Abstract
Previous studies have shown that the PDZ-binding motif of the E6 oncoprotein from the mucosal high-risk (HR) human papillomavirus (HPV) types plays a key role in HPV-mediated cellular transformation in in vitro and in vivo experimental models. HR HPV E6 oncoproteins have the ability to efficiently degrade members of the PDZ motif-containing membrane-associated guanylate kinase (MAGUK) family; however, it is possible that other PDZ proteins are also targeted by E6. Here, we describe a novel interaction of HPV type 16 (HPV16) E6 with a PDZ protein, Na+/H+ exchange regulatory factor 1 (NHERF-1), which is involved in a number of cellular processes, including signaling and transformation. HPV16 E6 associates with and promotes the degradation of NHERF-1, and this property is dependent on the C-terminal PDZ-binding motif of E6. Interestingly, HPV16 E7, via the activation of the cyclin-dependent kinase complexes, promoted the accumulation of a phosphorylated form of NHERF-1, which is preferentially targeted by E6. Thus, both oncoproteins appear to cooperate in targeting NHERF-1. Notably, HPV18 E6 is not able to induce NHERF-1 degradation, indicating that this property is not shared with E6 from all HR HPV types. Downregulation of NHERF-1 protein levels was also observed in HPV16-positive cervical cancer-derived cell lines, such as SiHa and CaSki, as well as HPV16-positive cervical intraepithelial neoplasia (CIN). Finally, our data show that HPV16-mediated NHERF-1 degradation correlates with the activation of the phosphatidylinositol-3′-OH kinase (PI3K)/AKT signaling pathway, which is known to play a key role in carcinogenesis.
INTRODUCTION
Approximately 15 mucosal human papillomavirus (HPV) types, referred to as high-risk (HR) HPV types, are linked to the development of cervical cancer and a subset of other anogenital cancers (28). HPV type 16 (HPV16) is the most common type within the HR HPV group, being responsible for approximately 50% of all cervical cancers worldwide (12). Biological studies have demonstrated that E6 and E7 play a key role in HPV-mediated carcinogenesis (5). Both viral oncoproteins display transforming properties in in vitro and in vivo experimental models, mainly by interacting with and neutralizing the functions of several cellular proteins, including products of tumor suppressor genes (5). E7 from HPV16 and other HR HPV types deregulates cell cycle control by targeting the retinoblastoma tumor suppressor protein (pRb1) and its related proteins p107 and p130, also termed pocket proteins (5). pRb1 negatively regulates, via direct association, the activity of several transcription factors, including members of the E2F family. HPV16 E7 is able to directly interact with pRb1, promoting its degradation via the proteasome pathway. This event results in the release and constitutive activation of E2F complexes that, in turn, initiate the transcription of genes encoding positive cell cycle regulators, such as cyclin E and cyclin A. Like E7, the HR HPV E6 oncoprotein associates with another tumor suppressor, p53, leading to its degradation via the ubiquitin pathway (5). The role of p53 is to safeguard the integrity of the genome by inducing cell cycle arrest or apoptosis in response to genotoxic stress. Therefore, its inactivation by the E6 protein can lead to chromosomal instability and increase the probability of an HPV-infected cell evolving toward malignancy. Although the inactivation of pRb and p53 by E6 and E7, respectively, are crucial steps in HPV-mediated carcinogenesis, it is also known that additional E6 functions play an important role in promoting cellular transformation. HR HPV E6 oncoproteins associate with several members of the membrane-associated guanylate kinase (MAGUK) family, i.e., the human homologues of the Drosophila disc large protein (DLG), hDLG, Scribble, MUPP1, MAGI-1, MAGI-2, and MAGI-3 (22). MAGUK family members associate with various membrane and cytoplasmic proteins, acting as scaffolds for the formation of multiprotein complexes, and they are involved in the regulation of such cellular processes as cell-cell contact and cell polarity. They contain various protein/protein interaction domains, including PSD-90/Dlg/ZO-1 homology (PDZ) domains. The high-risk HPV E6 oncoproteins have a 4-amino-acid PDZ-binding motif at the C terminus that mediates the interaction with the MAGUK family members (22). The E6/MAGUK association leads to degradation of the cellular proteins, with consequent loss of cell-cell contact and cell polarity. Deletion of the PDZ motif abolishes HPV16 E6's ability to associate with hDLG without influencing its interaction with p53. However, loss of the PDZ-binding domain results in a strong reduction of the HPV16 E6 transforming properties in in vitro and in vivo experimental models (14, 24). In addition, the PDZ-binding motif is not present in E6 proteins from the low-risk (LR) HPV types, further suggesting its importance in HPV-induced carcinogenesis.
In addition to the members of the MAGUK family, other proteins involved in crucial cellular events contain PDZ domains. Na+/H+ exchanger regulatory factor 1 (NHERF-1) is a multidomain scaffolding protein, which has two PDZ domains in the N-terminal region and which regulates the trafficking and signaling of several G protein-coupled receptors (GPCRs) (4). The C-terminal region (CT; residues 242 to 358) also has properties that regulate the binding activity of the second PDZ domain and also includes a motif of approximately 14 amino acids that mediates the association with the actin-binding protein ezrin. Interactions between NHERF-1 and ezrin contribute to several cellular signaling events, including intracellular trafficking and assembly of protein complexes involving receptors and ion channels (2, 9). In addition, NHERF-1 complexes regulate cell shape and migration and are implicated in cancer development via the interaction with platelet-derived growth factor receptor (PDGFR), merlin (NF-2), β-catenin, and PTEN (4, 10).
The key role played by NHERF-1 in carcinogenesis and the fact that only the high-risk HPV E6 proteins possess PDZ-binding motifs suggested that NHERF-1 might be a PDZ domain-containing target of HR HPV E6 proteins, a possibility that had not previously been investigated. We show here that NHERF-1 is, indeed, a target of HPV16 E6. Furthermore, our data suggest that HPV16 E7 may cooperate with E6 in promoting the destabilization of NHERF-1 by inducing the accumulation of a phosphorylated form of NHERF-1 that is preferentially targeted by E6.
MATERIALS AND METHODS
Plasmids.
NHERF-1 wild-type or mutant genes, as well as E6 and/or E7 wild-type or mutant genes from different HPV types, were cloned in the following vectors: pGEX2T for the production of bacterial recombinant fusion proteins and pcDNA-3 (Invitrogen) and the retroviral vector pBabe (11) or pLXSN (Clontech) for expression in eukaryotic cells. Wild-type or mutant NHERF-1 from genes cloned in pcDNA-3 was fused at the N terminus with a histidine epitope tag.
Cell procedures.
NIH 3T3 rodent fibroblasts, human embryonic kidney (HEK293) cells, and Phoenix cells were cultured in fetal calf serum (FCS)-supplemented Dulbecco modified Eagle medium (DMEM; Gibco) using standard culture conditions. Primary human keratinocytes were isolated from neonatal foreskin and grown together with NIH 3T3 feeder cells in FAD medium (3 parts Ham's F12:1 part DMEM, 5% fetal calf serum, insulin [5 μg/ml], epidermal growth factor [EGF; 10 ng/ml], cholera toxin [8.4 ng/ml], adenine [24 μg/ml], hydrocortisone [0.4 μg/ml]). Feeder cells were prepared by irradiating NIH 3T3 cells (137Cs; 80 Gy). High-titer retroviral supernatants (>5 × 106 IU/ml) were generated by transient transfection into Phoenix cells and used to infect primary human keratinocytes as described previously (3). Transient-transfection experiments with HEK293 cells or human keratinocytes were performed using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche) transfection reagents according to the manufacturers' protocols.
HPV16 E6/E7, and HPV18 E6/E7 gene silencing in HPV16 E6- and E7-expressing human keratinocytes or cervical cancer-derived cell lines (SiHa and HeLa) was achieved by transient transfection using the small interfering RNAs (siRNAs) si16E6/E7 (5′ UUAAAUGACAGCUCAGAGG 3′) and si18E6/E7 (5′ CAUUUACCAGCCCGACGAG 3′), which were custom made by Dharmacon. siRNA E6AP, the On Target Plus Smart Pool siRNA against UBE3A, was also provided by Dharmacon.
The proteasome pathway was inhibited using the inhibitor MG132 (CBZ-Leu-Leu-Leu-al) (50 μM) (Sigma) for 4, 6, and 8 h. Inhibition of the cyclin-dependent kinase 1/2 (CDK1/2) complexes was achieved using roscovitine (Sigma) at 1, 5, and 10 μM.
Immunofluorescence staining of monolayer cultured keratinocytes was performed on cells grown on cover slides, fixed with formaldehyde, and stained by an anti-NHERF-1 antibody (ERM-binding phosphoprotein; BD 611160; Transduction Laboratories) and secondary antibody Alexa Fluor 532–goat anti-mouse IgG (H+L). The slides were mounted using mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) (H-1200; Vectashield) and analyzed with an immunofluorescence Axioplan2 microscope from Zeiss (Leica).
RT-PCR.
Total cellular RNA was extracted from cells using the Absolutely RNA miniprep kit (Stratagene). Reverse transcriptase PCR (RT-PCR) analyses were carried out as described previously (1) using primers 5′ TCACCAATGGGGAGATACAG 3′ (forward) and 5′ GTCTTGGGAATTCAGCTCCT 3′ (reverse) for NHERF-1 and 5′ AAGGTGGTGAAGCAGGCGT 3′ (forward) and 5′ GAGGAGTGGGTGTCGCTGTT 3′ (reverse) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
In vitro degradation assay.
Proteins were transcribed and translated in vitro in rabbit reticulocyte lysate using the Promega TNT system according to the manufacturer's instructions. The HPV E6 proteins and NHERF-1 proteins were radiolabeled with [35S]cysteine. Degradation assays were performed as previously described (20).
GST pulldown assay.
Glutathione S-transferase (GST) fusion protein synthesis in Escherichia coli BL21 and protein purification were performed as described in the Pharmacia handbook. Approximately 0.6 mg of C33A cellular extracts was incubated with GST/E6 proteins immobilized on glutathione beads (2 μg). After 1 h at 4°C, the beads were washed five times in a buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, and 10 mM NaF. The beads were resuspended in SDS-PAGE sample buffer, and the eluted proteins were run on an SDS-polyacrylamide gel, followed by immunoblotting with anti-GST or anti-NHERF-1 antibody.
Immunoblotting.
Total protein extracts were prepared, and SDS-PAGE and immunoblotting were performed as described in reference 1. Antibodies for the following proteins were used: β-actin (C4; MP Biomedicals), α-actinin (Santa Cruz; SC15335), NHERF-1 (ERM-binding phosphoprotein; BD Transduction Laboratories; 611160), β-tubulin (Sigma; T5201), His tag (Cell Signaling; 2365), AKT (Cell Signaling; 9272), phospho-AKT Ser 473 (Cell Signaling; 9271), glutathione S-transferase (Cell Signaling; 2622), E6AP (Sigma; E6AP-330), and p53 (Dako; DO-7).
Immunohistochemistry.
Immunohistochemical staining was performed on sections of HPV16-positive cervical intraepithelial neoplasia grade III (CINIII). The cervical specimens were collected in the context of the Tata Memorial Centre Rural Cancer Project, Nargis Dutt Memorial Cancer Hospital, Barshi, Solapur, Maharashtra, India, and were kindly provided by B. M. Nene. Processing of the specimens was performed in accordance with the ethical guidelines for handling of human material. Staining was carried out on 3-μm sections and performed using an anti-NHERF-1 antibody (ERM-binding phosphoprotein; BD Transduction Laboratories; 611160). Immunohistochemical signal was revealed with the Vectastain EliteABC kit (Vector Laboratories; PK 6102) according to the manufacturer's protocol. Images (magnitudes of ×10 and ×40) were taken with a Nikon Eclipse E600 camera.
RESULTS
HPV16 E6 binds NHERF-1 and induces its degradation in vitro.
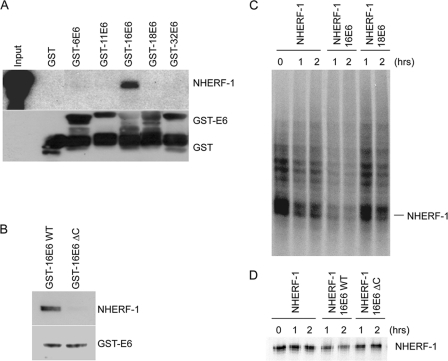
Since the HR HPV E6 proteins have a PDZ-binding motif, with which they are known to target certain PDZ-containing cellular proteins, it was possible that they might interact with the PDZ domain-containing NHERF-1 protein. To determine whether this might be the case, we performed a GST pulldown assay using GST-E6 fusion proteins and extracts from C33A cells. After a washing, the bound proteins were separated by SDS-PAGE and detected by immunoblotting with an anti-NHERF-1 antibody; the results are shown in Fig. 1A. NHERF-1 was found to be associated with GST-HPV16 E6, but not with E6 from the low-risk HPV types 6, 11, and 32 (Fig. 1A). Surprisingly, we did not observe any interaction between HPV18 E6 and NHERF-1. HPV16 and -18 E6s have almost identical PDZ-binding motifs (148ETQL151 in HPV16 E6 and 155ETQV158 in HPV18 E6). However, previous studies have shown that the two viral proteins have different affinities for different MAGUK proteins (21). In addition, the L151V mutation in HPV16 E6 conferred the HPV18 E6 binding properties for a specific set of PDZ proteins, while the opposite was observed for the V158L HPV18 E6 mutant (21). In this case, however, the V158L HPV18 E6 mutation did not increase the affinity of the viral oncoprotein for NHERF-1, suggesting that other HPV16 E6 domains, in addition to the PDZ-binding motif, may be involved in complex formation (data not shown), although deletion of the PDZ-binding motif in HPV16 E6 strongly affected its efficiency in binding NHERF-1 in a GST pulldown assay (Fig. 1B).
Fig. 1.
HPV16 E6 binds NHERF-1 and promotes its degradation in in vitro assays. (A and B) C33A cell extracts were incubated with the different GST fusion proteins indicated and loaded onto SDS-PAGE gel. The amounts of NHERF-1 associated with the different GST E6 proteins were determined by immunoblotting using a specific anti-NHERF-1 antibody. One-tenth of the total cellular extract (60 μg) used in the GST pulldown assay (input) was used for SDS-PAGE. (C and D) In vitro-translated NHERF-1 was incubated at 30°C for 0, 1, or 2 h either alone or with in vitro-translated HPV16 or HPV18 E6 (C) or wild-type (WT) or ΔC mutant HPV16 E6 (D). The remaining targeted proteins were visualized by SDS-PAGE and autoradiography.
Having shown that the NHERF-1 protein is bound by HPV16 E6, we were interested to know what might be the effect of the interaction upon the NHERF-1 protein. HR HPV E6 proteins have been shown to induce the degradation of a number of their cellular targets, such as p53 and the MAGUK proteins. Therefore, we performed an in vitro degradation assay, shown in Fig. 1C, in which in vitro-translated HPV16 or HPV18 E6 proteins were incubated with NHERF-1 for the indicated times. In agreement with the GST pulldown assay, HPV16 E6 induced rapid degradation of NHERF-1, while HPV18 E6 did not significantly influence NHERF-1 stability (Fig. 1C). In addition, an HPV16 ΔC E6 mutant lacking the PDZ-binding motif was not able to induce NHERF-1 degradation (Fig. 1D). Together, these data show that NHERF-1 is a target of HPV16 E6 in vitro.
HPV16 E6 and E7 cooperate in promoting NHERF-1 degradation.
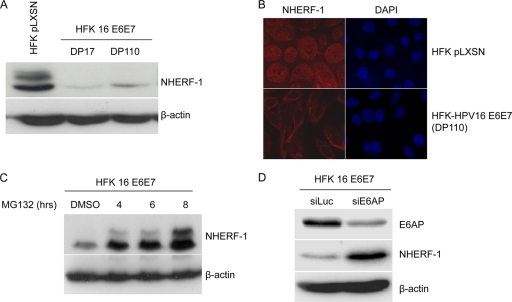
To confirm our in vitro data, we next wanted to determine whether HPV16 E6 could induce NHERF-1 degradation when expressed in primary human foreskin keratinocytes (HFKs). Cells were transduced with empty (pLXSN) or HPV16 E7/E6-expressing retrovirus, and NHERF-1 levels were determined by immunoblotting and by immunofluorescent staining. In control HFKs (HFK pLXSN), we consistently observed that the anti-NHERF-1 antibody recognized two proteins bands (Fig. 2A). Most likely, the slower-migrating band corresponds to the hyperphosphorylated form of NHERF-1, which is known to be targeted by several cellular kinases (4, 8, 25), including cyclin-dependent kinases (CDKs) (6). Coexpression of HPV16 E6 and E7 in HFKs strongly decreased the levels of both forms of NHERF-1 (Fig. 2A and B). NHERF-1 levels were affected immediately after the retroviral transduction in early doubling populations (DPs) before the immortalization of primary keratinocytes and continued to remain low in high DPs (Fig. 2A). Addition of the proteasome inhibitor MG132 to the culture medium led to the accumulation of NHERF-1 in HFKs expressing HPV16 E6 and E7 (Fig. 2C), indicating that, as previously observed for other E6 cellular targets, HPV16 E6 induces NHERF-1 degradation via the proteasome pathway. In addition, gene silencing of the ubiquitin ligase E6AP by siRNA resulted in an increase of NHERF-1 levels (Fig. 2D), indicating that this ubiquitin ligase is involved in E6-induced NHERF-1 degradation.
Fig. 2.
NHERF-1 protein levels are downregulated in HFKs expressing HPV16 E6 and E7. (A) Protein extracts of primary keratinocytes transduced with empty retrovirus vector (HFK pLXSN) or keratinocytes expressing HPV16 E6 and E7 genes (HFK 16 E6E7) were analyzed by immunoblotting using anti-NHERF-1 and anti-β-actin antibodies. β-Actin staining was included as a loading control. (B) Primary HFKs transduced with empty retrovirus (pLXSN) and HFKs expressing HPV16 E6 and E7 were seeded on coverslips. After immunofluorescent staining for NHERF-1 with specific antibodies, fluorescent signal was visualized using fluorescence microscopy. (C) HFKs expressing HPV16 E6 and E7 were treated with the protease inhibitor MG132 for the times indicated. NHERF-1 protein levels were determined by SDS-PAGE and immunoblotting. (D) HFKs expressing HPV16 E6 and E7 were transiently transfected with the indicated small interfering RNAs. After preparation of total protein extracts, NHERF-1, E6AP, and β-actin protein levels were determined by immunoblotting using specific antibodies.
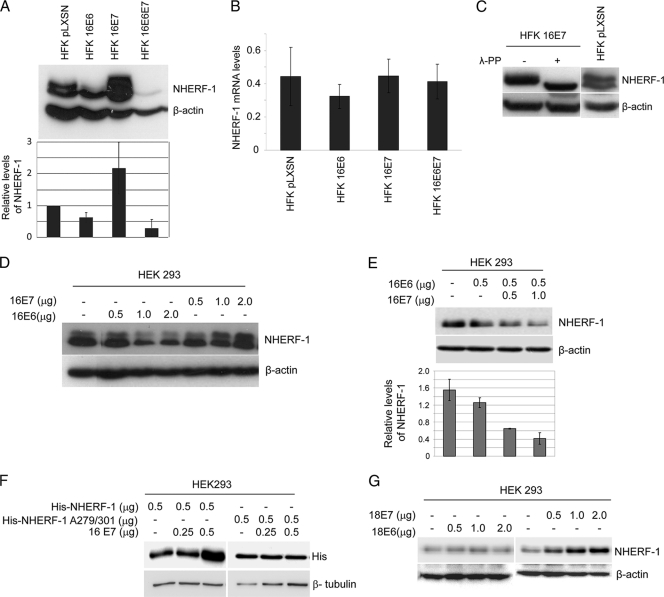
A comparison of NHERF-1 levels in HFKs expressing HPV16 E6 and/or E7 showed that the level of NHERF-1 was significantly reduced when both viral oncoproteins were present (Fig. 3A). In the absence of E7, E6 appeared to preferentially target the slower-migrating NHERF-1 band (Fig. 3A). In contrast, E7 alone induced a strong accumulation of NHERF-1 protein (Fig. 3A), which was not due to an increase in its mRNA levels (Fig. 3B). Immunoblotting using low levels of total protein extracts revealed that HPV16 E7-expressing HFKs mainly expressed the NHERF-1 form with slow migration in SDS-PAGE (Fig. 3C). Treatment of the total protein extract from HPV16 E7-expressing HFKs with lambda protein phosphatase (λ-PP) led to a change in NHERF-1 mobility, indicating that E7 promotes NHERF-1 phosphorylation (Fig. 3C).
Fig. 3.
HPV16 E6 and E7 cooperate in NHERF-1 degradation. (A) Protein extracts of primary keratinocytes transduced with empty retrovirus vector (HFK pLXSN) or recombinant retroviruses, as indicated, were analyzed by immunoblotting using anti-NHERF-1 and anti-β-actin antibodies (top). The intensities of the protein bands in three independent experiments were quantified (bottom). (B) Total RNA was also extracted from the indicated cells, and NHERF-1 or GAPDH mRNA levels were measured by quantitative RT-PCR. y axis numbers represent arbitrary units of NHERF-1 mRNA levels in indicated cells standardized to the GAPDH levels. The data are the means of three independent experiments. The differences between the NHERF-1 levels in the different cells are not statically significant. (C) Protein extracts from HFKs transduced with empty (pLXSN) or HPV16 E7 retrovirus were incubated in the presence and absence of λ-PP and analyzed by immunoblotting using anti-NHERF-1 and anti-β-actin antibodies. (D) HEK293 cells were transfected with increasing concentrations of pLXSN HPV16 E6 or E7 as indicated. After 24 h, protein extracts were analyzed by immunoblotting. (E) HEK293 cells were transfected with pLXSN expressing HPV16 E6 together with increasing concentrations of pLXSN expressing HPV16 E7 as indicated. After 24 h, protein extracts were analyzed by immunoblotting (top). The intensities of the bands were quantified in three independent experiments and are represented in the histogram (bottom). (F) HEK293 cells were transfected with pcDNA3 expressing His-tagged NHERF-1 (His-NHERF-1), wild type or A279/A301 mutant, together with increasing concentrations of pLSXN expressing HPV16 E7 as indicated. After 24 h, protein extracts were analyzed by immunoblotting. (G) HEK293 cells were transfected with increasing concentrations of pLXSN expressing HPV18 E6 or E7 as indicated. After 24 h, protein extracts were analyzed by immunoblotting.
To determine whether or not HPV16-mediated NHERF-1 degradation is a cell-specific event, we also determined the level of NHERF-1 in human embryonic kidney (HEK293) cells expressing the viral oncoproteins. As shown in Fig. 3D, HPV16 E6 decreases the endogenous levels of NHERF-1, while HPV16 E7 promotes the accumulation of the slow-migrating form of NHERF-1. In addition, increasing concentrations of E7 significantly enhanced E6 ability to induce NHERF-1 degradation (Fig. 3E).
It has been previously shown that NHERF-1 is phosphorylated by cdc2 (CDK1) complexes at serines 279 and 301 (6), and HPV16 E7 is known to have the ability to activate CDK complexes (5, 7, 23). The E7-mediated NHERF-1 accumulation was abolished by the mutation of serine 279 and serine 301 to alanine (Fig. 3F), indicating that E7 stimulates NHERF-1 phosphorylation.
As expected, NHERF-1 was not affected by HPV18 E6, while HPV18 E7 promoted its accumulation (Fig. 3G). The latter event is in agreement with the fact that HPV18 E7 shares with HPV16 E7 the ability to activate CDK complexes.
In summary, these data show that HPV16 E7 cooperates with E6 in NHERF-1 degradation by promoting its phosphorylation via CDK activation.
NHERF-1 phosphorylation by CDK stimulates its E6-mediated degradation.
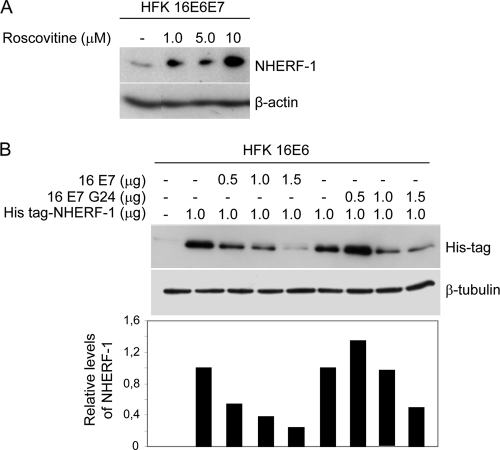
To corroborate our data showing the involvement of CDK activity in E6-mediated NHERF-1 degradation, we next determined whether the CDK inhibitor roscovitine could affect HPV16-mediated NHERF-1 degradation. As shown in Fig. 4A, increasing concentrations of roscovitine increased NHERF-1 levels in HPV16 E6- and E7-expressing HFKs. In agreement with these data, we observed that the HPV16 E7 mutant (C24G), which harbors a mutation in the pocket protein-binding motif and is not able to increase cyclin E and cyclin A transcription, was less efficient than the wild-type E7 in cooperating with E6 to promote NHERF-1 degradation in HFKs (Fig. 4B). These findings indicate that the ability of E7 to stimulate E6-induced degradation of NHERF-1 is mediated by its property of activating CDK complexes.
Fig. 4.
CDK-mediated NHERF-1 phosphorylation is required for its degradation induced by HPV16 E6. (A) HFKs expressing HPV16 E6 and E7 were treated with the CDK inhibitor roscovitine at the indicated concentrations, and NHERF-1 protein levels were determined by SDS-PAGE and immunoblotting. (B) HFKs expressing HPV16 E6 were transfected with pLXSN expressing His-NHERF-1, together with increasing concentrations of pLSXN expressing HPV16 E7, wild type or the glycine 24 mutant (G24), as indicated. After 24 h, protein extracts were analyzed by immunoblotting (top), and the intensities of the protein bands were quantified (bottom).
NHERF-1 inhibits AKT activation induced by HPV16.
It has been previously shown that NHERF-1 is able to mediate the interaction of platelet-derived growth factor (PDGF) receptor with tumor suppressor PTEN, which in turn attenuates the activation of phosphatidylinositol-3′-OH kinase (PI3K) signaling (19). PI3K is known to play an important role in processes involved in carcinogenesis by activating the serine/threonine protein kinase AKT (26).
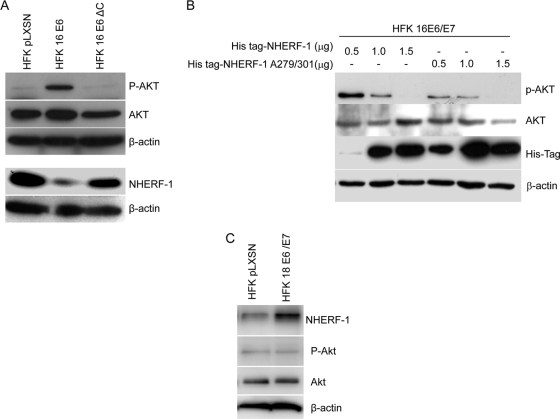
HPV16 E6 expression in HFKs resulted in the accumulation of the phosphorylated and active form of AKT (Fig. 5A). In contrast, the HPV16 E6 deletion mutant that lacks the PDZ-binding motif in the C terminus was not able to activate AKT HFKs (Fig. 5A), suggesting that AKT activation in E6-expressing cells may be due to NHERF-1 degradation. In accordance with this hypothesis, when we increased the expression levels of NHERF-1, the AKT activation in HPV16 E6/E7-expressing HFKs was proportionally decreased. This phenomenon was even more evident when we overexpressed in the same cells the NHERF-1 A279/301 mutant, which is not efficiently targeted by HPV16 E6 and E7 (Fig. 5B). In agreement with the inability of HPV18 E6 to promote NHERF-1 degradation, AKT was not found to be phosphorylated in human keratinocytes expressing HPV18 E6 and E7 (Fig. 5C). Thus, these studies showed that one of the consequences of E6-mediated NHERF-1 degradation is the activation of the PI3K/AKT signaling pathway.
Fig. 5.
HPV16 E6-induced NHERF-1 degradation leads to activation of AKT signaling. (A) Protein extracts of primary keratinocytes transduced with empty retrovirus vector (pLXSN), or HPV16 E6/E7-expressing retrovirus were analyzed by immunoblotting using anti-NHERF-1, anti-AKT, anti-phospho-AKT, and anti-β-actin antibodies. (B) HFKs expressing HPV16 E6 and E7 were transfected with increasing concentrations of pcDNA3 expressing His-NHERF-1, wild type or A279/A301 mutant. After 24 h, protein extracts were analyzed by immunoblotting. (C) Protein extracts of primary keratinocytes transduced with empty retrovirus vector (pLXSN) or HPV18 E6/E7-expressing retrovirus were analyzed by immunoblotting using anti-NHERF-1, anti-AKT, anti-phospho-AKT, and anti-β-actin antibodies.
NHERF-1 is downregulated in cervical cancer-derived cell lines and HPV16-positive cervical lesions.
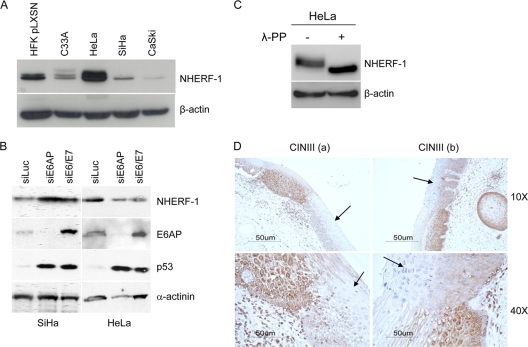
To corroborate our data obtained in HFKs transduced with recombinant retroviruses, we determined the NHERF-1 expression levels in cervical cancer-derived cell lines. As shown in Fig. 6A, NHERF-1 levels were very low in SiHa and CaSki cells. CaSki cells have a higher copy number of HPV16 DNA integrated in the host genome and express higher levels of E6 and E7 genes in comparison to SiHa cells (17). Thus, the slight difference in NHERF-1 levels between the two cell lines might be explained by the different intracellular concentrations of E6 and E7 proteins (Fig. 6A). Inhibition of HPV16 E6 and E7 expression by siRNA resulted in an increase of NHERF-1 levels in SiHa cells, while HPV18 E6 and E7 downregulation did not induce any changes in the levels of NHERF-1 (Fig. 6B). In addition, siRNA gene silencing of E6AP in SiHa, but not in HeLa cells, significantly alleviated the E6-mediated NHERF-1 degradation (Fig. 6B), indicating that this ubiquitin ligase is involved in the event.
Fig. 6.
NHERF-1 levels are downregulated in HPV16-positive cervical cancer cell lines and in cervical lesions. (A) Protein extracts of different cell lines as indicated were prepared and analyzed by immunoblotting using anti-NHERF-1 and anti-β-actin antibodies. (B) SiHa and HeLa cells were transiently transfected with the indicated small interfering RNAs. After preparation of total protein extracts, NHERF-1, E6AP, p53, and α-actinin protein levels were determined by immunoblotting using specific antibodies. (C) Protein extracts from HeLa cells were incubated in the presence and absence of λ-PP and analyzed by immunoblotting using anti-NHERF-1 and anti-β-actin antibodies. (D) Sections of HPV16-positive cervical intraepithelial neoplasia grade III (CINIII) from two different women (a and b) were stained with a specific anti-NHERF-1 antibody. Arrows indicate the dysplastic regions. The images are shown at two magnifications (×10 and ×40).
In agreement with in vitro data which indicated that NHERF-1 is not targeted for degradation by HPV18 E6, high levels of NHERF-1 were detected in HPV18-positive HeLa cells (Fig. 6A), similar to what we observed in HPV18 E6/E7-expressing HFKs (Fig. 5C). Lambda phosphatase treatment of HeLa protein extract showed that NHERF-1 was heavily phosphorylated in these cells (Fig. 6C). Finally, the HPV-negative cervical cancer cell line C33A expressed both forms of NHERF-1, but at lower levels than in HFKs (Fig. 6A).
We next examined the expression level of NHERF-1 protein in cervical premalignant lesions, i.e., cervical intraepithelial neoplasia grade III (CINIII) (n = 4). In all CINIII lesions NHERF-1 was weakly expressed. NHERF-1 was downregulated exclusively in these lesions, but not in the surrounding fully differentiated tissue. Figure 6D shows two representative stained sections. Together, these data provide evidence that HPV16 can also target NHERF-1 in the context of a natural infection.
DISCUSSION
Several independent studies have shown that E6 protein from the HR HPV types can interact with and induce the degradation of certain MAGUK family members, in a PDZ-binding-dependent manner, thus affecting the regulation of cell polarity and cell proliferation control in HR HPV-infected cells (22). The E6 protein from the LR HPV types lacks a PDZ-binding motif, indicating that the E6 interaction with the PDZ proteins may play an important role in HPV-mediated carcinogenesis. These conclusions are further supported by the fact that the deletion of the PDZ-binding domain strongly affects the HPV16 E6's transforming properties (14, 24). Here, we describe a new interaction of HPV16 E6 with a PDZ cellular protein, NHERF-1, which is involved in a number of cellular processes, including cellular signaling and transformation (4). Similar to the previously described interactions of E6 with several PDZ proteins, the HPV16 E6/NHERF-1 association resulted in the degradation of the cellular protein via the proteasome pathway. This event appears to be mediated by the ubiquitin ligase E6AP, since E6AP downregulation in HPV16 E6/E7 keratinocytes, as well as in the cervical cancer cell line SiHa, resulted in an increase of NHERF-1 levels. In addition, we present several lines of evidence suggesting that HPV16 E7 cooperates with E6 in targeting NHERF-1. In fact, HPV16 E7, through inactivation of the pocket proteins and consequent stimulation of the CDKs, promotes the accumulation of phosphorylated NHERF-1, which appears to be preferentially degraded by HPV16 E6. Accordingly, an HPV16 E7 mutant that is unable to bind the pocket proteins and increase the CDK1/2 activity did not synergize with E6 in promoting NHERF-1 degradation. Similarly, inhibition of CDK1/2 activity or mutations of the specific serines of NHERF-1 that are phosphorylated by CDK1/2 significantly affected the E6-mediated degradation of NHERF-1.
As far as we are aware, this is the first example of E6 and E7 being formally shown to cooperate in targeting the same cellular protein, although previous studies have suggested that HR HPV E6s also preferentially target a phosphorylated form of hDLG (13). In fact, the CDK1/2 phosphorylated form of hDLG is completely absent in the cervical cancer cell lines CaSki and HeLa, while it is easily detected in the HPV-negative immortalized epithelial HaCaT cells (13). Thus, as shown for NHERF-1, it is likely that the two oncoproteins from HR HPV types cooperate in the inactivation of hDLG and possibly other PDZ proteins.
The ability of HPV16 E6 to target NHERF-1 requires the presence of an intact PDZ motif. However, this is a highly specific form of PDZ recognition since HPV18 E6, which has a slightly different PDZ-binding motif, it is not able to interact with NHERF-1 or induce its degradation. Indeed, it has previously been shown that additional amino acid residues surrounding the canonical PDZ recognition motif in E6 are differently involved in PDZ domain recognition, with very minor changes having a profound effect on the interaction (21, 27). It is also worth noting that, although HPV16 E6 efficiently targets NHERF-1 in cellular experimental models, the strength of interaction between HPV16 E6 and NHERF-1 in vitro is actually quite weak and certainly not as strong as that for some other PDZ domain-containing targets such as MAGI-1 (M. Thomas, personal observations). One possible explanation for this phenomenon is that NHERF-1/E6 association may be greatly increased by posttranslational modifications of the cellular protein, which cannot occur in an in vitro GST pulldown assay. This possibility is currently explored in ongoing studies. It is interesting to speculate why HPV16, but not HPV18, might need to inactivate NHERF-1 during its life cycle, although it is also possible that HPV18 may target the NHERF-1-associated pathway through another component, as has been shown with the interactions of HR HPV E6s and the hDlg/hScrib complex (21).
In agreement with the data observed in artificial experimental models, NHERF-1 levels are very low also in the HPV16-positive cervical cancer-derived cell lines SiHa and CaSki, as well as in premalignant cervical lesions. In SiHa cells, NHERF-1 degradation induced by HPV16 E6 appears to be mediated by E6AP, since inhibition of its expression significantly increased NHERF-1 levels, while no changes in NHERF-1 expression were observed in the HPV18-positive cervical cancer cell line HeLa upon E6AP gene silencing. In HeLa cells, we detected high levels of NHERF-1, similar to those in primary keratinocytes expressing only HPV16 E7. In agreement with previous data (6), our data show that NHERF-1 is highly phosphorylated in HeLa cells. The biological significance of NHERF-1 phosphorylation mediated by CDK1/2 is still unclear. It has been reported that NHERF-1 phosphorylation by CDK1/2 affects its ability to oligomerize and thus possibly to interact with specific cellular proteins (6); however, whether these events alter the tumor suppressor functions of NHERF-1 remains to be investigated.
A previous study found that HPV16 E6 induces AKT activation (18). Our data provide evidence that this event is a consequence of E6-induced NHERF-1 degradation and activation of the PI3K signaling pathway. In agreement with this conclusion, it has been shown that knocking down the NHERF-1 gene in a mouse model led to AKT activation (15). Many studies have demonstrated the importance of this pathway in several key processes involved in carcinogenesis, including cellular proliferation and apoptosis. Interestingly, previous studies have shown that HPV16 has an additional mechanism of AKT activation. In fact, HPV16 E7 associates with and inhibits protein phosphatase 2A (PP2A), which is responsible for dephosphorylating and inactivating AKT. In HPV16 E7-expressing cells, due to the PP2A inhibition, AKT is maintained in its phosphorylated and activated form (16).
Thus, HPV16 has developed at least two independent mechanisms to activate the AKT-mediated signaling pathway: on one side E6 and E7 cooperate to promote NHERF-1 degradation and AKT phosphorylation; on the other side, E7 prevents AKT dephosphorylation by inhibiting PP2A. These apparently redundant mechanisms of AKT activation underline the importance of this pathway in HPV-mediated carcinogenesis.
ACKNOWLEDGMENTS
We are grateful to all the members of our laboratories for their cooperation, B. M. Nene for providing the cervical specimens, C. Carreira Romao for the immunohistochemical staining, and Valeria Casavola for her scientific advice.
The study was in part supported by grants from La Ligue Contre le Cancer (Comité du Rhône) to M.T. and B.S.S., the Association pour la Recherche sur le Cancer to M.T., Association for International Cancer Research to M.T. and L.B., and the Associazione Italiana per la Ricerca sul Cancro to L.B. R.R. was supported by a fellowship of the University of Bari, Italy (D.R. no. 7889 03.06.2008-L.392 30.11.89).
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Accardi R., et al. 2006. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 7:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bretscher A., Edwards K., Fehon R. G. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3:586–599 [DOI] [PubMed] [Google Scholar]
- 3. Caldeira S., et al. 2003. The E6 and E7 proteins of cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 77:2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Georgescu M. M., Morales F. C., Molina J. R., Hayashi Y. 2008. Roles of NHERF1/EBP50 in cancer. Curr. Mol. Med. 8:459–468 [DOI] [PubMed] [Google Scholar]
- 5. Ghittoni R., et al. 2010. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40:1–13 [DOI] [PubMed] [Google Scholar]
- 6. He J., Lau A. G., Yaffe M. B., Hall R. A. 2001. Phosphorylation and cell cycle-dependent regulation of Na+/H+ exchanger regulatory factor-1 by Cdc2 kinase. J. Biol. Chem. 276:41559–41565 [DOI] [PubMed] [Google Scholar]
- 7. He W., Staples D., Smith C., Fisher C. 2003. Direct activation of cyclin-dependent kinase 2 by human papillomavirus E7. J. Virol. 77:10566–10574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J., et al. 2007. Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J. Biol. Chem. 282:27086–27099 [DOI] [PubMed] [Google Scholar]
- 9. Mahon M. J., Donowitz M., Yun C. C., Segre G. V. 2002. Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature 417:858–861 [DOI] [PubMed] [Google Scholar]
- 10. Morales F. C., et al. 2007. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol. 27:2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgenstern J. P., Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munoz N., et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 13. Narayan N., Subbaiah V. K., Banks L. 2009. The high-risk HPV E6 oncoprotein preferentially targets phosphorylated nuclear forms of hDlg. Virology 387:1–4 [DOI] [PubMed] [Google Scholar]
- 14. Nguyen M. L., Nguyen M. M., Lee D., Griep A. E., Lambert P. F. 2003. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6's induction of epithelial hyperplasia in vivo. J. Virol. 77:6957–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Y., Weinman E. J., Dai J. L. 2008. Na+/H+ exchanger regulatory factor 1 inhibits platelet-derived growth factor signaling in breast cancer cells. Breast Cancer Res. 10:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pim D., Massimi P., Dilworth S. M., Banks L. 2005. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830–7838 [DOI] [PubMed] [Google Scholar]
- 17. Rajeevan M. S., et al. 2005. Epidemiologic and viral factors associated with cervical neoplasia in HPV-16-positive women. Int. J. Cancer 115:114–120 [DOI] [PubMed] [Google Scholar]
- 18. Spangle J. M., Munger K. 2010. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol. 84:9398–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi Y., Morales F. C., Kreimann E. L., Georgescu M. M. 2006. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 25:910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas M., Glaunsinger B., Pim D., Javier R., Banks L. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas M., Massimi P., Navarro C., Borg J. P., Banks L. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222–6230 [DOI] [PubMed] [Google Scholar]
- 22. Thomas M., et al. 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27:7018–7030 [DOI] [PubMed] [Google Scholar]
- 23. Tommasino M., et al. 1993. HPV16 E7 protein associates with the protein kinase p33CDK2 and cyclin A. Oncogene 8:195–202 [PubMed] [Google Scholar]
- 24. Watson R. A., Thomas M., Banks L., Roberts S. 2003. Activity of the human papillomavirus E6 PDZ-binding motif correlates with an enhanced morphological transformation of immortalized human keratinocytes. J. Cell Sci. 116:4925–4934 [DOI] [PubMed] [Google Scholar]
- 25. Weinman E. J., et al. 2010. Cooperativity between the phosphorylation of Thr95 and Ser77 of NHERF-1 in the hormonal regulation of renal phosphate transport. J. Biol. Chem. 285:25134–25138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang S., Yu D. 2010. PI(3)king apart PTEN′s role in cancer. Clin. Cancer Res. 16:4325–4330 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y., et al. 2007. Structures of a human papillomavirus (HPV) E6 polypeptide bound to MAGUK proteins: mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J. Virol. 81:3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350 [DOI] [PubMed] [Google Scholar]