No. of unique reflections | 17,632 | 30,020 | 18,512 |
| Redundancy | 3.5 (2.4) | 5.4 (4.0) | 5.5 (5.7) |
| Completeness (%) | 96.3 (78.9) | 98.1 (84.9) | 99.9 (99.8) |
| 〈I〉/〈σ〉 | 12.4 (2.15) | 26.16 (4.7) | 8.2 (12.6) |
| Rmerge (%) | 14 (35.6) | 5.5 (20.6) | 9.0 (13.2) |
| Refinement statistics | |||
| Rcryst (%) | 0.1813 | 0.1413 | |
| Rfree (%) | 0.2372 | 0.1874 | |
| Est. coord. error (Å) | 0.26 | 0.25 | |
| Est. phase error (°) | 24.00 | 16.90 | |
| RMSD from ideal | |||
| Bond length (Å) | 0.008 | 0.005 | |
| Bond angle (°) | 1.033 | 0.838 | |
| Wilson B-factor (Å2) | |||
| Protein | 24.51 | 19.42 | |
| Water | 30.69 | 37.38 | |
| Other ligands | 74.00 | ||
| No. of atoms | 2,776 | 1,552 | |
| No. of water molecules | 179 | 265 | |
| No. of other molecules | 2 Ni | 0 | |
| Ramachandran plot | |||
| Favored (%) | 98.71 | 100 | |
| Allowed (%) | 100 | 100 | |
Statistics for data collection and phases are shown.
Est. coord. error, estimated coordinate error; RMSD, root mean square deviation.
The values for the interferons found by using the iodide derivative and native data set are shown. The values in parentheses are the values for the highest-resolution shells.
(ii) IFNφ1.
Initial screening was done using the index Screen system (Hampton Research) at both 4°C and RT. Optimal crystals were grown by mixing equal volumes of protein solution with reservoir solution containing 0.2 M NaCl, 0.1 M Bis-Tris (pH 5.5), and 25% PEG 3350 at RT. Upon optimization of these conditions, it was established that 0.2 M NaCl, 20% PEG 3350, 0.1 M HEPES (pH 6.5), 3% Jeffamine M-600 gave the best crystals. Needle-shaped crystals appeared after 2 to 5 days. They were flash frozen in a cryo buffer containing 0.2 M NaCl, 25% PEG 3350, 0.1 M HEPES (pH 6.5), 3% Jeffamine M-600, and 17.5% glycerol. Data were collected at the I911-3 beam line at the MAX laboratory at 0.8742 Å. The data were indexed using HKL2000 (26) and scaled using scalepack to 2.1 Å. The crystals belonged to space group P21 with unit cell lengths a = 46.766 Å, b = 60.743 Å, and c = 49.679 Å and angles α = 9°, β = 9°, and γ = 93.341°.
The phases were obtained by molecular replacement in Phaser (21) using a poly(A) model of IFNφ2 lacking helix F. The initial structure was built using Phenix (1), and successive cycles of building and refinement were done in coot (8) and Phenix. The final R and Rfree values were 0.2372 and 0.1813, respectively, at a resolution of 2.1 Å. See Table 1 for details.
Phylogenetic analysis.
Automated alignment was done using Clustal W2.0.12. Signal peptides and N- and C-terminal portions of the cytokines were removed leaving approximately 130 amino acids. All sequences were found at the NCBI data bank (NCBI accession numbers given after the cytokine sequence names) (human IFN-α2 [huIFNA2], NP_000596.2; huIFNA5, NP_002160; huIFNA8, NP_002161.2; human IFN-β [huIFNB], NP_002167; Gallus gallus (chicken) IFN-α1 [gaIFNA1], CAA63214; gaIFNA2, CAA63216; gaIFNA3, CAA63216.1; gaIFNB, CAA63217; Xenopus tropicalis IFN1 [xtIFN1], CAO03085; xtIFN2, CAO03086; xtIFN3, CAO03087; xtIFN4, CAO03088; xtIFN5, CAO03089; Gallus gallus IL-10 [gaIL-10], NP_001004414; xtIL-10, NP_001165400; huIL-10, NP_000563.1; Danio rerio (zebrafish) IL-10 [drIL-10], NP_001018621.2; human IFN-λ2 [huIFNL2], NP_742150; huIFNL3, NP_742151; huIFNL1, NP_742152; gaIFNL, NP_001121968.1; xtIFNL3, ACV32136.1; xtIFNL2, ACV32135.1; xtIFNL1, ACV32134.1; drIFN1, NP_997523.1; drIFN2, NP_001104552.1; drIFN3, NP_001104553.1; drIFN4, NP_001155212.1; drIFNG, NP_998029.1; huIFNG, NP_000610.2; gaIFNG, NP_990480.1; xtIFNG, ABU54059.1).
Structure-guided alignment was performed manually with the help of available 3D structures (PDB accession numbers or entry codes given after the cytokines; IL-10, 2H24; IFN-α2, 1RH2; IFN-λ3, 3HHC; IFN-γ, 1EKU). IL-10 and IFN-γ exist as dimers where the E and F helices of one subunit occupy the position of the other subunit; an artificial monomer was then generated by keeping regions A to D of one subunit and regions E and F of the other subunit. 3D structures were superposed two by two using coot, and matching amino acids were identified with the help of MacPyMol.
After generating a multiple alignment either using Clustal or guided by the structure, phylogenetic analysis was performed on the Mobyle portal (http://mobyle.pasteur.fr). Parsimony analysis was done with the protpars program. Neighbor-joining analysis was done by first running the protdist program, followed by the neighbor program. Maximum likelihood was done with the phyml program, using the SPR option for tree topology search. Sets containing 1,000 bootstraps were analyzed in each case; in all other cases, default parameters were used.
RESULTS
Structure of recombinant IFNφ1 and IFNφ2.
Recombinant zebrafish IFNs were produced in Escherichia coli, and their bioactivity was tested as reported previously (2) (see Fig. S1 in the supplemental material). IFNφ1 and IFNφ2, which could be produced at high purity and were highly biologically active, were selected for crystallization and structural determination. Crystallization was carried out as described in Materials and Methods. Due to the high quality of the IFNφ2 crystals and the derived diffraction data, we solved the IFNφ2 structure first and used this structure as a partial model for obtaining the phases for IFNφ1.
(i) Structure of IFNφ2.
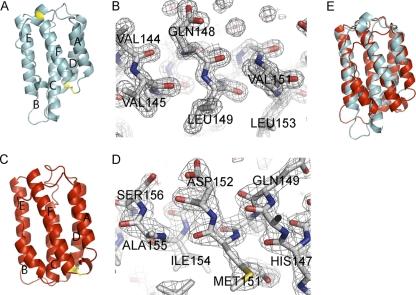
Cryo-protected IFNφ2 crystals diffracted to 1.49 Å, which to our knowledge is the highest resolution obtained for any class II helical cytokine. The phases were determined using an iodine derivative, and an initial model was built automatically by Resolve (40). This model was completed manually and refined; it includes residues 6 to 156. The structure of IFNφ2 is shown in Fig. 1A and B. Details of data collection, phasing, and refinement are given in Table 1. The final structure reveals a typical type I IFN architecture with 6 structural elements designated A through F (Fig. 1A), and with the exception of element B, they are all α-helical. The core of the structure is made up of helices A, C, D, and F that are oriented in the up-up-down-down orientation typical for a class II helical cytokine. Helix F is long and straight, giving the molecule an elongated shape resembling an American football, which is the hallmark of type I IFNs. As expected, two disulfide bridges are present, one linking the N terminus to helix D and one linking the AB loop to helix F, which is analogous to the disulfides found in mammalian IFN-α. Element B is well defined in the crystal structure but does not adopt a defined secondary structure.
Fig. 1.
Structure of the zebrafish interferons. (A) Cartoon representation of the structure of IFNφ2 (PDB accession number or entry code 3PIW). The structural elements are labeled A through F. The two disulfides are shown in yellow. (B) Electron density (2σ) of a fraction of the F helix in IFNφ2 is shown. Residues from helix F are labeled. (C) Cartoon representation of the structure of IFNφ1 (PDB entry code 3PIV). The structural elements are labeled A through F. The single disulfide is shown in yellow. (D) Electron density (2σ) of a fraction of the F helix in IFNφ1 is shown. Residues from helix F are labeled. (E) Comparison of the structures of IFNφ1 (red) and IFNφ2 (cyan). The two molecules have been superimposed in coot using SSM superimpose.
(ii) Structure of IFNφ1.
The crystals were cryo-protected with glycerol and diffracted to 2.08 Å. The structure was solved with Molecular Replacement, using a polyalanine model of IFNφ2, excluding helix F, as a search model. Helix F was excluded in the search model because it is the defining difference between type I and III IFNs, and we did not want to introduce any bias. The model was completed manually and includes residues 4 to 160 of the mature peptide. Details of data collection and refinement are given in Table 1; the structure is shown in Fig. 1C and D. IFNφ1 is structurally highly similar to IFNφ2, adopting the same basic type I IFN architecture with 6 structural elements designated A through F. As expected, the structure contains one disulfide bridge, connecting the N terminus to helix D.
(iii) Comparing the structures of IFNφ1 and IFNφ2.
The two fish IFN structures overlay well with a root mean square deviation (RMSD) value of 1.85 Å, where the most noticeable differences are the lengths of helices C and F (Fig. 1E). Helix C is longest in IFNφ2, consisting of 23 amino acids as opposed to only 19 in IFNφ1. When looking at helix F, that of IFNφ1 is the longest containing 25 amino acids, whereas in IFNφ2, it spans 19 amino acids. The available structural and mutational data from human type I and III IFNs indicate that receptor binding is primarily located on helices A and F (10, 25). However, as IFNφ1 and IFNφ2 display roughly equal biological activity, the different lengths of helices C and F do not appear to affect receptor binding. This is probably because the differences in helices C and F are found in the opposite end of the molecule as the expected receptor binding site. This has, however, not been experimentally confirmed. In other words, despite their limited sequence homology (19% identical amino acids) and the fact that they bind different receptors, the 3D structures of IFNφ1 and IFNφ2 are very similar.
(iv) Comparison of zebrafish IFNs to mammalian members of the class II cytokine family.
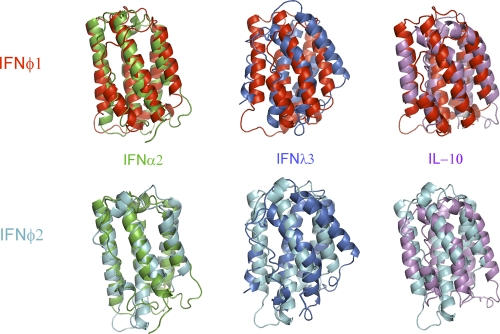
Structurally, type I IFNs constitute their own distinct group within the class II helical cytokine family, typified by a straight helix F. This is opposed to the characteristic bending of helix F in other class II helical cytokines, including class III IFNs (11). Figure 2, top panel, shows the comparison of IFNφ1 with human IFN-α, IFN-λ, and IL-10; IFNφ2 is superimposed on the same cytokines in the bottom panel of Fig. 2. From this comparison, it is clear that structurally, both IFN-φ1 and IFNφ2 belong to type I IFNs. The RMSD values support this view, as the differences between the value for IFNφ1 versus the values for IFN-α and IFN-β are 2.14 Å and 1.60 Å, respectively, whereas the values for distance to IL-10 and IFN-λ are 3.04 Å and 3.40 Å, respectively. The same is true for IFNφ2; the RMSD values versus IFN-α and IFN-β are 2.00 Å and 1.75 Å, respectively, whereas the distances to IL-10 and IFN-λ are 3.51 Å and 3.17 Å, respectively (Table 2). On the sequence level, type I IFNs are characterized by the presence of a conserved motif (CAWE) in helix F, which is also found in IFNφ2. A closer examination of the structure reveals that this motif is part of a larger conserved structural motif formed around the EF loop and the junction of helices C, E, and F (Fig. 3). In the following, the amino acid numbering is according to IFNφ2, and the capital letters in parentheses refer to the helix within which the given amino acid is found. Trp142(F) and Trp80(D) form a hydrophobic core, Glu143(F) of helix F interacts with both helices C and E, while Val145(F) and Arg146(F) interact with both the D helix and the AB loop. In addition, Leu132(E) and Leu128(E) found at the end of helix E form a hydrophobic pocket with Val145(F) and Leu70(C). These interactions are conserved in the two groups of fish IFNs and among mammalian type I IFNs as well (allowing for conservative substitutions only). Extending the motif to the middle of helix E, we observed that position 125 is always an F or Y in type I IFNs. Looking at the structures, it can be seen that Y/F125 of type I IFNs and fish IFNs can interact with residues of helix F (Fig. 3).
Fig. 2.
Comparison of the zebrafish interferons to mammalian type II cytokines. (Top) Comparison of IFNφ1 to human IFN-α2 (PDB accession number or entry code 1RH2), IFN-λ3 (PDB entry code 3HHC), and IL-10 (PDB entry code 2H24). (Bottom) Comparison of IFNφ2 to human IFN-α2, IFN-λ3, and IL-10. The molecules have been superimposed in coot using SSM superimpose.
Table 2.
Root mean square deviations of the alignment of selected type I cytokine structuresa
| Cytokine | RMSD value for cytokine pair: |
||||||
|---|---|---|---|---|---|---|---|
| IFNφ1 | IFNφ2 | IFN-α | IFN-β | IFN-λ | IL-10 | IFN-γ | |
| IFNφ1 | 1.85 | 2.14 | 1.60 | 3.40 | 3.04 | 3.43 | |
| IFNφ2 | 2.00 | 1.75 | 3.17 | 3.51 | 3.90 | ||
| IFN-α | 2.29 | 3.32 | 2.98 | 4.01 | |||
| IFN-β | 3.23 | 3.79 | 3.56 | ||||
| IFN-λ | 2.80 | 2.56 | |||||
| IL-10 | 3.06 | ||||||
The structures of IFNφ1, IFNφ2, IFN-α, IFN-β, IFN-λ3, IL-10, and IFN-γ were superimposed in coot using SSM superimpose. The root mean square deviation (RMSD) values generated by coot are shown.
Fig. 3.
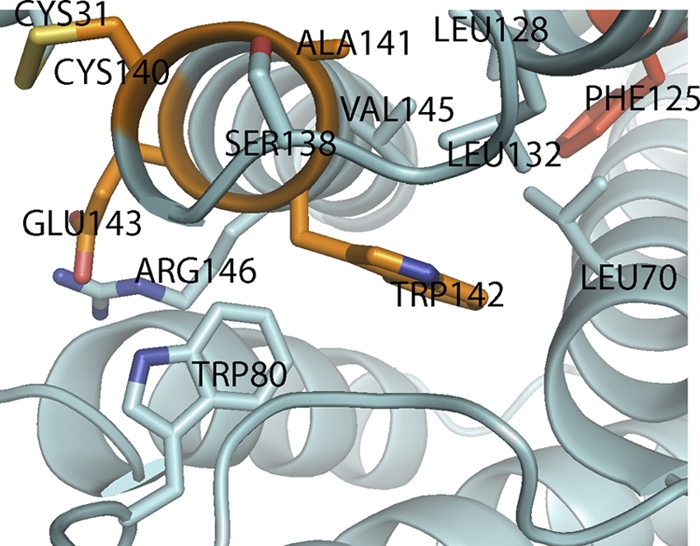
The CAWE motif of IFNφ2. Conserved residues in type I interferons in the vicinity of the CAWE motif are shown as sticks. The residues in the CAWE motif are colored orange. The highly conserved aromatic residue (Phe) is colored red. Visible helices and loops are labeled. See text for details.
The overall structural motif described here seems conserved among all type I IFNs and locks the N-terminal end of helix F to the other helixes in the four-helix bundle and is most likely responsible for the characteristic fold of type I IFNs. Among the vast number of known type I IFNs, there are examples lacking one or more of the described residues. However, one has to keep in mind that the specific activities of these subtypes are largely unknown and thus also the functional consequence of these substitutions. The only exception seems to be Y/F125, which appears to be universally conserved.
IFNφ1 is lacking one of the two otherwise conserved disulfide bridges connecting the beginning of helix F to the AB loop and is thus missing the “C” of the CAWE motif. Nevertheless, IFNφ1 retains a robust antiviral activity (2, 17, 19) (see Fig. S1 in the supplemental material). Our crystallographic data indicate that the interactions around the CAWE motif are of sufficient strength to maintain the fold even in the absence of this disulfide bridge.
Phylogeny of IFN.
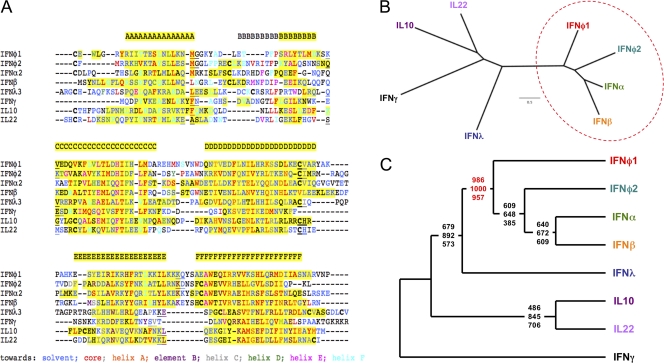
The phylogeny of fish IFNs has been debated in recent years. Until now, traditional sequence alignment and subsequent calculation of phylogenetic trees has not given a clear answer (28, 32, 38, 39). Indeed, we reached a similar conclusion when, using protein sequences of IL-10 and type I, type II, and type III IFNs of zebrafish (Danio rerio), clawed frog (Xenopus tropicalis), chicken (Gallus gallus), and human (Homo sapiens), we generated a multiple alignment using Clustal W (see Fig. S2 in the supplemental material) and constructed a phylogenetic tree (see Fig. S3 in the supplemental material). In this tree, IFN-γ and IL-10 both form monophyletic groups. Type III IFNs from clawed frogs, chickens, and humans are also clearly monophyletic, but with no clear homologs in fish. Zebrafish group I IFNs appear as a sister group; however, the bootstrap values are low, and thus, there is no firm statistical support for this. Finally, the type I IFN side of the tree is almost useless, as no relationship between two proteins from different species is supported by a significant bootstrap value. Zebrafish group II IFNs cluster with human and chicken type I IFNs, but only very loosely. This is surprising, given the clear structural similarity that we observe between fish IFNs and mammalian type I IFNs. However, phylogenetic reconstruction is entirely dependent on the assumed sequence alignments, and automatic alignment procedures such as the one used here (Clustal W) become unreliable with proteins that share less than 30% identity (14). Furthermore, Clustal W assumes a certain phylogeny prior to alignment, which in our case could lead to circular arguments. Thus, we generated a protein alignment guided by our structures (Fig. 4A) and used this alignment to infer the phylogeny of the IFN family members. The resulting phylogenetic tree (Fig. 4B) shows that both IFNφ1 and IFNφ2 group with mammalian type I IFNs. Different tree-generating algorithms (parsimony, distance, and maximum likelihood) yielded similar results (Fig. 4C). In all cases, the existence of a “type I IFN clade” that encompasses fish IFNs and mammalian type I IFNs was found to be supported by high bootstrap values. This result was based on similarities over the whole sequence and not only on the shape of specific regions (i.e., helix F and the CAWE motif) and reinforces our previous conclusion: fish IFNs of both groups belong to the type I IFN family, both structurally and phylogenetically.
Fig. 4.
Structure-guided alignment of class II cytokines. (A) Multiple alignment of zebrafish IFNφ1 and IFNφ2 with human class II cytokines, established from superimposing crystallographic structures. Amino acids have been colored according to the principal orientation of the side chain, using the code at the bottom of the alignment; if the orientation of the side chain is ambiguous or unknown, the amino acid was left in black. Alpha-helical regions are shown on a yellow background. Cysteines engaged in disulfide bridges are depicted in bold type. Exon boundaries are represented by underlined letters (the last amino acid coded by an exon and the first amino acid coded by the next exon). Gaps introduced to maximize alignment are indicated by dashes. (B) Phylogenetic tree obtained using a distance (neighbor-joining) method, deduced from the alignment in panel A. The “type I IFN clade” is circled. Bar, 0.5 amino acid substitutes. (C) Consensus cladograms obtained from the alignment in panel A, with the IFN-γ sequence chosen as an outgroup. For each node, the bootstrap support values (out of 1,000 replicates) are given for distance, parsimony, and maximum likelihood methods by the top, middle, and bottom numbers, respectively.
DISCUSSION
Here we present the crystal structures of IFNφ1 and IFNφ2 from zebrafish. The IFNφ1 and IFNφ2 structures are very similar to one another as well as to the structures of mammalian type I IFNs. This was a surprise to us, as we had previously argued that fish IFNs were orthologs of mammalian type III IFNs (17). However, aligning IFN sequences from distantly related species using standard alignment tools is problematic, due to the low level of sequence conservation. Solving the 3D structure of fish IFNs thus yielded unique additional information that could be used to generate an improved protein alignment. Although this approach is not without its drawbacks—such as a possible subjective bias, or excessive influence of convergent evolution—we believe that the phylogenetic relationships inferred from this alignment are much more reliable than the previous ones.
Evolutionary trees, calculated using different algorithms but all based upon the structural alignment, place zebrafish IFNs of both groups firmly within the type I IFN clade. However, because of low bootstrap values at the IFNφ1/IFNφ2 node, our data cannot help determine which of the two groups of fish IFNs (if any) would be the true ancestral gene of tetrapod type I IFNs. This could well be due to the fact that the last common ancestor of zebrafish and mammals had only one IFN gene, and we suspect that the duplication events leading to the two groups of fish IFNs happened after the split between the ancestor of tetrapods and that of teleost fish. In such a case, it is meaningless to discuss which of the zebrafish IFNs is more closely related to tetrapod type I IFNs. At any rate, our data strongly suggest that fish IFNs and mammalian type I IFNs share a ancestor, which had the archetypal structure of a type I IFN.
If both group I and II fish IFNs are members of the type I IFN family, what about the origin of type III IFN? A recent study by Qi et al (28) clearly demonstrated that tetrapod type III IFNs form a monophyletic group but shed little light on their origin. If structure can be considered an instrumental criterion, then we have to consider that type III IFNs are derived from an IL-10-related cytokine. This origin is also suggested by the fact that the type III IFN receptor complex is made up of IL-10R2 and IFNLR1 with the latter being encoded by a gene that lies in tandem with IL-22R1 in all sequenced tetrapod genomes. This genomic position suggests that IFNLR1 and IL-22R1 have been generated by tandem duplication of a parental gene. Thus, we consider a scenario where type III IFN would be derived from the IL-10/IL-22 family as being the most likely.
The question now arises why bony fish and tetrapods both independently diversify their IFN systems? By “diversification of their IFN systems,” we are referring to acquisition of multiple IFN systems with different receptor complexes, not to the diversification of the ligands in the context of a fixed receptor. A first reason for this duplication would be fighting viral escape. Viruses often evolve inhibitors of the IFN systems (29), and evolving multiple receptor-ligand systems is an obvious counterstrategy. However, we consider this interpretation to be oversimplified, particularly since type I and type III IFN systems share substantial parts of their intracellular signaling machinery, which could be targeted by viral inhibitors (44).
Then why has it been evolutionary advantageous to evolve two independent IFN systems, as clearly happened in both the bony fish and amniote lineages? First, the mechanism is clearly different, as in fish, a straightforward duplication of the parental IFN receptor gene leads to the creation of the two CRFB1 and CRFB2 receptor chains, which still share the CRFB5 receptor subunit. Simultaneously, duplication of the genes encoding the ligand was then followed by separate coevolution of receptor-ligand interaction, resulting in the current situation with two separate but related systems. The origin of the IFN-λ family is less clear and discussed above, but it is unlikely that IFN-λ is directly evolutionarily related to the type I IFNs. However, the underlying functionality which drove evolution toward two IFN systems might be very similar. Most viruses enter our body via the epithelium, and thus epithelial cells suffer from high rates of viral infection. In mammals, IFN-λ appears to be a first-line defense IFN, with primary receptor expression in cells of epithelial origin (27, 36). Thus, due to the limited tissue tropism, type III IFN expression can protect high-risk tissues without the toxicity associated with a full activation of the type I IFN system. This view is supported by recent studies, providing strong evidence for a critical role of IFN-λ in controlling viral infection in both lung and gut epithelia (22, 23). However, at present, it is unclear whether a similar division of labor exists between the two IFN systems found in fish. Experiments are under way to establish the distribution of the CRFB1 and CRFB2 receptor subunits in zebrafish.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hans Henrik Gad for discussions on structural analysis and critical reading of the manuscript. We are grateful to Susanne Vends and Gitte K. Hartvigsen for expert technical assistance. We are also grateful to the staff of beam line I911 at the MAX laboratory (Lund, Sweden) for help in data collection.
Beam line access was funded partly via the Danscatt consortium. O.J.H. received a junior research fellowship from the Danish Cancer Society. Research in the authors' laboratories was supported by grants from the Danish Cancer Society, Danish Medical Research Council (grant 22-04-0704), Novo Nordisk Foundation, Carlsberg Foundation, and Agence Nationale de la Recherche (ANR-MIME-2007 and ANR-10-MIDI 010).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Adams P. D., et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 2. Aggad D., et al. 2009. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 183:3924–3931 [DOI] [PubMed] [Google Scholar]
- 3. Altmann S. M., Mellon M. T., Distel D. L., Kim C. H. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 77:1992–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang C., et al. 2003. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J. Biol. Chem. 278:3308–3313 [DOI] [PubMed] [Google Scholar]
- 5. de Kinkelin P., Dorson M. 1973. Interferon production in rainbow trout (Salmo gairdneri) experimentally infected with Egtved virus. J. Gen. Virol. 19:125–127 [DOI] [PubMed] [Google Scholar]
- 6. de la Fortelle E., Bricogne G. 1997. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276:472–494 [DOI] [PubMed] [Google Scholar]
- 7. Dellgren C., Gad H. H., Hamming O. J., Melchjorsen J., Hartmann R. 2009. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 10:125–131 [DOI] [PubMed] [Google Scholar]
- 8. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 9. Fox B. A., Sheppard P. O., O'Hara P. J. 2009. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS One 4:e4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gad H. H., et al. 2009. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J. Biol. Chem. 284:20869–20875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gad H. H., Hamming O. J., Hartmann R. 2010. The structure of human interferon lambda and what it has taught us. J. Interferon Cytokine Res. 30:565–571 [DOI] [PubMed] [Google Scholar]
- 12. Galabov A. S. 1981. Induction and characterization of tortoise interferon. Methods Enzymol. 78:196–208 [DOI] [PubMed] [Google Scholar]
- 13. Isaacs A., Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267 [PubMed] [Google Scholar]
- 14. Kececioglu J., Kim E., Wheeler T. 2010. Aligning protein sequences with predicted secondary structure. J. Comput. Biol. 17:561–580 [DOI] [PubMed] [Google Scholar]
- 15. Kotenko S. V., et al. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77 [DOI] [PubMed] [Google Scholar]
- 16. Krause C. D., Pestka S. 2005. Evolution of the class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacol. Ther. 106:299–346 [DOI] [PubMed] [Google Scholar]
- 17. Levraud J. P., et al. 2007. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 178:4385–4394 [DOI] [PubMed] [Google Scholar]
- 18. Long S., et al. 2004. Identification of a cDNA encoding channel catfish interferon. Dev. Comp. Immunol. 28:97–111 [DOI] [PubMed] [Google Scholar]
- 19. Lopez-Munoz A., Roca F. J., Meseguer J., Mulero V. 2009. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J. Immunol. 182:3440–3449 [DOI] [PubMed] [Google Scholar]
- 20. Lutfalla G., et al. 2003. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCoy A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mordstein M., et al. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mordstein M., et al. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84:5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagem R. A., et al. 2002. Crystal structure of recombinant human interleukin-22. Structure 10:1051–1062 [DOI] [PubMed] [Google Scholar]
- 25. Nudelman I., et al. 2010. Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY. Biochemistry 49:5117–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 27. Pulverer J. E., et al. 2010. Temporal and spatial resolution of type I and III interferon responses in vivo. J. Virol. 84:8626–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi Z., Nie P., Secombes C. J., Zou J. 2010. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 184:5038–5046 [DOI] [PubMed] [Google Scholar]
- 29. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 30. Robertsen B., Bergan V., Rokenes T., Larsen R., Albuquerque A. 2003. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 23:601–612 [DOI] [PubMed] [Google Scholar]
- 31. Robertsen B., Zou J., Secombes C., Leong J. A. 2006. Molecular and expression analysis of an interferon-gamma-inducible guanylate-binding protein from rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 30:1023–1033 [DOI] [PubMed] [Google Scholar]
- 32. Schultz U., Kaspers B., Staeheli P. 2004. The interferon system of non-mammalian vertebrates. Dev. Comp. Immunol. 28:499–508 [DOI] [PubMed] [Google Scholar]
- 33. Senda T., et al. 1992. Three-dimensional crystal structure of recombinant murine interferon-beta. EMBO J. 11:3193–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sheldrick G. M. 2008. A short history of SHELX. Acta Crystallogr. A 64:112–122 [DOI] [PubMed] [Google Scholar]
- 35. Sheppard P., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 [DOI] [PubMed] [Google Scholar]
- 36. Sommereyns C., Paul S., Staeheli P., Michiels T. 2008. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 38. Stein C., Caccamo M., Laird G., Leptin M. 2007. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 8:R251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun B., Robertsen B., Wang Z., Liu B. 2009. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev. Comp. Immunol. 33:547–558 [DOI] [PubMed] [Google Scholar]
- 40. Terwilliger T. C. 2003. Improving macromolecular atomic models at moderate resolution by automated iterative model building, statistical density modification and refinement. Acta Crystallogr. D Biol. Crystallogr. 59:1174–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter M. R., et al. 1995. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature 376:230–235 [DOI] [PubMed] [Google Scholar]
- 42. Young H. A., Bream J. H. 2007. IFN-gamma: recent advances in understanding regulation of expression, biological functions, and clinical applications. Curr. Top. Microbiol. Immunol. 316:97–117 [DOI] [PubMed] [Google Scholar]
- 43. Zdanov A., et al. 1995. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure 3:591–601 [DOI] [PubMed] [Google Scholar]
- 44. Zhou Z., et al. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 81:7749–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zou J., Tafalla C., Truckle J., Secombes C. J. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 179:3859–3871 [DOI] [PubMed] [Google Scholar]
- 46. Zou J., et al. 2004. Identification of an interferon gamma homologue in Fugu, Takifugu rubripes. Fish Shellfish Immunol. 17:403–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.