Abstract
Avian influenza viruses of the H9N2 subtype have seriously affected the poultry industry of the Far and Middle East since the mid-1990s and are considered one of the most likely candidates to cause a new influenza pandemic in humans. To understand the genesis and epidemiology of these viruses, we investigated the spatial and evolutionary dynamics of complete genome sequences of H9N2 viruses circulating in nine Middle Eastern and Central Asian countries from 1998 to 2010. We identified four distinct and cocirculating groups (A, B, C, and D), each of which has undergone widespread inter- and intrasubtype reassortments, leading to the generation of viruses with unknown biological properties. Our analysis also suggested that eastern Asia served as the major source for H9N2 gene segments in the Middle East and Central Asia and that in this geographic region within-country evolution played a more important role in shaping viral genetic diversity than migration between countries. The genetic variability identified among the H9N2 viruses was associated with specific amino acid substitutions that are believed to result in increased transmissibility in mammals, as well as resistance to antiviral drugs. Our study highlights the need to constantly monitor the evolution of H9N2 viruses in poultry to better understand the potential risk to human health posed by these viruses.
INTRODUCTION
Avian influenza viruses of the H9N2 subtype are endemic in poultry populations across Asia and the Middle East. These viruses fall into a number of genetically defined lineages, with the majority of those circulating in Asia belonging to two lineages—G1 and Y280—represented by the prototype viruses A/quail/Hong Kong/G1/97 and A/duck/Hong Kong/Y280/97, respectively, both of which became established in domestic poultry during the mid-1990s (16, 32, 50). Although H9N2 viruses are classified as low-pathogenicity avian influenza viruses, they sometimes cause significant disease in poultry, occasionally accompanied by high mortality and a marked reduction in egg production (4). To reduce the economic impact of the H9N2 infection in poultry, vaccination programs are commonly undertaken in several Asian countries (2, 4, 23).
Since 1997, H9N2 viruses have been reported in multiple avian species throughout Asia, the Middle East, Europe, and Africa (1, 2, 6, 14, 15, 16, 17, 18, 22, 23, 28, 44, 50) and have occasionally been transmitted from poultry to mammalian species, including humans and pigs (5, 24, 32). Further evidence of an expanded mammalian host range includes efficient replication of H9N2 in experimental mice without adaptation (9). Numerous recent H9N2 isolates contain the amino acid leucine at position 226 (L226) in their hemagglutinin (HA) receptor-binding site (RBS), which exhibits preferential binding to human-like α2-6-linked sialic acid (SAα2-6) receptors and is regarded as one of the key elements in the successful infection of humans (14, 18, 24, 45, 50). Recent research using ferrets demonstrated that L226-containing H9N2 viruses were more likely to be transmitted, although no aerosol transmission was observed (44). However, a chimeric H9N2 influenza virus carrying the surface glycoproteins of an avian H9N2 virus and the six internal genes of a human H3N2 virus was found to possess increased transmissibility via respiratory droplets (42).
It is well established that reassortment between isolates from different host species can generate viruses with pandemic potential. The detection of avian influenza H9N2 viruses in domestic pigs and humans clearly creates opportunities for such reassortment events (5, 49). In addition, the extensive cocirculation of H9N2 viruses with other avian influenza viruses is likely to generate appropriate conditions for the development of reassortant viruses. Indeed, intersubtype reassortment between cocirculating H9N2 virus and highly pathogenic H5N1 or H7N3 virus has been detected in China (16, 51) and Pakistan (6, 18). The circulation of H9N2 viruses throughout Eurasia, along with their ability to infect mammals and humans and the potential for future reassortment, clearly raises concern about their pandemic potential.
While the genetic and antigenic evolution of H9N2 viruses in China is well documented, information on the genetic properties of those H9N2 viruses circulating in Central Asia and the Middle East is limited to studies of a few strains collected in a single country (1, 15, 17, 18, 26, 33, 41). To explore the genetic characteristics, as well as the spatial and evolutionary dynamics, of the H9N2 lineages that cocirculate in Central Asia and the Middle East, we conducted a phylogenetic analysis of whole-genome sequences from H9N2 viruses sampled between 1998 and 2010 from nine Asian countries: Iran, Iraq, Israel, Jordan, the United Arab Emirates (UAE), Pakistan, Afghanistan, Saudi Arabia, and Qatar. The central aim of our study was to reveal the extent of inter- and intrasubtypic reassortment, as well as the frequency and pattern of viral gene flow, in order to identify those geographical regions that are likely to serve as key sources of H9N2 viruses. Additionally, we examined the emergence of amino acid mutations and their significance in terms of drug resistance, adaptation to different host species, and pandemic potential.
MATERIALS AND METHODS
Viruses included in this study.
We sequenced the complete genomes of 29 avian influenza H9N2 viruses isolated from poultry in Afghanistan, Jordan, Saudi Arabia, Iraq-Kurdistan, Iran, and the UAE from 2004 to 2010, as well as the partial genomic sequences of five additional H9N2 viruses from Iraq (HA and NA), Qatar (PB2, HA, NP, NA, M, and NS), Jordan (PB2, PB1, HA, NP, NA, M, and NS), the UAE (PB2, PB1, PA, HA, NA, M, and NS), and Saudi Arabia (PA, HA, NP, NA, M, and NS). These sequences were analyzed together with all publicly available sequence data for H9N2 viruses isolated between 1998 and 2008 from Central Asian and Middle Eastern countries, including Israel, Iran, Saudi Arabia, the UAE, Pakistan, and Jordan. Overall, 107, 109, 111, 178, 109, 138, 103, and 119 full-length H9N2 sequences were analyzed for the PB2, PB1, PA, HA, NP, NA, M, and NS segments, respectively.
Nucleotide sequencing.
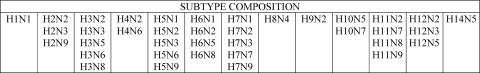
For the isolates sequenced in this study, viral RNA was extracted from the infected allantoic fluid of specific-pathogen-free fowls' eggs using the Nucleospin RNA II kit (Macherey-Nagel, Duren, Germany) and reverse transcribed with the SuperScript III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA). PCR amplifications were performed by using specific primers (sequences are available on request). The complete coding sequences were generated using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were cleaned up using the PERFORMA DTR Ultra 96-Well kit (Edge BioSystems, Gaithersburg, MD) and sequenced in a 16-capillary ABI PRISM 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). Sequence data were assembled and edited with SeqScape software v2.5 (Applied Biosystems). Sequences from all eight gene segments were aligned and compared with all publicly available H9N2 sequences of viruses from Central Asian and Middle Eastern countries, and with Eurasian H9N2 sequences representative of distinct H9N2 lineages. Sequences of the six internal genes were also compared with representative sequences of viruses of different subtypes available in GenBank (Fig. 1).
Fig. 1.
Influenza virus subtypes included in the phylogenetic analysis of internal gene segments.
Phylogenetic analysis.
For each of the eight genome segments, maximum-likelihood (ML) trees were estimated using the best-fit general time-reversible (GTR) + I + Γ4 model of nucleotide substitution using PAUP* (47). Parameter values for the GTR substitution matrix, base composition, gamma distribution of the rate variation among sites (with four rate categories, Γ4), and proportion of invariant sites (I) were estimated directly from the data using MODELTEST (37). A bootstrap resampling process (1,000 replications) using the neighbor-joining (NJ) method was used to assess the robustness of individual nodes of the phylogeny, incorporating the ML substitution model defined above.
Substitution rates and times to common ancestry.
Rates of nucleotide substitution per site per year and the time to the most recent common ancestor (tMRCA) of the sampled data were estimated using the BEAST program, version 1.5.3 (11), which employs a Bayesian Markov chain Monte Carlo (MCMC) approach. For each analysis, the Bayesian skyline coalescent tree prior was used, as this is clearly the best descriptor of the complex population dynamics of influenza A virus (12). Two molecular clock models—strict (constant) and uncorrelated lognormal relaxed clock—were compared by analyzing values of the coefficient of variation (CoV) in Tracer (11), such that CoV values of >0 are evidence of non-clock-like evolutionary behavior. We also compared two nucleotide substitution models: a GTR + Γ4 model similar to that described above and a codon-based SRD06 model. In all cases, the SRD06 model performed better than the GTR + Γ4 model, as previously demonstrated for a large set of RNA viruses (39). In all cases, uncertainty in the data is reflected in the 95% highest probability density (HPD) values for each parameter estimate, and in each case, chain lengths were run for sufficient time to achieve coverage as assessed using the Tracer v1.5 program (11). Finally, maximum clade credibility (MCC) phylogenetic trees were estimated from the posterior distribution of trees generated by BEAST using the program TreeAnnotator v1.5.3 (11) after the removal of an appropriate burn in (10% of the samples). The MCC trees were visualized using the program FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/), which allowed us to estimate the tMRCA of each individual node of the trees.
Phylogeography of H9N2 in Central Asia and the Middle East.
To determine the extent and pattern of geographical structure in the H9N2 viruses sampled here, we first grouped the sequences of viruses isolated in Central Asia and the Middle East into eight geographic regions, each representing one country or two adjacent countries. These geographic regions were as follows: Saudi Arabia/Qatar (the latter represented by a single sequence), Jordan, Israel, Iran, Iraq, Afghanistan, Pakistan, the UAE, eastern Asia, and Europe. To assess the overall degree of geographical structure among H9N2 viruses sampled from the Central Asian and Middle Eastern regions analyzed here, we used the BaTS program (31) to estimate values of the association index (AI) and parsimony score (PS) statistics of phylogeny-trait association with the traits (the geographical regions) defined above. Importantly, this method uses the posterior distribution of trees obtained from the BEAST analysis described above and thereby accounts for phylogenetic uncertainty in the data. The BaTS program also allowed us to assess the level of clustering in individual locations using the monophyletic clade (MC) size statistic (31). In all cases, 1,000 random permutations of tip locations were undertaken to create a null distribution for each statistic.
Analysis of selection pressures.
Gene- and site-specific selection pressures for all segments of Central Asian and Middle Eastern H9N2 viruses were measured as the ratio of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions per site. In all cases, dN/dS ratios were estimated using the single-likelihood ancestor counting and fixed-effects likelihood ML methods available at the Datamonkey online version of the Hy-Phy package (36). All analyses utilized the GTR model of nucleotide substitution and employed NJ phylogenetic trees.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available from GISAID under accession numbers EPI301452 to EPI301655 and EPI223068 to EPI223123.
RESULTS
Phylogenetic relationships among H9N2 viruses in Central Asia and the Middle East from 1998 to 2010: multiple introductions and extensive reassortment events.
To investigate the genetic diversity of avian influenza H9N2 viruses in Central Asia and the Middle East, we analyzed the eight genome segments of all of the H9N2 viruses from this geographic region that are available in GenBank (the number of available sequences ranged from 71 for the M gene to 145 for the HA gene) combined with 34 genome sequences newly generated in this study. Phylogenetic relationships between these viruses and representative sequences from European and Asian countries, along with the G1 and Y280 established Eurasian H9N2 lineages, were determined. The topologies of all of the eight ML phylogenetic trees revealed substantial genetic diversity, exemplified by multiple cocirculating H9N2 lineages and frequent reassortment events, which could be schematically identified as either intersubtype (between different HA subtypes) or intrasubtype (between different H9 genetic clusters) exchanges.
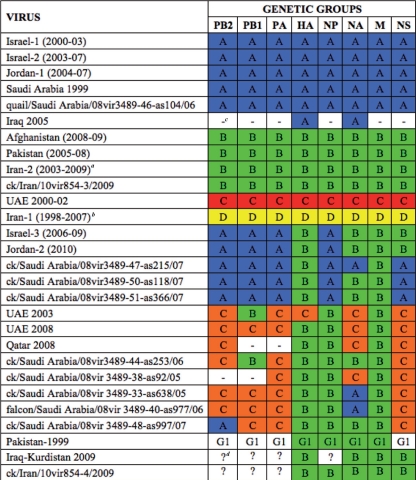
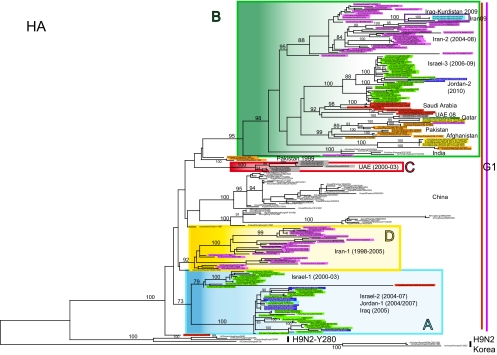
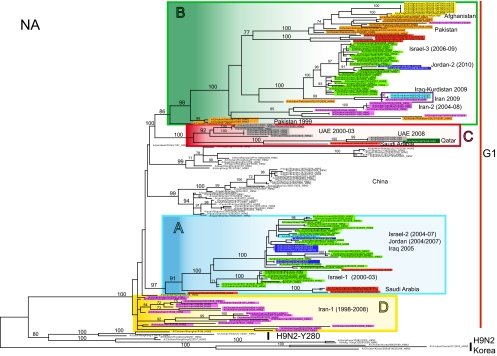
Analysis of the HA phylogeny identifies four monophyletic groups of H9N2 viruses in Central Asia and the Middle East—designated A, B, C, and D (Table 1)—each of which is defined by high bootstrap values (>70%) and average percent pairwise nucleotide distances within and between groups of <5% and >5%, respectively (Fig. 2). The phylogenetic groups identified in the HA tree (A to D) are maintained in most of the remaining gene segments (Fig. 3; see Fig. S1 to S6 in the supplemental material). Importantly, the topologies of the eight segment phylogenies show that each group (A to D) is characterized by a unique genomic constellation, indicative of extensive mixing of gene segments from multiple avian influenza virus subtypes.
Table 1.
Genome constellations of the H9N2 viruses collected in Central Asia and the Middle East
Cluster represented in all phylogenies by ck/Iran/854v10-5/2008.
Cluster represented in all phylogenies by ck/Iran/11T/99 and ck/Iran/854v10-1/02.
—, No sequences available.
?, Genetic group different from A, B, C, or D.
Fig. 2.
ML phylogenetic tree for the HA gene segment of H9N2 avian influenza viruses from the Middle East and Asia. Sequences of H9N2 viruses analyzed in this study are highlighted with different colors, according to the countries of origin, as follows: purple for Iran, green for Israel, red for Saudi Arabia, gray for the UAE, yellow for Afghanistan, orange for Pakistan, blue for Jordan, and light blue for Iraq. Genetic groups are colored as follows: group A is blue, group B is green, group C is red, and group D is yellow. The numbers at the nodes represent bootstrap values.
Fig. 3.
ML phylogenetic tree for the NA gene segment of H9N2 avian influenza viruses from the Middle East and Asia. The color scheme is the same as that used in Fig. 2.
Of particular note, our phylogenetic analysis suggests that the HA, NA, and M genome segments of the four genetic groups (A to D), as well as the NP gene of group B, derive from the G1 lineage, while the remaining gene segments have been replaced through intersubtype reassortment and do not show any relationship with the previously identified G1 and Y280 Eurasian H9N2 lineages, generating many different genotypes (Fig. 2 and 3; see Fig. S1 to S6 in the supplemental material). As recently described by Iqbal et al., some H9N2 strains collected in Pakistan from 2005 to 2006 have acquired the NS gene segment from highly pathogenic avian influenza (HPAI) virus H5N1 (clade 2.2) and Pakistani HPAI H7N3 virus (18). However, in most cases, the origin of the H9N2 segments cannot be established.
Viruses possessing the gene segment constellation A or B appear to have circulated extensively in Central Asia and the Middle East (Table 1), while groups C and D were detected only in the UAE between 2000 and 2002 and in Iran from 1998 to 2007, respectively. Group A contains viruses isolated in Israel (Israel-1 and Israel-2 group in the phylogenetic trees and in Table 1) and Jordan (Jordan-1) between 2000 and 2007 and A/quail/Saudi Arabia/08vir3489-46-as104/2006. Of note, A/ck/Saudi Arabia/532/1999 contains the same gene composition and could be considered the progenitor of these viruses, as it is located near the root of cluster A in all of the phylogenetic trees (Fig. 2 and 3; see Fig. S1 to S6 in the supplemental material), except the HA, where it does not belong to any identified cluster.
Viruses from Pakistan (2006 to 2008) and Afghanistan (2008 to 2009), A/ck/Iran/10vir854-5/2008, and A/ck/Iran/10vir854-3/2009 were identified as group B. Our phylogenetic analysis suggests that this group arose after acquiring the HA, NA, M, and NP gene segments from viruses isolated in Pakistan in 1999 (entirely belonging to the G1 lineage), as Pakistani strains tended to cluster near the root of the cluster B in the four phylogenies. In contrast, the remaining genes have been obtained by intersubtype reassortment events.
Cluster D consists only of viruses from Iran (Iran-1) that, based on the HA phylogeny, circulated from 1998 to 2007. Due to the lack of sequence data for the internal genes of most of the Iranian viruses previously deposited in GenBank, this group is represented only by A/ck/Iran/11T/99 and A/ck/Iran/10vir854-1/02 in the PB2, PA, and NP phylogenies. Interestingly, the NP gene appears to cluster with A/duck/Hong Kong/Y439/97, suggesting that the Y439 H9N2 lineage circulating in Asia may be the source of this gene.
The occurrence of intersubtype reassortment is evident also for three H9N2 viruses from Iraq-Kurdistan and Iran. Interestingly, four gene segments (HA, NA, M, and NS) of two 2009 Kurdistani strains and five gene segments (HA, NA, M, NP, and NS) of A/chicken/Iran/10vir854-4/2009 fall within genotype B, while the remaining genes do not belong to any H9N2 groups (A to D) and show low sequence similarity to H9N2 Asian viruses, indicative of intersubtype gene mixing (Table 1).
Besides these intersubtype reassortment events, extensive intrasubtype reassortments among H9N2 viruses belonging to the four genetic groups (A to D) seem to have played an important role in the genesis of viruses with unique genome constellations (Table 1). In particular, reassortant strains containing a mixture of genes from groups A and B have been isolated in Israel (Israel-3) from 2006 to 2009, in Jordan (Jordan-2) in 2010, and in Saudi Arabia in 2007 (Table 1). Interestingly, these viruses show three different types of reassortments: Israel-3 and Jordan-2 contain four genes from group A and four from group B, two Saudi Arabian isolates possess five genes from group A and three from group B, and one virus from Saudi Arabia has acquired another gene from cluster A.
Distinct reassortments between groups B and C are identified in the UAE in 2003 and 2008, in Qatar in 2008, and in Saudi Arabia in 2005 and 2006 (Table 1). In addition, two different triple reassortant viruses, containing gene segments belonging to the clusters A, B, and C, have been detected in Saudi Arabia from 2005 to 2007. The 2007 isolate was probably generated by further reassortment events involving the PB2 and NA genes of the Saudi Arabian strains circulating in 2005 and 2006.
More generally, these data suggest that a unique reassortant virus became predominant in Israel and Jordan and spread in these two countries between 2006 and 2010. In the other countries, reassortant viruses circulated for shorter periods and underwent further reassortment events, generating strains with distinct gene compositions. For example, in Saudi Arabia, six distinct reassortant viruses were detected: two in 2005, two in 2006, and two in 2007 (Table 1).
Rates of nucleotide substitution and time scale of viral evolution in Central Asia and the Middle East.
Rates of nucleotide substitution and tMRCAs were estimated for each segment of the entire population of H9N2 viruses from Central Asia and the Middle East. For all genes, the lower 95% HPDs of CoV values of the relaxed (uncorrelated lognormal) molecular clock were >0, so a relaxed molecular clock model, which allows for rate variation across lineages, was used in preference to a strict molecular clock.
The mean substitution rates for all segments were found to be within the range typically observed for avian influenza viruses (8). The highest rate of evolution was observed in the NA segment (4.26 × 10−3 substitutions/site/year; 95% HPD, 3.70 × 10−3 to 4.80 × 10−3), while the lowest rate was observed in the M segment (2.22 × 10−3 substitutions/site/year; 95% HPD, 1.78 × 10−3 to 2.69 × 10−3) (Table 2).
Table 2.
Estimated rates of nucleotide substitution and tMRCA for H9N2 viruses from Central Asia and the Middle East
| Gene | Evolutionary rates (10−3) (95% HPD) | Mean tMRCA (95% HPD) |
|---|---|---|
| PB2 | 3.20 (2.32–4.12) | 1968 (1945–1985) |
| PB1 | 3.13 (2.35–4.02) | 1976 (1957–1991) |
| PA | 3.14 (2.31–3.95) | 1974 (1952–1992) |
| HA | 4.14 (3.54–4.75) | 1993 (1990–1996) |
| NP | 2.84 (2.24–3.46) | 1981 (1970–1990) |
| NA | 4.26 (3.70–4.80) | 1994 (1991–1996) |
| NS | 3.82 (2.76–4.87) | 1979 (1960–1993) |
| M | 2.22 (1.78–2.69) | 1989 (1982–1995) |
To establish when H9N2 virus genes were introduced in Central Asia and the Middle East, we estimated the tMRCA for the entire population of H9N2 viruses circulating in this area (the time-scaled MCC trees for each gene segment, showing nodal divergence times, are provided in Fig. S7 in the supplemental material). As expected given the different origins of the internal gene segments, the tMRCAs calculated for the PB2, PB1, PA, NP, and NS genes were relatively distant in time (mean tMRCAs ranged from 1968 to 1981; Table 2), while the mean tMRCAs estimated for the segments belonging to the G1 lineage (HA, NA, and M) ranged from 1989 to 1994. Notably, the tMRCAs estimated for the HA and NA genes were very similar—1993 (1990 to 1996, 95% HPD) and 1994 (1991 to 1996, 95% HPD), respectively—and approximately 5 and 4 years before the detection of H9N2 viruses in Central Asia and the Middle East (Table 2).
Dating the time of emergence of each gene segment of lineage A and B showed different tMRCAs, supporting the results of the phylogenetic analysis depicting multiple reassortment events among gene segments derived from different sources. In particular, the mean tMRCAs estimated for the genes of lineage A ranged from 1987 to 1997, while the tMRCAs obtained for the genes of lineage B ranged from 1991 to 2001, suggesting that this lineage may have emerged in 2001, when H9N2 viruses were already circulating in Central Asia and the Middle East.
Phylogeography of H9N2 in Central Asia and the Middle East.
Our phylogenetic analysis also revealed that most of the H9N2 genes circulating in Central Asia and the Middle East clustered with viruses from eastern Asia, suggesting that birds in this area may be an important source of H9N2 genetic diversity. In addition, in all phylogenetic trees, the Jordanian and Afghani H9N2 viruses showed a close relationship to the H9N2 sequences from Israel and Pakistan, respectively, which may reflect a considerable trade in commercial poultry between these countries.
Although our phylogenetic analysis revealed some mixing of H9N2 sequences among localities, most notably Saudi Arabia, which is indicative of at least some widespread viral gene flow, most of the viruses sampled from individual countries tended to cluster together, which is strongly suggestive of major geographic subdivisions among H9N2 viruses. To examine the extent and pattern of geographical structure in these data in a more quantitatively rigorous manner, we used a series of phylogeny-trait association tests, in which each H9N2 virus was assigned to a different geographic region. Accordingly, our Bayesian MCMC analysis of geographical association revealed a very strong geographic clustering of H9N2 strains by country of origin (P = 0 for both AI and PS statistics in all gene segments). Similarly, when the extent of phylogenetic clustering of individual countries was tested, significant population subdivision was observed in all countries. Such localized clustering was especially strong in Jordan and Afghanistan, in which the MC statistic was strongly significant in all gene segments (P < 0.0009; see Table S1 in the supplemental material), while in Pakistan and Iran, lower significance values were observed for four and three genes, respectively (P > 0.001; see Table S1 in the supplemental material), indicative of some gene flow involving birds from these localities. Notably, the MC statistic was significant for every country analyzed, indicating that H9N2 genetic diversity is shaped primarily by within-country evolution rather then extensive migration between countries.
Diversifying selection in the H9N2 genes and identification of potential additional glycosylation sites.
Our analysis of selection pressures revealed that most codons in all genes were subject to purifying selection (mean dN/dS ratios varied between 0.07 and 0.61; Table 3). Despite this, we identified several individual codons in the HA, NA, PB2, PA, NS1, and M1 genes that may be subject to positive selection (Table 3). Specifically, three positively selected residues in the HA gene at positions 168, 198, and 234 (160, 190, and 226 in H3 numbering) are located at the receptor binding site or on the tip of the HA (25). Substitutions in these positions may affect virus interactions with cell surface receptors. Furthermore, site 234 (226 in H3 numbering) falls within epitope II of H9 and plays a role in the receptor binding specificity of HA (20). In addition, additional potential glycosylation sites are detected at position 145 (137 in H3 numbering) in two viruses from Israel (2007 and 2008), at position 206 (198 in H3 numbering) in the HA protein of A/quail/Saudi Arabia/08vir3489-46-as104/06 and A/chicken/Iran/SS1/1998, and at position 218 (210 in H3 numbering) in four viruses from the UAE (2000 to 2003), three from Pakistan (1999), and three from Iran (1998 to 2007). Interestingly, sites 137 and 198 have previously been demonstrated to be antigenically relevant amino acid positions in H9 hemagglutinin (20, 35) and glycosylation in these two sites could play a role in antigenic variation. Similarly, in the NA gene, we identified seven residues under positive selection pressure, six of which were located in the NA globular head (25). Mutations at these positions may influence the antigenic specificity of these viruses.
Table 3.
Amino acid sites under putative positive selection and mean dN/dS ratio for each genea
| Gene product | Site(s) under positive selectionb | Mean dN/dS ratio |
|---|---|---|
| HA | 168 (160), 198 (190), 234 (226), 282 (274), 283 (275) | 0.23 |
| NA | 42, 43, 50, 111, 141, 356, 463 | 0.26 |
| PB2 | 451 | 0.08 |
| PB1 | 0.07 | |
| PA | 237, 272 | 0.08 |
| NP | 0.07 | |
| NS1 | 171, 197, 215 | 0.38 |
| NS2 | 0.31 | |
| M1 | 74 | 0.13 |
| M2 | 0.61 |
P < 0.05.
Position numbers are for H9 HA; those for H3 HA are in parentheses.
Molecular characterization. (i) Amino acid mutations with possible implications for pandemic emergence.
Recent studies have demonstrated that the amino acid leucine (L) at position 226 in the HA RBS plays a key role in human virus-like receptor specificity and promotes the transmission of H9N2 in ferrets (46). Sorrel et al. showed that the combination of four key amino acid residues at the RBS of the HA molecule (H183, A189, E190, and L226) is essential for the respiratory droplet transmission of a reassortant virus carrying the surface proteins of an avian H9N2 virus in a human H3N2 backbone (42). Our analysis of H9N2 from Central Asia and the Middle East reveals that 23/42 (54.8%) viruses of lineage A, 91/97 (93.8%) viruses of lineage B, 6/7 (85.7%) viruses of lineage C, and 16/29 (55.1%) viruses of lineage D contain the amino acid leucine at position 226 (H3 numbering) at the RBS. Moreover, seven viruses show three of the four key amino acid residues at the RBS. In particular, the A/ostrich/Eshkol/619/02, A/turkey/Israel/619/02, A/chicken/Iran/SS1/98, A/chicken/Iran/TH77/98, A/quail/Dubai/301/00, and A/quail/Dubai/302/00 viruses possess H183, E190, and L226, while A/quail/Saudi Arabia/08vir3489-46-as104/06 contains H183, A189, and E190. Interestingly, an alanine at position 189 is rarely observed among H9N2 viruses (only 6 of 847 Eurasian H9 sequences available in GenBank show A189) and the Saudi Arabian isolate is the only virus from the G1 lineage that carries this substitution. In addition, position 189, located on the tip of the globular head of HA1, is considered a critical antigenic site and has been implicated in H9 escape mutants (42).
To understand the pandemic risk posed by the H9N2 viruses, we also investigated the species-associated signature positions thought to be characteristic of human influenza A viruses. Residues located at the avian-human signature positions previously described by Chen et al., Finkelstein et al., and Pan et al. and identified in the H9N2 genes are listed in Table S2 in the supplemental material (7, 13, 30). Although the predominant amino acid found is consistent with avian influenza viruses at most marker locations, in a small proportion of H9N2 sequences, the amino acid prevalent in human-hosted viruses has been acquired. Accordingly, we found 21 host markers in the PB2, PB1-F2, PA, NP, M2, and NS2 proteins (see Table S2). However, no single H9N2 isolate contains more than 4 of these 21 sites.
(ii) Amino acid mutations associated with virulence.
Our analysis of the NS1 protein revealed the existence of two mutations, at positions 92 and 227, which have previously been demonstrated to modulate avian influenza virus pathogenicity. Mutation D92E is observed in three viruses isolated in Pakistan in 1999, while E227K is found in 47 of 119 H9N2 viruses analyzed here. The glutamic acid residue at position 92 of the NS gene appears to be implicated in influenza virus resistance to host antiviral activity exerted by interferons and tumor necrosis factor alpha (38).
Mutation E227R/K is a persistent host marker (see Table S2 in the supplemental material) located in the extreme C-terminal region of NS1 (7, 13, 30) and has been shown to modulate avian influenza virus pathogenicity through mechanisms not yet completely clarified (19, 29). In particular, the four C-terminal residues of the NS1 gene correspond to a PDZ ligand domain (PL), which is a protein-protein recognition module that organizes diverse cell-signaling assemblies (40). Viruses containing the C-terminal four-residue PL sequence from the 1918 H1N1 virus (KSEV) and from H5N1 HPAI virus (ESEV or EPEV) showed a significantly increased virulence and pathogenicity in infected mice (19). Interestingly, 82 of the 119 H9N2 viruses analyzed in this study contain one of these motifs. In addition, all H9N2 viruses belonging to group A contained the substitution N66S in the PB1-F2 protein. This mutation, also found in the Hong Kong 1997 H5N1 viruses and in the 1918 H1N1 pandemic strain, has been shown to be associated with high-pathogenicity phenotypes in mice (10).
(iii) Amino acid substitutions associated with resistance to antiviral drugs.
Sequence analysis of the M2 ion channel protein revealed that 22 of the H9N2 viruses analyzed here contained amino acid changes at two positions previously associated with adamantane resistance (43). Specifically, 12 isolates (1 from Iran, 6 from the UAE, and 5 from Israel) had the substitution V27A, while 10 viruses (6 from Iran, 3 from the UAE, and 1 from Qatar) possessed the mutation S31N, as already described by Aamir et al. in 7 isolates from the UAE (1).
DISCUSSION
The H9N2 influenza viruses currently endemic in Asian poultry populations are considered one of the most likely candidates to cause a new influenza pandemic in humans (3). The detection of increasingly large numbers of H9N2 strains showing human-like receptor specificity, combined with the growing evidence of reassortment involving this subtype, emphasizes its potential to emerge and establish itself in the human population.
This study represents the largest evolutionary analysis of H9N2 viruses undertaken to date. We show that H9N2 influenza viruses isolated from 1998 to 2010 in Central Asia and the Middle East comprise four distinct and cocirculating groups (A, B, C, and D), each of which has undergone widespread inter- and intrasubtype reassortments. Unlike groups C and D, which were detected in only one country, groups A and B have circulated extensively in Central Asia and the Middle East and have been identified in six Asian countries from 1999 to the present day. Although it is difficult to determine the precise origin of the H9N2 lineages, our phylogenetic analysis revealed that the HA, NA, and M genes of H9N2 viruses from Central Asia and the Middle East share the same progenitor, as they are all included within the G1 lineage. This indicates whether these gene segments have ever undergone intersubtype reassortment events since 1998, when the first isolates of this lineage were detected in this region. Of note, the G1 lineage was responsible for human infection cases in Hong Kong in 1999 (24) and at the end of 2009 (48), suggesting that this variant is still potentially infectious for humans.
Despite the frequent reassortment events and the identification of multiple cocirculating viruses in each country, we also observed a statistically significant geographical subdivision of viral strains. Hence, after entering a specific country, H9N2 viruses circulate among poultry populations in that locality, giving rise to well-defined genetic groups. This was particularly apparent for most of the viruses isolated from Israel, Jordan, the UAE, and Afghanistan. In contrast, Saudi Arabian H9N2 sequences are dispersed throughout the phylogenetic trees and are generated by multiple reassortment events among lineages A, B, and C. The Arabian peninsula is one of the most import markets for poultry in the world and has seen a rapid increase in imports of live poultry in the last 10 years (27). Birds from this locality may act as an ecological sink, receiving viruses from different areas, but also as a regional source population. Indeed, our results show that the precursor of group A, which includes viruses from Israel, Jordan, and Iraq, is a Saudi Arabian strain (A/chicken/Saudi Arabia/532/99).
Our phylogeny-based analysis of geographical association identifies avian populations from Pakistan and Iran as two possible sources of the H9N2 virus in Central Asia and the Middle East, although across the data set as a whole, by far the strongest signal was for population subdivision. This is consistent with our phylogenetic analysis that identifies Pakistani viruses as the main source of genes for group B, while Iranian strains appear to be a source of genes (PB1 and M) for the UAE and Saudi Arabian viruses. However, all inferences regarding the geographical source of H9N2 clearly need to be confirmed with a larger and less biased sample of viruses. Indeed, the long branches in all of the phylogenetic trees that divide viruses collected from different countries confirm that surveillance and sampling in Central Asian and Middle Eastern countries are currently inadequate.
The trade in poultry and poultry products seems to play the major role in the spread of H9N2 viruses among Central Asian and Middle Eastern countries, as all H9N2 isolates collected to date have been from domestic birds, mainly chickens. Unfortunately, our data do not shed light on the means by which the virus was introduced into Central Asia and the Middle East, as neither of the two main means of spread—through wild birds or trade of poultry—can be definitely excluded on the basis of these data. However, our phylogenetic analysis does suggest that eastern Asia plays the main role in the introduction of H9N2 viruses into this geographic region. To date, the mixing of HPAI H5N1 and H9N2 viruses has been detected only in China (16) and Pakistan (18). However, the extensive cocirculation of these two subtypes in some countries could lead to further reassortment events, generating viruses with a range of phenotypic properties.
Also of note was our identification of amino acid residues under positive selection in the antigenic sites of the HA molecule, such that some H9N2 viruses could differ substantially in antigenicity. This finding is compatible with the idea that the mass vaccination of poultry adopted in many of these countries is a major factor driving the evolution of the H9N2 viruses. Among the amino acid substitutions in H9 HA, 76.4% of the viruses analyzed contain the amino acid leucine at position 226 that is responsible for human virus-like receptor specificity and critical for replication and direct transmission of H9N2 viruses in ferrets (46). Furthermore, seven viruses contain three of the four residues at the RBS of the HA molecule that are considered critical for human-to-human transmission (42). In particular, A/quail/Saudi Arabia/08vir3489-46-as104/2006 contains three of these point mutations essential for respiratory droplet transmission and, more importantly, possesses the change T189A, which has been shown to dramatically alter the antigenicity of viruses (42).
The data generated in this study provide the most comprehensive insight into the epidemiology and evolution of H9N2 virus in Central Asia and the Middle East. Despite these efforts, there remains a lack of information concerning H9N2 in this very large geographic region. Additional study and timely surveillance of H9N2 are clearly needed to identify any increments in viral adaptation to humans and to constantly monitor the mixing of avian H9N2 with other influenza virus subtypes, including those circulating in humans, that may favor the emergence of influenza viruses with pandemic potential.
Supplementary Material
ACKNOWLEDGMENTS
We thank Boehringer Ingelheim Vetmedica for its assistance in facilitating exchange of information, data sharing, sample submissions, and financial support. Part of this work was financially supported by the Italian Ministry of Health through RC IZS VE 14/09 and by the Food & Agricultural Organization of the United Nations (LoA PR n. 41097) and has been conducted in the framework of EU project FLUTRAIN. Edward C. Holmes was funded in part by NIH grant GM080533-04.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Aamir U. B., Wernery U., Ilyushina N., Webster R. G. 2007. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 361:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander D. J. 2003. Report on avian influenza in the Eastern Hemisphere during 1997-2002. Avian Dis. 47(3 Suppl.):792–797 [DOI] [PubMed] [Google Scholar]
- 3. Alexander P. E., De P., Rave S. 2009. Is H9N2 avian influenza virus a pandemic potential? Can. J. Infect. Dis. Med. Microbiol. 20(2):e35–e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown I. H., et al. 2006. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev. Biol. (Basel) 124:45–50 [PubMed] [Google Scholar]
- 5. Butt K. M., et al. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 43:5760–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cameron K. R., et al. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36–41 [DOI] [PubMed] [Google Scholar]
- 7. Chen G. W., et al. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 12:1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen R., Holmes E. C. 2006. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 7:214. [DOI] [PubMed] [Google Scholar]
- 9. Choi Y. K., et al. 2004. Continuing evolution of H9N2 influenza viruses in southeastern China. J. Virol. 78:8609–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conenello G. M., Zamarin D., Perrone L. A., Tumpey T., Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drummond A. J., Rambaut A., Shapiro B., Pybus O. G. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192 [DOI] [PubMed] [Google Scholar]
- 13. Finkelstein D. B., et al. 2007. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 81:10292–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge F. F., et al. 2009. Genetic evolution of H9 subtype influenza viruses from live poultry markets in Shanghai, China. J. Clin. Microbiol. 47:3294–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Golender N., et al. 2008. Genetic characterization of avian influenza viruses isolated in Israel during 2000-2006. Virus Genes 37:289–297 [DOI] [PubMed] [Google Scholar]
- 16. Guan Y., et al. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372–9380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Homayounimehr A. R., Dadras H., Shoushtari A., Pourbakhsh S. A. 2010. Sequence and phylogenetic analysis of the haemagglutinin genes of H9N2 avian influenza viruses isolated from commercial chickens in Iran. Trop. Anim. Health Prod. 6:1291–1297 [DOI] [PubMed] [Google Scholar]
- 18. Iqbal M., Yaqub T., Reddy K., McCauley J. W. 2009. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One 4(6):e5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson D., Hossain M. J., Hickman D., Perez D. R., Lamb R. A. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105:4381–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaverin N. V., et al. 2004. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J. Virol. 78:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a. Kosakovsky Pond S. L., Frost S. D. W. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 21. Kosakovsky Pond S. L., Frost S. D. W. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 22. Lee Y. J., et al. 2007. Continuing evolution of H9 influenza viruses in Korean poultry. Virology 359:313–323 [DOI] [PubMed] [Google Scholar]
- 23. Li C., et al. 2005. Evolution of H9N2 influenza viruses from domestic poultry in mainland China. Virology 340:70–83 [DOI] [PubMed] [Google Scholar]
- 24. Lin Y. P., et al. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U. S. A. 97:9654–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matrosovich M. N., Krauss S., Webster R. G. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156–162 [DOI] [PubMed] [Google Scholar]
- 26. Monne I., et al. 2007. Genetic comparison of H9N2 AI viruses isolated in Jordan in 2003. Avian Dis. 51(1 Suppl.):451–454 [DOI] [PubMed] [Google Scholar]
- 27. Nicita A. 2008. Avian influenza and the poultry trade. Policy research working paper 4551. World Bank Development Research Group, Washington, DC: http://siteresources.worldbank.org/INTTOPAVIFLU/Resources/AI_PoultryTrade_WorkingPaper.pdf [Google Scholar]
- 28. Nili H., Asasi K. 2003. Avian influenza (H9N2) outbreak in Iran. Avian Dis. 47(3 Suppl.):828–831 [DOI] [PubMed] [Google Scholar]
- 29. Obenauer J. C., et al. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580 [DOI] [PubMed] [Google Scholar]
- 30. Pan C., et al. 2010. Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One 5(3):e9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parker J., Rambaut A., Pybus O. G. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239–246 [DOI] [PubMed] [Google Scholar]
- 32. Peiris J. S., et al. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perk S., et al. 2006. Genetic characterization of the H9N2 influenza viruses circulated in the poultry population in Israel. Comp. Immunol. Microbiol. Infect. Dis. 29:207–223 [DOI] [PubMed] [Google Scholar]
- 34. Perk S., et al. 2009. Phylogenetic analysis of hemagglutinin, neuraminidase, and nucleoprotein genes of H9N2 avian influenza viruses isolated in Israel during the 2000-2005 epizootic. Comp. Immunol. Microbiol. Infect. Dis. 32:221–238 [DOI] [PubMed] [Google Scholar]
- 35. Ping J., et al. 2008. Single-amino-acid mutation in the HA alters the recognition of H9N2 influenza virus by a monoclonal antibody. Biochem. Biophys. Res. Commun. 371:168–171 [DOI] [PubMed] [Google Scholar]
- 36. Reference deleted.
- 37. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 38. Seo S. H., Hoffmann E., Webster R. G. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950–954 [DOI] [PubMed] [Google Scholar]
- 39. Shapiro B., Rambaut A., Drummond A. J. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9 [DOI] [PubMed] [Google Scholar]
- 40. Sheng M., Sala C. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1–29 [DOI] [PubMed] [Google Scholar]
- 41. Soltaniavar M., Shoushtari H., Bozorgmehrifard M., Charkhkar S., Eshratabadi F. 2010. Molecular characterization of hemagglutinin and neuraminidase genes of H9N2 avian influenza viruses isolated from commercial broiler chicken in Iran. J. Biol. Sci. 10(2):145–150 [Google Scholar]
- 42. Sorrell E. M., Wan H., Araya Y., Song H., Perez D. R. 2009. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 106:7565–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki H., et al. 2003. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J. Infect. Chemother. 9(3):195–200 [DOI] [PubMed] [Google Scholar]
- 44. Toroghi R., Momayez R. 2006. Biological and molecular characterization of avian influenza virus (H9N2) isolates from Iran. Acta Virol. 50:163–168 [PubMed] [Google Scholar]
- 45. Wan H., Perez D. R. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. 81:5181–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan H., et al. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3(8):e2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilgenbusch J. C., Swofford D. 2003. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics Chapter 6:Unit 6.4 [DOI] [PubMed] [Google Scholar]
- 48. World Health Organization 2010. Antigenic and genetic characteristics of influenza A(H5N1) and influenza A(H9N2) viruses and candidate vaccine viruses developed for potential use in human vaccines. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/disease/avian_influenza/guidelines/201002_H5_H9_VaccineVirusUpdate.pdf [Google Scholar]
- 49. Xu C., Fan W., Wei R., Zhao H. 2004. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect. 6:919–925 [DOI] [PubMed] [Google Scholar]
- 50. Xu K. M., et al. 2007. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J. Virol. 81:2635–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu K. M., et al. 2007. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 81:10389–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.