Abstract
Background:
Sodium bicarbonate cotransporter (NBC) genes encode proteins that execute coupled Na+ and HCO3- transport across epithelial cell membranes. We report the discovery, characterization, and genomic context of a novel human NBC-like gene, SLC4A9, on chromosome 5q31.
Results:
SLC4A9 was initially discovered by genomic sequence annotation and further characterized by sequencing of long-insert cDNA library clones. The predicted protein of 990 amino acids has 12 transmembrane domains and high sequence similarity to other NBCs. The 23-exon gene has 14 known mRNA isoforms. In three regions, mRNA sequence variation is generated by the inclusion or exclusion of portions of an exon. Noncoding SLC4A9 cDNAs were recovered multiple times from different libraries. The 3' untranslated region is fragmented into six alternatively spliced exons and contains expressed Alu, LINE and MER repeats. SLC4A9 has two alternative stop codons and six polyadenylation sites. Its expression is largely restricted to the kidney. In silico approaches were used to characterize two additional novel SLC4A genes and to place SLC4A9 within the context of multiple paralogous gene clusters containing members of the epidermal growth factor (EGF), ankyrin (ANK) and fibroblast growth factor (FGF) families. Seven human EGF-SLC4A-ANK-FGF clusters were found.
Conclusion:
The novel sodium bicarbonate cotransporter-like gene SLC4A9 demonstrates abundant alternative mRNA processing. It belongs to a growing class of functionally diverse genes characterized by inefficient highly variable splicing. The evolutionary history of the EGF-SLC4A-ANK-FGF gene clusters involves multiple rounds of duplication, apparently followed by large insertions and deletions at paralogous loci and genome-wide gene shuffling.
Background
The human sodium bicarbonate cotransporters (NBCs), along with the inorganic anion exchangers, comprise the SLC4A subfamily of proteins, a part of the solute carrier (SLC) superfamily. The coupled transport of Na+ and HCO3- across the plasma membrane of epithelial cells is involved in the regulation of intracellular pH, intracompartmental pH, and intercompartmental pH gradients in many organ systems, as suggested by expression of NBCs in the kidney, pancreas, heart, retina, skeletal muscle and other organs [1,2,3]. Basolateral HCO3- cotransport is necessary for proper buffering of digestive enzymes secreted by the pancreas [4]. NBCs are also responsible for electrogenic transepithelial bicarbonate cotransport in kidney proximal tubules [4,5].
Five human NBC transcripts (SLC4A4-SLC4A8) have been previously cloned and mapped [1,2,3,4,6,7,8,9]. Most recently, NBC4 [10] and SLC4A10 [11] have been cloned. We report the discovery and a genomic analysis of a sixth member of this family, SLC4A9, a novel and alternatively spliced NBC-like gene expressed at high levels in normal adult kidney. We also present an in silico analysis of the genomic structure of NBC4 and evaluate conserved paralogous clustering of SLC4A genes with the members of the ankyrin, epidermal growth factor (EGF), and fibroblast growth factor (FGF) gene families in the human genome.
Results
Isolation and genomic structure of SLC4A9
As part of a positional cloning project, we became interested in a region of 5q31 between D5S393 and D5S2927. We annotated all draft and finished genomic sequence from this region using SeqHelp [12].
Presubmission contig h174.3 of bacterial artificial chromosome (EAC) clone CTC-329D1 (now GenBank AC008438) included four regions of high translated sequence similarity with known mammalian NBCs. GeneFinder, Genie and GRAIL 1.3 predicted exons throughout the contig, including, but not limited to, the regions of NBC homology. We named the gene with the HUGO-approved symbol SLC4A9. It is currently represented by ten kidney clones and one testis clone from the IMAGE consortium (Unigene: Hs.166669). Two expressed sequence tag (EST) clones (IMAGE: 1734773 and 1533693) were sequenced to completion, yielding a 1,036 base pair (bp) cDNA contig. The ESTs were later found to cover exons 15-18 and 20B-D (1734773) and 20C-E (1533693) of SLC4A9 when exons are numbered as in Figure 1. The assembled cDNA sequence matched an NBC-like portion of 329D1 and additional new sequence elsewhere on the genomic contig. The presumptive gap in the draft was closed by designing primers c67F and c67R from confirmed sequence and sequencing an approximately 1.8 kilobase (kb) PCR fragment from genomic DNA. The resulting 30,161-bp contig included known sequence from two pieces of 329D1, as well as 659 bp of new sequence.
Figure 1.
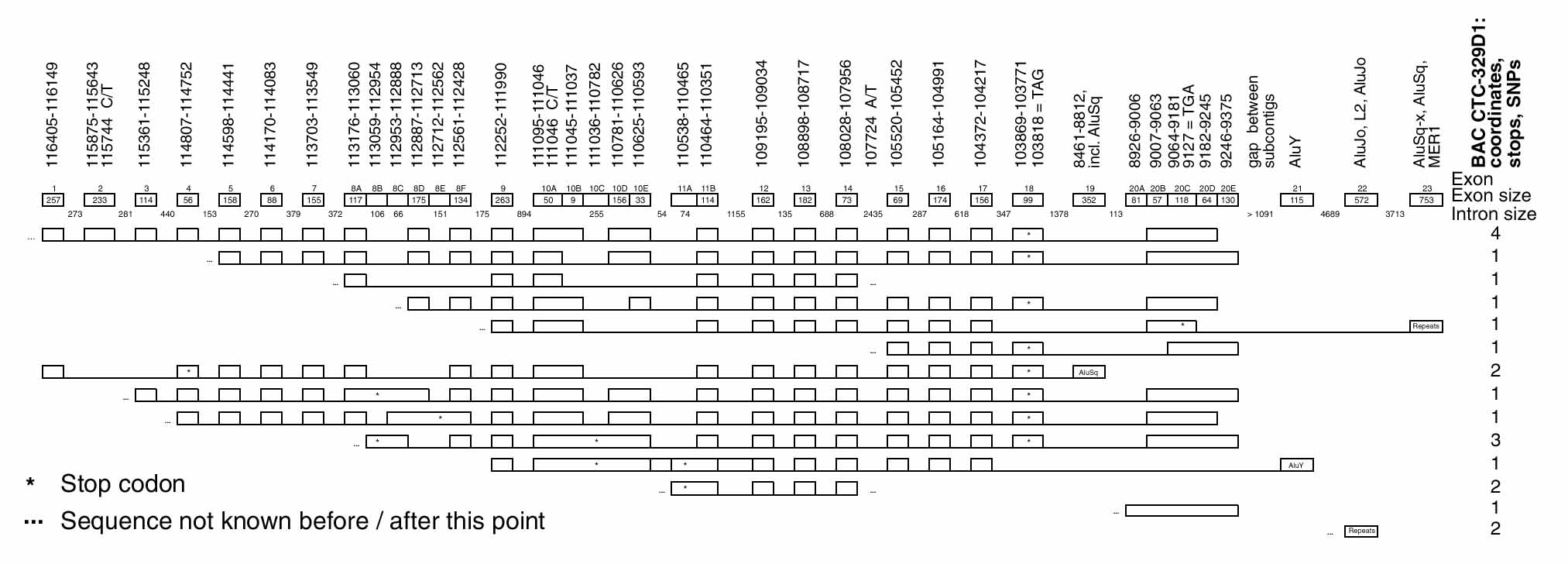
Genomic organization and alternative splicing of SLC4A9. Sequence coordinates refer to nucleotide positions on two different draft-phase contigs of BAC CTC-329D1, GenBank AC008438.1 [gi: 5686628]. The first six rows after the heading row present all the putative coding isoforms, in order of decreasing known length. The bottom eight rows present the noncoding isoforms (including clones whose coding potential could not be determined because of insufficient available sequence). The sixth coding isoform is an IMAGE clone from the NCI CGAP kid11 kidney cDNA library (IMAGE: 2696136). For the noncoding isoforms, the first and second clones from the bottom are IMAGE clones from the NCI CGAP kid11 kidney cDNA library (IMAGE: 2384256 and 2379164, respectively). The fourth clone from the bottom was isolated from the Clontech long-insert fetal brain cDNA library. All other clones were isolated from the Clontech long-insert adult kidney cDNA library.
Putative exons on the 12-kb h174.3 contig were defined by a consensus of multiple-algorithm exon predictions, NBC homologies, and IMAGE clone coverage. Primers were designed from flanking intronic sequences. Exons and adjoining splice sites were amplified by PCR from genomic DNA. Snonymous coding sequence polymorphisms 111046 C→T and 115744 C→T and intronic single-nucleotide polymorphism (SNP) 107724 A→T were identified (all 329D1 sequence coordinates refer to positions on GenBank AC008438.1, GI no. 5686628).
To extend the known 5' portion of the gene's coding sequence, an adult human kidney library (Clontech, cat. no. HL5031t) and a λTriplEx2 long-insert fetal brain cDNA library (Clontech, cat. no. HL5504u) were probed with the two IMAGE clones. The libraries were also amplified with primers 5-LDA and oi-E (Table 1).
Table 1.
Selected primers used for the amplification, cloning and sequencing of SLC4A9
| Primer name | Sequence (5' to 3') |
| oi-A | CGGGGCTCAGGACCTTTTAATG |
| oi-B | ACTCCTACTCCAGCTAATTC |
| oi-C | CTCTGAATCTTCACTGTCAC |
| oi-D | TGAAGAGGTGGACCCTGGTC |
| oi-E | AGATGGAGGCTCCTGTAAGG |
| oi-F | ACTCATTCAGTGGAAGTGAC |
| 6797up | CCAGGACTGAGGAGAAGTCG |
| 5219up | TTGGCAGAGGCACCCGTAGG |
| 345F | GGTCCTCTGCCTACGGGTGCC |
| 345R | GGATGACTGCTGTGATCTGTTG |
| 5-LDA | CTCGGGAAGCGCGCCATTGTGTTGGT |
| 3-LDA | ATACGACTCACTATAGGGCGAATTGGCC |
| Unigene F | CTGCAAGGCGATTAAGTTGGGTAAC |
| Unigene R | GTGAGCGGATAACAATTTCACACAGGAAACAGC |
| c67F | CATTTCTGTGAATTAGCTGGAGTAG |
| c67R | CGACTGAGCATCTGGAAGTTAAG |
Figure 1 illustrates the genomic structure and splicing variation of SLC4A9. SLC4A9 exon sizes vary from 56 bp (exon 4) to 263 bp (exon 9). Intron phase is distributed quite randomly in the 5' half of the sequence, although toward the 3' end of the gene, phase o introns become prevalent. The SNP in exon 10A is immediately adjacent to an alternatively used 5' splice site, but no correlation between the presence of exon 10B in cDNA clones and C or T at nucleotide 111,046 was observed. All introns conform to the GT-AG rule.
Cloning the 5' end of the SLC4A9 transcript
Two observations suggested to us that none of the known SLC4A9 mRNA isoforms is full length: the open reading frames (ORFs) of all the isoforms start at the very first or second base at the 5' end of the clones, and the complete inserts of the clones are ≤ 3.6 kb, whereas the SLC4A9 transcripts on northern blots are 4.3 and 6.0 kb (Figure 2). We therefore undertook a comprehensive effort to find the 5' end of SLC4A9.
Figure 2.
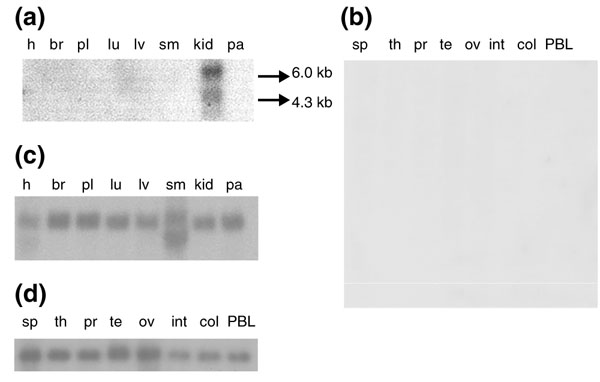
Multiple-tissue northern (MTN) blot analysis of SLC4A9 expression. (a,b) Hybridization with the SLC4A9 345F/R fragment as a probe. (c,d) Hybridization with the control β-actin cDNA as a probe. (a,c) MTN1; h, heart; br, brain; pl, placenta; lu, lung; lv, liver; sm, skeletal muscle; kid, kidney; pa, pancreas. (b,d) MTN2; sp, spleen; th, thymus; pr, prostate; te, testis; ov, ovary; int, small intestine; col, colon without mucosa; PBL, peripheral blood leukocytes. Not shown are the experiments with MTN3 (stomach, thyroid, spinal cord, lymph nodes, trachea, adrenal glands and bone marrow). No SLC4A9 expression was detected and no control hybridization was conducted on MTN3 (data not shown). (b) The entire blot is shown (reduced scale). (a,c,d) Signal areas only are shown. All MTN blots were obtained from Clontech.
The lack of full-length clones in the Clontech adult kidney cDNA library was not surprising, because the library was dT-primed and mostly contained inserts of under 3.8 kb in length (manufacturer's data). Therefore, we used PCR-based approaches to determine the sequence of the 5' end of the mRNA. Nested RACE-PCR (rapid amplification of cDNA ends with PCR) on Marathon kidney cDNA (Clontech) with four different primer combinations (three within known exons and one within a GeneFinder-predicted exon 5' of exon 1) and appropriate nested adaptor primers produced smears and multiple bands over several attempts. Analysis of the RACE products by sequencing the gel-extracted bands and random TA clones revealed 100% mispriming, even though the gene-specific RACE primers did not have any homologies to non-SLC4A9 human sequence. The sequenced TA clones most frequently corresponded to mitochondrial DNA sequences and to the FBN2 gene, which coincidentally maps to 5q23 centromeric of SLC4A9.
In addition to RACE on the Marathon cDNA, we used the Advantage 2 PCR technique (Clontech) on undiluted aliquots of the kidney and fetal brain long-insert phage libraries multiple times with all possible primer combinations of one vector primer (either forward or reverse) and one appropriately oriented gene-specific primer. All gene-specific RACE primers and all internal sequencing primers used during the determination of the complete sequences of the IMAGE clones were tried, one by one. With the exception of the three TA clones obtained with the oi-E primer, all such experiments resulted in 100% mispriming. This result was identical to that obtained when both gene-specific and random-primed reverse transcription, followed by RACE with the same multiple gene-specific primers as above, were performed on a non-Clontech sample of total RNA freshly extracted from a kidney biopsy. In summary, we have been unable to obtain a full-length SLC4A9 transcript with current commercial RACE and RT-PCR (reverse transcription-PCR) protocols.
The 5'-adjoining region of SLC4A9 on 5q31
Despite the failure of experimental attempts to characterize the 5' end of SLC4A9, in silico analyses of the region expected to contain this portion of the gene have been informative. The gene immediately centromeric to, and 12,840 bp from, SLC4A9 is HEGFL, which encodes a heparin-binding member of the EGF family. Genomic DNA sequence between the 5' end of HEGFL and exon 1 of SLC4A9 provides some clues as to the structure of the 5' end of SLC4A9. A putative promoter on the SLC4A9-encoding strand was predicted by the Lawrence Berkeley Laboratories (LBL) neural network promoter prediction algorithm [13], with a score of 1.0 at bp 129,291-129,242 of AC008438.1. This sequence has been shown to have promoter activity [14] on the strand opposite to the coding strand of SLC4A9. As HEGFL and SLC4A9 are transcribed in opposite orientations and have 5' ends facing each other, the promoter may be bidirectional. Four possible exons are predicted 5' of SLC4A9 by GeneFinder. However, neither protein homologies nor consensus Kozak sequences are seen in the region.
SLC4A9 expression, ortholog comparison and protein sequence analysis
Northern blot analysis reveals that expression of SLC4A9 is extremely restricted (Figure 2). Transcripts of 4.3 and 6.0 kb are seen at high levels in kidney but not in any other tissues tested. This is consistent with the kidney origin of 10 of the 11 public ESTs corresponding to SLC4A9. The consistently smeary background, observed regardless of the probe and hybridization stringency, may be due to the presence of low levels of alternatively spliced SLC4A9 mRNA variants.
While this manuscript was undergoing revision, the first mammalian SLC4A9 ortholog, that in the rabbit, was published [15]. The rabbit gene encodes a sodium-independent anion exchanger; this underscores the importance of not assigning functions to NBC-like genes in the absence of experimental evidence. Similarly to human SLC4A9, the rabbit gene is alternatively spliced. Both the RACE-verified complete rabbit cDNA and our incomplete human cDNA are approximately 3.2 kb long. In rabbit this is, however, consistent with the size of the major transcript on the northern blots, and no transcripts over 3.8 kb are seen. In human, the known cDNA size is much less than the 4.3-kb and 6.0-kb signals on the northern blots. In the absence of major differences in coding sequence, this strongly suggests rapid evolution of species-specific 5' and 3'-untranslated regions (UTRs), which are longer in the human gene.
Table 2 summarizes the properties of six human NBC and NBC-like genes. NBC4 and HNBC7 were discovered during our in silico annotation of genomic sequences. NBC4 has since been described in detail [10]. SLC4A9 has the most restricted expression pattern, evidenced by high tissue specificity and low dbEST representation.
Table 2.
Human Na+/HCO3- cotransporter and cotransporter-like genes
| Gene name (and common aliases) | Longest coding mRNA sequence (in nucleotides) (with GenBank accession number) | Genomic sequence (GenBank accession number) | Chromosomal location | Number of known ESTs* | Tissues/organs/cell types where expression is shown by ESTs nad/or northern blots (order: normal tissues; tumors; fetal tissues) | mRNA isoforms due to alternative splicing and/or alternative polyadenylation | Reference† |
| SLC4A4 (SLC4A5, HNBC1) | 7586 nt NM_003759.1 | AC019089.3 | 4q21 | 109 | Kidney, pancreas (northern, ESTs); brain, liver, prostate, colon, stomach, thyroid, spinal cord; fetal lung, fetal testis (ESTs); no tumor ESTs | 6 | [6] |
| SLC4A6 (SLC4A7, HNBC2) | 7785 nt AF047033.1 | AC025392.2, AC024936.3 | 3p22 | 34 | Adult heart (northern), skeletal muscle (northern, ESTs); retina, neuronal precursors, neurons, stomach, colon, uterus, testis, brain, trabecular bone; Gessler-Wilms tumor, liposarcoma, HeLa cells; various fetal tissues (ESTs) | 4 | [2] |
| SLC4A8 (HNBC3, KIAA0739) | 4079 nt AB018282.1 | AC025097.9, AC027750.3, AC021343.1 | 12q13 | 13 | Brain (northern, ESTs), skeletal muscle, kidney, thyroid, spinal cord, trachea, adrenal gland (northern), testis (ESTs); germ-cell tumors (ESTs); weak expression in many other organs and various fetal tissues (ESTs) | 4 | [9] |
| SLC4A9 | > 3258 nt(submission in progress) | AC008438.1 | 5q31 | 11 | Kidney (northern, ESTs); testis (single EST), fetal brain (long-insert cDNA); no tumor ESTs | 14 | This paper |
| NBC4 | 6082 nt AF207661 | AC005041, AC006030.2, AC073263 | 2p11.2 | 16 | Brain, heart, liver, lung, placenta, spleen, stomach (northern); colon, kidney, testis (northern, ESTs); pancreas, uterus, germinal center B cells (ESTs); germ-cell tumors, mantle cell lymphoma, adenocarcinoma; weak expression in other organs (northern) and various fetal tissues (ESTs) | ≥ 2 (not enough data) | [10] |
| HNBC7 | > 1383 nt (no acc. #) | AC064816.1, AC018411.3, AL139426.2 | 1p32-31 | 10 | Kidney, prostate, multiple sclerosis lesions, frontal cortex (ESTs); no tumor or fetal ESTs | ≥ 3 (not enough data) | This paper (partial sequence: Unigene Hs.211115) |
Tissues are adult unless denoted as fetal. SLC4A-designated genes and NBC4 have been experimentally documented. Except for SLC4A9, northern blot data are from published results referenced in the text for each gene under consideration. SLC4A10 was omitted because of the lack of a full-length public mRNA sequence. *Human ESTs matching (100%) experimentally documented exons and NBC-homologous predicted exons, as of 31 May 2000. †Independent investigators often describe the same SLC4A gene. The reference listed for each gene reflects the first time that gene was published.
The amino-acid sequence of the 990 amino acid SLC4A9 inferred from the major splice isoform was subjected to secondary structure and hydropathy analysis by TMPRED [16]. Consistent with the results for NBC4 [10], SLC4A9 is predicted to be a 12-transmembrane protein with a relatively long (amino acids 1-265) cytoplasmic amino terminus and a shorter, also cytoplasmic, carboxyl terminus (amino acids 929-990). NetPhos v.2.0 [17] predicted several serine and threonine phosphorylation sites. The relative lengths of the cytoplasmic domains and the distribution of phosphorylation sites were strikingly similar to those observed for NBC4 [10]. The predicted transmembrane segments and phosphorylation sites are indicated in Figure 3.
Figure 3.

ClustalW protein sequence alignment of SLC4A9 with paralogous proteins described in the literature or discovered in silico in this report, in order of decreasing similarity. Proteins shown: SLC4A9 (isoform I); SLC4A8/HNBC3 (BAA34459.1); SLC4A4/SLC4A5/HNBC1 (AAD42020.1); SLC4A6/SLC4A7/HNBC2 (AAD38322.1); NBC4 (AAG18492) and HNBC7 (translated Unigene cluster Hs.211115 and translations of SeqHelp-identified NBC-homologous exons from AC018411.3, AL139426.1, and AC064816.1). The known 990 amino acid carboxy-terminal portion of SLC4A9 isoform I is shown, preceded by spaces. The longest publicly available protein sequence was selected for SLC4A4/A5, SLC4A6/A7 and SLC4A8. Spaces indicate undetermined amino-terminal portions of SLC4A9 and undetermined amino-terminal and carboxy-terminal portions in the currently available HNBC7 sequence. Dashes indicate known gaps or known absence of sequences at indicated positions. White on black background, identity across most or all of the proteins shown. Black on dark gray background, strong similarity in the type of amino acid at position shown. Black on light gray background, weak similarity in the type of amino acid at position shown. Vertical bars indicate exon boundaries on the SLC4A9 sequence. Exon numbers and exon fragment designations are immediately to the right of each bar. Each bar is directly over the first amino acid of the exon or fragment. For phase 1 introns, the amino acid whose codon is broken by the intron or breakpoint is considered to follow the intron or breakpoint; for phase 2, it is considered to precede it. On the SLC4A9 sequence, the 12 transmembrane segments are in red. Intracellular serine, threonine and tyrosine residues predicted by NetPhos v.2.0 [16] as putative phosphorylation sites with a score of 0.5 or above are in blue.
The predicted SLC4A9 protein aligns both to human (Figure 3) and rat (data not shown) NBCs. SLC4A9 is most similar to SLC4A4 (49% identity) and SLC4A6 (48%), followed by NBC4 (44%) and SLC4A8 (43%). The exact extent of protein sequence similarity of HNBC7 to SLC4A9 cannot be determined since too little HNBC7 sequence can be inferred.
SLC4A9 as a part of an ancient, multiply duplicated EGF-SLC4A-ANK-FGF gene cluster
To identify gene clusters containing NBC-like SLC4A genes, we consolidated information from human radiation hybrid (RH) maps and the GSS and HTGS databases for seven SLC4A NBCs, 19 FGF family members, and 10 EGF family members [18]. The electronic mapping strategy involved anchoring genomic sequences that matched each gene to the GB4 RH map via RH-mapped sequence-tagged sites (STSs) or gene-based markers. The possibility that ankyrins might also be a part of this cluster was suggested by the presence of a novel ankyrin gene, ANKfc (from fetal cochlea), less than 100 kb distal to SLC4A9 on 5q31. We were able to determine map positions for all five SLC4A, all four ANK, 9 of 10 EGF, and 17 of 19 FGF family members. All cases where members of at least two of the four families cluster are shown in Figure 4.
Figure 4.
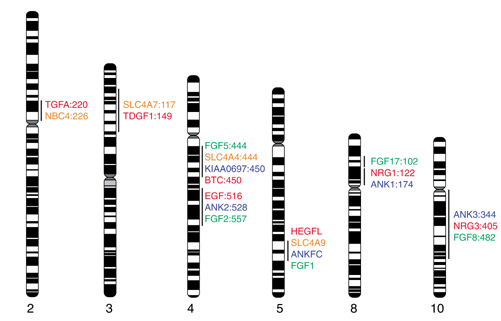
Mapping of conserved paralogous gene clusters containing selected members of the EGF, SLC4A, ANK and FGF families. Numbers refer to GB4 RH map positions of genes or their closest mapped markers, if such positions are known or can be inferred from HTGS and STS data. On 5q31-3, RH-inferred gene order was superseded by sequence data, and is not shown. On 8p21-12, the break in the line indicates a possible intracluster rearrangement. For clusters that do not contain genes representing each of the four families, additional genes belonging to the unrepresented families are predicted. EGF family members shown: TGFA, TDGF1, BTC (betacellulin), EGF, HEGFL, NRG1, NRG3; EGF genes not shown: AREG and EREG (4q13-21), NRG2 (5q31). SLC4A genes not shown: SLC4A8 (12q13), SLC4A10 (2q31), HNBC7 (1p32-31). KIAA0697 (a long-insert cDNA from RIKEN large-scale gene discovery projects) is highly homologous to the unique middle domain of ANKfc and is therefore included as a candidate novel ankyrin-family gene despite the absence of ankyrin repeats from its cDNA. FGF gene not shown: FGF20 (8p13-22).
We used SLC4A family members to test the hypothesis that the origin and repeated duplication of the EGF-FGF cluster predated the human-mouse divergence. Four of the seven known SLC4A genes were found near either an EGF gene or an FGF gene, or both. The genomic location of EGF and FGF family members on human chromosome 5q conforms to the syntenic relationship with mouse chromosome 18 [19].
Ten human genes belong to the EGF family [18]. HEGFL on 5q31, EGF on 4q25, TGFA (transforming growth factor α) on 2p13, and AREG (amphiregulin), EREG (epiregulin), and BTC (betacellulin) on 4q13-q21 are EGF paralogs. TDGF1 (teratocarcinoma-derived growth factor), approximately 20.5 megabases (Mb) proximal to SLC4A7 on 3p22, shares structural similarities with TGFA [20]. Distance approximations are based on the sum of draft clone lengths and estimated gap sizes obtained from the Draft Human Genome Browser [21]. Three neuregulin genes (NRG1-3) are also in the EGF family [18]. All these genes have orthologs in the mouse, suggesting that multiple duplications of an ancestral EGF-like gene predated the mouse-human divergence. Nineteen known loci encode members of the FGF family, of which at least five map near EGF paralogs: FGF1 on 5q31 (approximately 1.5 Mb distal of HEGFL), FGF2 on 4q25 (approximately 14.7 Mb distal to EGF), FGF8 on 10q25 (cytogenetically close to NRG3), FGF17 on 8p21 (approximately 11.0 Mb distal to HGL), and FGF5 on 4q21 (approximately 5.0 Mb from BTC). Mouse genes Btc and Fgf5 are located close to each other in the region of mouse chromosome 5 syntenic to human 4q13-q21. The cluster size in the mouse is unknown because of the lack of sequence data.
As expected, a novel NBC-like gene [10] is located at 2p11-12 proximal to TGFA. The distance between the two genes is approximately 4.0 Mb. The protein sequence of NBC4 is similar to that of SLC4A9 (see Figure 3), but the genomic structures of the two genes differ. Exon boundaries are only partly conserved, and NBC4 includes an intron of 20.5 kb, longer than the genomic sequence containing exons 1-20 of SLC4A9. Portions of the NBC4 mRNA are completely identical to the expressed regions of two human genes on 2p13: DCTN1 (dynactin, a homolog of the Drosophila p150Glued gene) and MTHFD2 (methylene tetrahydrofolate dehydrogenase) (Figure 5). Specifically, the 5'-UTR of NBC4 matches 1,646 nucleotides of coding sequence and 87 nucleotides of the 3'-UTR of DCTN1, and the 3' UTR of NBC4 matches 92 nucleotides of coding sequence and 75 nucleotides of the 3'-UTR of the published MTHFD2 sequence. In addition, a 241 nucleotide overlap of NBC4 and MTHFD2 ESTs in the antisense orientation is inferred from annotation of GenBank AC073263. These ESTs correspond to alternate, extended 3'-UTR forms of the two genes, which are different from the 3'-UTRs of the published full-length mRNAs.
Figure 5.
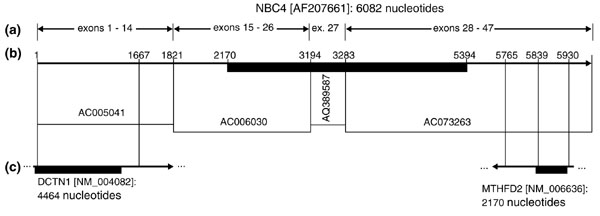
Genomic and cDNA database matches for NBC4 [10]. Arrows indicate the direction of transcription. Solid black rectangles show the location of ORFs. (a) The AF207661 NBC4 cDNA with selected exon locations and sequence coordinates marked. (b) Fully sequenced (AC005041, AC006030), BAC-end (AQ389587), and HTGS draft (AC073263) genomic GenBank accessions matching portions of the cDNA are shown. (c) cDNA sequence matches, with sequence coordinates shown along each match. The DCTN1 match is contiguous, whereas the MTHFD2 match is not. In both cases, an untranslated region of AF207661 matches a portion of the other cDNA's ORF and some of its 3' UTR.
We used public genomic resources to determine whether chromosomal locations of genes from any one of the four families (EGF, SLC4A, ANK and FGF) can be used to predict the genomic location of novel members of the remaining families. Ankyrins mapped near several known EGF ligand and/or FGF genes (Figure 4). In particular, on chromosome 4q25, ANK2 is located between EGF and FGF2, and EGF is proximal to FGF2; this gene ordering is supported both by the Human BAC Accession Map [22] and direct HTGS-to-GB4 RH mapping. It was therefore not surprising to discover a novel ankyrin, ANKfc, immediately distal to SLC4A9, and thus distal to the EGF paralog HEGFL, on 5q31.
Searching the HTGS database with human NBC and NBC-like cDNA queries yielded draft-phase genomic sequences (AL139426, AC018411, AC064816) similar to some, but identical to none, of the five HNBC genes described above (Table 2). NBC-homologous exons from these sequences were combined with Unigene cluster Hs.211115 to predict yet another novel sodium bicarbonate cotransporter-like gene, HNBC7. This gene maps to 1p31-32, where no EGF-FGF cluster is currently known to exist. Similarly, SLC4A8 is at 12q13, where no EGF-FGF cluster is yet known.
To test our hypothesis that the dispersed paralogous gene clusters are a product of multiple ancient duplications, we conducted a phylogenetic analysis of the EGF, SLC4A, and FGF genes we believe to fall in the clusters, along with their non-human orthologs. Neighbor-joining and maximum parsimony methods were used to construct phylogenetic trees for each of the gene families (Figure 6a-c). This enabled us to infer the most likely history of the cluster duplications (Figure 6d).
Figure 6.
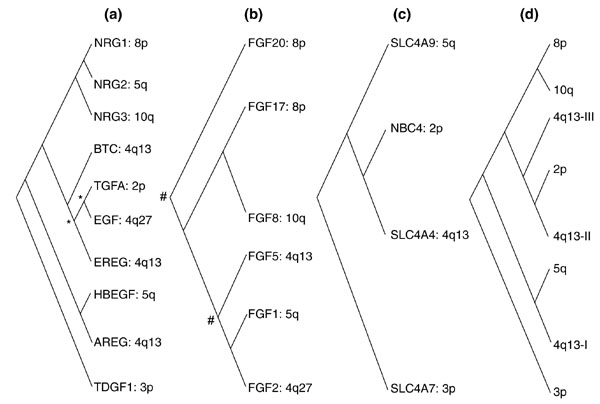
Results of a phylogenetic analysis of seven human EGF-SLC4A-FGF gene clusters. (a) EGF family; (b) FGF family; (c) SLC4A family; (d) inferred history of chromosomal duplications. Phylogenetic trees were constructed for each of the three gene families using four methods per family: neighbor-joining with uncorrected p, neighbor-joining with gamma correction, neighbor-joining with Poisson correction, and maximum parsimony. Non-bootstrap and 100-bootstrap inputs were used for each of the four methods. Therefore, 24 phylogenetic trees were constructed altogether. All phylogenetically informative orthologs known for each of the human genes were used. Human genes in the three families that did not map to any of the seven cluster loci are not shown. Alignments in MSF (text) format and the trees produced by PHYLIP from them can be found as additional data files available with the online version of this article. An intuitive interpretation of the unrooted trees is presented in a simplified cladogram format. Unlabeled nodes were supported by all four methods used. In (d), I, II, and III refer to three consecutive rounds of local duplication at the locus ancestral to 4q13. Nodes indicated by an asterisk (*) are supported solely by the three neighbor-joining trees. Nodes indicated by a hash mark (#) are supported by neighbor-joining with either gamma or Poisson correction only.
Two irregularities in Figure 6d are interesting from the standpoint of genomic history of duplicated genes. NRG2 at 5q31 is phylogenetically closer to the 8p gene NRG1 than to any other EGF gene, yet that relationship makes little sense if the duplication giving rise to clusters at 4q13 and 5q13 is far more ancient than that giving rise to the 8p and l0q clusters, as the EGF and FGF data suggest. The location of NRG2 at 5q31 is therefore noteworthy because only single members of the other families are present there and because NRG2 is phylogenetically very distant from the 5q EGF gene (HBEGF), making either multigene or single-gene tandem duplications within 5q highly unlikely. This product of a very recent duplication involving the 8p NRG1 gene may have been deposited at 5q31 as a random insertion of a newly duplicated gene away from its ancestral locus, in a process similar to that which deposited some SLC4A genes outside of their ancestral paralogous clusters. In addition, the history of the 2p cluster is somewhat obscure, as the 2p EGF gene is closest to the EGF gene at 4q27, whereas the 2p SLC4A gene is closest to the SLC4A at 4q13 (as no SLC4A gene is currently known to exist at 4q27). With the exception of these irregularities, the duplication history in Figure 6d is very well supported by the gene family trees in Figure 6a-c and the full PHYLIP trees (data not shown).
Discussion
Alternative splicing of SLC4A9
The existence of multiple cDNA sequences as a result of alternative splicing is the most interesting feature of SLC4A9. Most SLC4A9 alternative splicing is accounted for by the selective inclusion or exclusion of portions of exons 8, 10, 11, 19 and 20. In contrast to these alternatively spliced portions of the gene, the use of exons 4-7, 9 and 12-17 appears to be a constant feature of the various isoforms. No splice variation is observed for these exons in any cDNA clones or dbEST entries examined, with the exception of the fetal brain clone, in which exon 12 contains extra sequence from the 5'-adjoining intron.
All intron-exon junctions of SLC4A9 feature consensus splice site sequences. Therefore, alternative splicing of SLC4A9 is not consistent with the hypothesis that weak or nonconsensus splice sites lead to intron retention or alternative splicing. Instead, yet-undetermined cis-acting intronic sequences may be responsible. The recurrent noncoding SLC4A9 transcripts may escape the normal mechanisms of nonsense decay [23] responsible for degradation of incorrectly spliced mRNAs with disrupted ORFs.
Of all alternative splicing in humans, only 20% occurs within coding regions [24]. SLC4A9 may be a member of a class of genes characterized by highly variant and inefficient splicing, a class first suggested by a comparison of ESTs to genomic sequences [25]. The high degree of alternative splicing of SLC4A9 may be the result of inefficient spliceosomal processing. One possible outcome of such inefficiency, IMAGE clone 2130425, is not included in the 14 isoforms on Figure 1. This nonlinearly spliced clone includes unique exons dissimilar to any exons of any other SLC4A9 cDNAs. A unique fragment in the exon 10-11 region is followed by a correctly spliced exon 11B and a part of exon 12, which splices backwards from a unique donor site to a unique partial version of exon 9, and continues directly to a unique fragment of exon 20, terminating at the common late polyadenylation signal.
The 3'-UTR of SLC4A9 is fragmented into six alternatively spliced exons, of which no more than two appear to be used per isoform and four harbor polyadenylation signals within expressed repetitive elements. Two alternate 3'-terminal untranslated exons of SLC4A9, exons 21 and 23, consist entirely of repetitive elements, except for a 10-nucleotide spacer in exon 23. The two polyadenylation sites within exon 20 are used at roughly equal frequencies, both in experimentally derived clones and public EST sequences corresponding to SLC4A9. Alternative polyadenylation is observed in fewer than 29% of human genes, based on an analysis of 8,700 human 3'-UTRs [26].
The structurally invariant carboxy-terminal 591 amino acids of SLC4A9 include the 12 transmembrane domains characteristic of sodium bicarbonate transporters. The alternatively spliced amino-terminal portion of SLC4A9 contains hydrophilic domains of unknown function. It is possible that alternative splicing of these domains leads to different spatial or electrochemical specificity. For example, in chick cochlea, different transcripts produced by alternative splicing of the SLO gene (homolog of Drosophila slowpoke) generate kinetically distinct calcium-activated potassium channels [27]. It is therefore tempting to speculate that proteins encoded by the alternative SLC4A9 transcripts might differ in stoichiometry or in the minimum voltage potential threshold required to activate cotransporter function.
SLC4A9 protein sequence: comparison to paralogous genes
Four large blocks of highly conserved amino acid sequence characterize all known HNBCs (Figure 3). They correspond to SLC4A9 amino acid positions 68-210, 223-352, 384-578 and 629-960. At both ends of blocks 68-210 and 629-960, at the carboxy-terminal end of block 384-578, and throughout block 223-352, SLC4A9 has significant sequence differences from most or all of the paralogs. Non-SLC4A9 proteins in the alignment have considerably greater homology between themselves in the equivalent regions than they do with SLC4A9.
The exon 8 and exon 10-11 hypervariably spliced regions correspond to SLC4A9 amino acid positions 353-494 and 583-702, respectively. It is intriguing that short amino-terminal portions of both of these regions (amino acids 353-383 and amino acids 583-631) are located in areas where the sequences of the paralogs are quite diverged. Extensive alternative splicing in these areas has not been reported for the other paralogs.
Alternately used exon 10d contains an almost perfect 41 nucleotide polypyrimidine tract. This region consists exclusively of Cs and Ts, except for the A at position 110,655 of AC008438.1. It codes for FFSLLLFLTSFFF, a highly hydrophobic stretch predicted by TMPRED to be within the sixth transmembrane domain of the protein. Exon 10D is absent from three SLC4A9 cDNA isoforms (Figure 1) whose ORFs are not disrupted except for the deletion of the 53 amino acids corresponding to this fragment. TMPRED analysis suggests that absence of exon 10D abrogates the sixth transmembrane domain but does not affect the 11 remaining transmembrane segments. Consequently, isoforms lacking exon 10D would be predicted to have an extracellular carboxyl terminus. The biological viability and function, if any, of such a protein cannot be known without biochemical analyses. However, the extracellular exposition of the normally hidden carboxyl terminus might be relevant to autoimmunity.
Dispersed paralogous gene clusters containing SLC4A genes
Four human SLC4A genes are each included in a conserved gene cluster (Figure 4). On 5q31, SLC4A9 is located between genes encoding HEGFL and FGF1 (Figure 4). The murine orthologs of HEGFL and FGF1 are in close proximity on mouse chromosome 18, suggesting that an as-yet-undescribed mouse ortholog of SLC4A9 may be located in the same region. This putative mouse ortholog of SLC4A9, partly contained in the BAC clone RG-MBAC_173P21 (GenBank AC027276), has 82-96% similarity to the human gene over 1897 nucleotides. Almost all exon boundaries are conserved between the coding portions of the mouse gene and SLC4A9 isoform I. However, exon 4 of human SLC4A9 does not appear in mouse.
Novel human SLC4A9 paralogs may be predicted on the basis of the genomic locations of the EGF-FGF clusters. The clusters in Figure 4 that contain the members of at least two of the other three gene families may also contain yet-uncharacterized SLC4A genes. Only a deeper sequence coverage of human EST libraries and draft genomic sequences will help determine if this hypothesis is correct. Ancient conserved paralogous clusters involving multiple functionally unrelated genes have been previously suggested to exist in the human genome [28,29,30]. However, the existence of some SLC4A genes outside conserved clusters suggests that intra-cluster rearrangements may have led to the expulsion of these genes from the conserved clusters. Yet other SLC4A duplication mechanisms may have complemented both the cluster duplication and the subsequent rearrangements.
Genomic implications of SLC4A9 splicing and structure
The genomic structure of SLC4A9 raises intriguing questions. What properties are unique to tissue-specific, repeat-expressing, alternatively spliced genes? Are introns containing repetitive elements spliced out more efficiently than introns without repeats, as appears to be the case for SLC4A9? What spliceosomal properties are responsible for frequent unconventional processing, in this case of four exons (8, 10, 11 and 20)? How did the repetitive elements 3' of the coding region become incorporated into the splicing framework of the gene?
SLC4A9 is a case study in the complexities of splicing. To identify such complexities, automated computational approaches to analyzing the structures of novel genes will have to incorporate full-length sequences of multiple long-insert cDNA clones. It is not known how many riddles similar to SLC4A9 there will be in the complete human genome sequence. Their very existence suggests, however, that individually characterizing and understanding numerous unconventional genes will be a major challenge.
Materials and methods
PCR-based screening of cDNA libraries
PCR with the Advantage 2 Polymerase Mix was performed in 50-μl volumes. Undiluted library lysate (1.0 μl to 5.0 μl) was used as template. PCR conditions were as suggested by the manufacturer (Clontech). The vector-specific 5-LDA or 3-LDA primers were the forward primers, and the 5'-directed primers oi-E, 6797up, and 5219up (see Table 1 for complete primer listing), designed from the 5'-most known part of the cDNA, were the reverse primers. Because of the lack of a priori knowledge about the anticipated size of SLC4A9 PCR products, if any, in the product mixture, TA cloning with the Original TA Cloning Kit and INVaF' host (Invitrogen) was performed directly on fresh unpurified total PCR products. Each unique TA clone (defined by a combination of Unigene F/R PCR product length and HinfIII restriction digest pattern) was amplified with the Unigene primers and sequenced.
Hybridization screening of cDNA libraries
A 345-bp portion in the 5' end of the insert of TA clone 3LD-oiE.TA.6 was amplified with primers 345F and 345R, gel-purified, 32P-labeled, and used to probe first-round filters of the adult kidney cDNA library in λTripleX. The filters were prehybridized for 1.5 h, hybridized overnight at 62.5°C, washed, and exposed to Biomax MR film (Kodak) for 18-72 h at -80°C. Cored plaques corresponding to positive clones were subjected to PCR as described below. For clones consistently yielding a smear or multiple bands, in vivo excision of the λTripleX insert into a pTripleX plasmid (using the Cre-Lox system in a BM25.8 recombinase-expressing host) was conducted and plasmid minipreps (Qiagen Spin Plasmid Kit) were obtained for PCR and sequencing.
PCR on phage clones
PCR with the Advantage 2 Polymerase Mix was performed in 50-μl volumes, using primer pairs 5-LDA/345R or 345F/3-LDA to amplify the entire insert as two overlapping products.
DNA sequencing
All sequencing except SNP detection, which is detailed below, was done with the BigDye terminator sequencing kit (PE Biosystems, Foster City, CA) using LongRanger premixed gels (FMC/BioWhittaker) on an Applied Biosystems 377-XL96 DNA sequencer.
SNP discovery
M13-21F and M13-28R-tagged primers were designed from intronic sequence to amplify every consensus exon of SLC4A9 plus at least 50 bp of the flanking introns. After Sephacryl HR-500 purification, amplicons were sequenced using the BigDye primer sequencing kit (PE Biosystems). SNPs were operationally defined as dual-color peaks half the height of the surrounding peaks, reproducible twice in both sequencing directions.
Northern blotting
SLC4A9 expression was first assayed by hybridization of two gene-specific probes, separately, to Clontech MTN blots I, II and III. The first probe was a mixture of the gel-purified, PCR-amplified inserts of IMAGE clones 1533693 and 1734773. The second was the 345F-345R PCR fragment of TA clone 3LD-oiE.TA.6. For Figure 2, membranes were prehybridized for 1 h and hybridized for 4 h at 62.5°C in QuikHyb solution (Stratagene). Positive control hybridization of a human β-actin cDNA probe (Clontech) to MTN 1 and 2 (Figure 2) confirmed the uniform loading of mRNA in each lane.
Sequence analysis
WU-BLAST [31] at EBI [32] and BLAST 2.0 [31] at NCBI [33] were used to search public databases. Other NCBI resources, in particular Pairwise BLAST, Entrez, MapView, and GeneMap '99, were used for the retrieval and analysis of sequence and map information pertaining to the genes whose structures and map positions are discussed in this report. SeqHelp 1.0b [12] was used for all sequence annotation. Protein feature display and alignments for Figure 3, and sequence preparation for Figures 1 and 5, were performed with Vector NTI Suite 5.5 (Informax Inc).
Phylogenetic analysis
The longest complete protein sequence was retrieved from GenPept (NCBI) for each human gene included in the analysis. The BLink feature of GenPept was then used to find nonhuman orthologs of each human EGF, SLC4A and FGF gene under consideration, and their longest sequences were retrieved as well. Sequences were first autoaligned using the AlignX feature of Vector NTI Suite 5.5. Each alignment was manually edited to eliminate divergent amino and carboxy termini and orphan-exon insertions, and to maximize the number of identical and highly conserved consensus positions. The manually edited alignments were exported to PHYLIP for distance calculation and tree construction.
Additional data files
Additional data files available with the online version of this article include:
For the EGF family:
Uncorrected p unbootstrapped neighbor-joining tree
Gamma-corrected unbootstrapped neighbor-joining tree
Poisson-corrected unbootstrapped neighbor-joining tree
Unbootstrapped maximum parsimony tree
Uncorrected p bootstrapped neighbor-joining tree
Gamma-corrected bootstrapped neighbor-joining tree
Poisson-corrected bootstrapped neighbor-joining tree
Bootstrapped maximum parsimony tree
For the FGF family:
Uncorrected p unbootstrapped neighbor-joining tree
Gamma-corrected unbootstrapped neighbor-joining tree
Poisson-corrected unbootstrapped neighbor-joining tree
Unbootstrapped maximum parsimony tree
Uncorrected p bootstrapped neighbor-joining tree
Gamma-corrected bootstrapped neighbor-joining tree
Poisson-corrected bootstrapped neighbor-joining tree
Bootstrapped maximum parsimony tree
For the SLC4A family:
Uncorrected p unbootstrapped neighbor-joining tree
Gamma-corrected unbootstrapped neighbor-joining tree
Poisson-corrected unbootstrapped neighbor-joining tree
Unbootstrapped maximum parsimony tree
Uncorrected p bootstrapped neighbor-joining tree
Gamma-corrected bootstrapped neighbor-joining tree
Supplementary Material
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.
Acknowledgments
Acknowledgements
This research was supported in part by the NIH (grant DC 01076). We thank John G. Quigley for a critical review of the manuscript. The sequence of the major splice isoform of SLC4A9 has been submitted to GenBank (accession number AF313465).
References
- Choi I, Romero MF, Khandoudi N, Bril A, Boron WF. Cloning and characterization of a human electrogenic Na+:HCO3- cotransporter isoform (hhNBC). Am J Physiol. 1999;276:C576–C584. doi: 10.1152/ajpcell.1999.276.3.C576. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Sasaki S, Marumo F. Molecular cloning of a new sodium bicarbonate cotransporter cDNA from human retina. Biochem Biophys Res Commun. 1998;246:535–538. doi: 10.1006/bbrc.1998.8658. [DOI] [PubMed] [Google Scholar]
- Pushkin A, Abuladze N, Lee I, Newman D, Hwang J, Kurtz I. Cloning, tissue distribution, genomic organization, and functional characterization of NBC3, a new member of the sodium bicarbonate cotransporter family. J Biol Chem. 1999;274:16569–16575. doi: 10.1074/jbc.274.23.16569. [DOI] [PubMed] [Google Scholar]
- Abuladze N, Lee I, Newman D, Hwang J, Boorer K, Pushkin A, Kurtz I. Molecular cloning, chromosomal localization, tissue distribution, and functional expression of the human pancreatic sodium bicarbonate cotransporter. J Biol Chem. 1998;273:17689–17695. doi: 10.1074/jbc.273.28.17689. [DOI] [PubMed] [Google Scholar]
- Romero MF, Boron WF. Electrogenic Na+:HCO3- cotransporters: cloning and physiology. Annu Rev Physiol. 1999;61:699–723. doi: 10.1146/annurev.physiol.61.1.699. [DOI] [PubMed] [Google Scholar]
- Burnham CE, Amlal H, Wang Z, Shull GE, Soleimani M. Cloning and functional expression of a human kidney Na+:HCO3- cotransporter. J Biol Chem. 1997;272:19111–19114. doi: 10.1074/jbc.272.31.19111. [DOI] [PubMed] [Google Scholar]
- Amlal H, Wang Z, Burnham C, Soleimani M. Functional characterization of a cloned human kidney Na+:HCO3- cotransporter. J Biol Chem. 1998;273:16810–16815. doi: 10.1074/jbc.273.27.16810. [DOI] [PubMed] [Google Scholar]
- Pushkin A, Abuladze N, Lee I, Newman D, Hwang J, Kurtz I. Mapping of the human NBC3 (SLC4A7) gene to chromosome 3p22. Genomics. 1999;58:321–322. [PubMed] [Google Scholar]
- Amlal H, Burnham CE, Soleimani M. Characterization of Na+/ HCO3- cotransporter isoform NBC-3. Am J Physiol. 1999;276:F903–F913. doi: 10.1152/ajprenal.1999.276.6.F903. [DOI] [PubMed] [Google Scholar]
- Pushkin A, Abuladze N, Newman D, Lee I, Xu G, Kurtz I. Cloning, characterization and chromosomal assignment of NBC4, a new member of the sodium bicarbonate cotransporter family. Biochim Biophys Acta. 2000;1493:215–218. doi: 10.1016/s0167-4781(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Yano H, Wang C, Yamashita S, Yokoyama Y, Yokoi N, Seino S. Assignment of the human solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 gene (SLC4A10) to 2q23→q24 by in situ hybridization and radiation hybrid mapping. Cytogenet Cell Genet. 2000;89:276–277. doi: 10.1159/000015633. [DOI] [PubMed] [Google Scholar]
- Lee MK, Lynch ED, King MC. SeqHelp: a program to analyze molecular sequences utilizing common computational resources. Genome Res. 1998;8:306–312. doi: 10.1101/gr.8.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neural Network Promoter Prediction http://www.fruitfly.org/seq_tools/promoter.html
- Raab G, Klagsbrun M. Heparin-binding EGF-like growth factor. Biochim Biophys Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]
- Tsuganezawa H, Kobayashi K, Iyori M, Araki T, Koizumi A, Watanabe SI, Kaneko A, Fukao T, Monkawa T, Yoshida T, et al. A new member of the HCO3- transporter superfamily is an apical anion exchanger of beta-intercalated cells in the kidney. J Biol Chem. 2000 doi: 10.1074/jbc.M004513200. http://www.jbc.org/cgi/reprint/M004513200v1 [DOI] [PubMed] [Google Scholar]
- TMPRED http://www.isrec.isb-sib.ch/software/TMPRED_form.html
- Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. http://www.cbs.dtu.dk/services/NetPhos [DOI] [PubMed] [Google Scholar]
- Phylchenkov AA. Cytokines of the EGF superfamily and oncogenesis. Exp Oncol [Russian] 1998;20:83–108. [Google Scholar]
- Pathak BG, Gilbert DJ, Harrison CA, Luetteke NC, Chen X, Klagsbrun M, Plowman GD, Copeland NG, Jenkins NA, Lee DC. Mouse chromosomal location of three EGF receptor ligands: amphiregulin (Areg), betacellulin (Btc), and heparin-binding EGF (Hegfl). Genomics. 1995;28:116–118. doi: 10.1006/geno.1995.1116. [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, Persico MG. Molecular characterization of a gene of the EGF family expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989;8:1987–1991. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draft Human Genome Browser. September 2000 release http://genome.ucsc.edu/goldenPath/septTracks.html
- Washington University Genome Sequencing Center: Human BAC Accession Map. September 5, 2000 freeze http://genome.wustl.edu:8021/pub/gsc1/fpc_files/freeze_2000_09_05/MAP/
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg TG, Landsman D. A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res. 1997;25:1626–1632. doi: 10.1093/nar/25.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan K, Michael TH, Jiang GJ, Hiel H, Fuchs PA. A molecular mechanism for electrical tuning of cochlear hair cells. Science. 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- Pebusque MJ, Coulier F, Birnbaum D, Pontarotti P. Ancient large-scale genome duplications: phylogenetic and linkage analyses shed light on chordate genome evolution. Mol Biol Evol. 1998;15:1145–1159. doi: 10.1093/oxfordjournals.molbev.a026022. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Phylogenetic tests of the hypothesis of block duplication of homologous genes on human chromosomes 6, 9, and 1. Mol Biol Evol. 1998;15:854–870. doi: 10.1093/oxfordjournals.molbev.a025990. [DOI] [PubMed] [Google Scholar]
- Jekely G, Friedrich P. The evolution of the calpain family as reflected in paralogous chromosome regions. J Mol Evol. 1999;49:272–281. doi: 10.1007/pl00006549. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- European Bioinformatics Institute http://www.ebi.ac.uk/blast2
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.
Alignment.
Uncorrected p unbootstrapped neighbor-joining tree.
Gamma-corrected unbootstrapped neighbor-joining tree.
Poisson-corrected unbootstrapped neighbor-joining tree.
Unbootstrapped maximum parsimony tree.
Uncorrected p bootstrapped neighbor-joining tree.
Gamma-corrected bootstrapped neighbor-joining tree.
Poisson-corrected bootstrapped neighbor-joining tree.
Bootstrapped maximum parsimony tree.


