Abstract
Cellular and molecular changes of the periodontium associated with a higher prevalence of oral diseases (e.g., chronic periodontitis) in aged populations have received little attention. Since impaired apoptosis during aging appears to be related to chronic inflammatory disorders, we hypothesized that the expression of genes associated with apoptotic processes are altered in aged healthy and periodontitis-affected gingival tissue. Ontology analysis of 88 genes related to apoptotic pathways was performed in gingival biopsies of healthy and periodontitis sites from young, adult, and aged non-human primates (Macaca mulatta), using the GeneChip® Rhesus Macaque Genome Array. Lower expression of anti-apoptotic and higher expression of pro-apoptotic genes were associated with healthy gingival tissue from young compared with aged animals. Few differences in gene expression were observed in healthy gingival tissue between adult and aged animals. Comparison between healthy and periodontitis gingival tissues showed that the up- or down-regulated apoptotic genes in diseased gingival tissue are different in adults compared with aged animals. These results suggest that apoptotic events normally occurring in gingival tissues could be reduced in aging,and unique aspects of apoptotic pathways are potentially involved in the pathophysiology of perio-dontal disease in adult vs. aged gingival tissues.
Keywords: apoptosis, gingival tissue, gerontology, gene expression, periodontitis
Introduction
Although previous studies have suggested a higher prevalence of periodontitis in elderly compared with younger populations (Streckfus et al., 1999; Albandar and Tinoco, 2002), it remains unclear if this observation is a natural consequence of aging. Consistent with age being a modifier for the prevalence or severity of periodontal disease, important biological differences in the composition and complexity of the oral microbial ecology, as well as the oral/systemic immune-inflammatory responses during aging, have been described in humans and animal models (Schlegel-Bregenzer et al., 1998; Nonnenmacher et al., 2001; Ebersole et al., 2008). Paradoxically, the reduced immunity in aging is accompanied by chronic inflammation (Back et al., 2007), which may reflect a perturbation of the innate immune mechanisms associated with age (Agrawal et al., 2007). Recent evidence supports an important role for apoptosis as an essential mechanism that regulates the inflammatory response through the generation of anti-inflammatory signals affecting phagocytes at the site of the infection, originally recruited to clear the infection (Fadok et al., 1998; Kimet al., 2004). Apoptosis is a mechanism of cell death without inflammation, and is crucial for the maintenance of tissue homeostasis at different stages of life, including embryonic development and normal tissue turnover during adulthood (Vaux and Korsmeyer, 1999). These programmed cell death events appear to occur in a tissue-specific manner with aging (i.e., increased apoptosis in skeletal muscles, neurons, and hepatocytes, and decreased in intestinal epithelial cells) (Holt et al., 1998; Higami and Shimokawa, 2000; Marzetti and Leeuwenburgh, 2006). Similar studies addressing the potential role of apoptosis in periodontitis have shown increased expression of apoptotic biomolecules, mainly by neutrophils and mononuclear cells (Gamonal et al., 2001), as well as down-regulation of pro-apoptotic molecules in lymphocytic cells associated with chronic adult periodontitis (Lucas et al., 2010). Nevertheless, additional reports have not confirmed a significant role for apoptosis in periodontitis (Koulouri et al., 1999). Most of these studies evaluated apoptotic changes in periodontitis using gingival biopsies from healthy and periodontitis-affected individuals within a broad age range, and for a limited number of molecules related to apoptosis.
There is limited evidence concerning biological changes of the gingival tissue that may help us better understand the higher prevalence and severity of oral diseases (e.g., periodontal disease) observed in an elderly population. It was recently described that the expression of pro-apoptotic markers related to both the intrinsic (i.e., cytochrome C) and extrinsic (i.e., caspase-3, tumor necrosis factor receptor-associated via death domain-TRADD and Bax) apoptotic pathways correlated with periodontitis in gingival biopsies from elderly patients (Das et al., 2009), which suggests that age could in fact determine a differential expression of apoptotic molecules within the periodontium (Das et al., 2009). With microarray gene expression technology and a non-human primate model (Struillou et al., 2010), age-related molecular gingival changes associated with the pathogenesis of periodontal disease were evaluated. Specifically, we developed an ontological analysis of apoptotic genes expressed in healthy gingival tissue with regard to age, as well as age-related variations of apoptotic gene expression in healthy compared with periodontitis-affected gingival tissues.
Materials & Methods
Animals and Diet
Rhesus monkeys (Macaca mulatta) (n = 15; 11 females and 4 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. Healthy animals (3/group) were distributed by age in 3 groups as follows: ≤ 3 yrs (young), 12-15 yrs (adult), and 18-22 yrs (aged).Only adult and aged animals (3 additional animals/group) with periodontitis were used, since periodontitis does not occur in young animals. The non-human primates were typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad, Madison, WI). The diet was supplemented with fruits and vegetables, and water was provided ad libitum in an enclosed corral setting.
Oral Clinical Parameters and Gingival Tissue Sample Collection
Following the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, anesthetized animals were examined by a single investigator using a Maryland probe on the facial aspect of the teeth, 2 proximal sites per tooth (mesio- and disto-buccal), excluding the canines and 3rd molars. The clinical examination included probing pocket depth (PD) and bleeding on probing (BOP; 0-3 scale) (Ebersole et al., 2008). Clinical characterization for each group (n = 3) included: young animals (mean PD < 2.0 mm, mean BOP < 0.5); adult healthy group (mean PD < 2.0 mm, mean BOP < 0.3); adult periodontitis group (mean PD > 3.4 mm, mean BOP > 1.0); aged healthy group (mean PD < 3.0 mm, mean BOP < 1); mean BOP < 1.0); and aged periodontitis group (mean PD > 3.4 mm, mean BOP > 1.5). With a standard gingivectomy technique, 2 adjacent buccal gingival samples from either healthy (PD ≤ 2 mm, no BOP) or periodontitis-affected tissue (PD > 4 mm, BOP ≥ 1) from the premolar/molar region of each animal were taken and handled individually to prepare RNA for microarray analysis.
RNA Extraction, Reverse Transcription,and Gene Chip Hybridizations
Total RNA was isolated from each gingival tissue with Trizol reagent (Invitrogen, Carlsbad, CA, USA). The extracted RNA was cleaned up with the Qiagen RNeasy mini kit (Qiagen, Valencia, CA, USA) and quantified by spectrophotometric analysis. Equal amounts of RNA were pooled from each sample for an individual animal, followed by a reverse transcription, and hybridization to the GeneChip® Rhesus Macaque Genome Array (Affymetrix, Santa Clara, CA, USA), similar to methods we have described previously (Meka et al., 2010). Probe arrays were scanned with the GeneChip Scanner 3000 for high-resolution scanning and GeneChip Operating Software MAS 5.0.
Data Analysis
The average gene expression for 88 genes associated with apoptosis pathways in the database of the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.genome.jp) (Kanehisa et al., 2010) was compared between different age groups and between healthy and periodontitis gingival tissues by the Student’s t test. We performed ontology analysis by mapping the statistically differentially expressed genes (p ≤ 0.05) between age groups into the KEGG pathways as previously described (Demmer et al., 2008). Using a p-value cutoff of p ≤ 0.05, we expected only up to 4 or 5 false-positives by chance out of 88 genes when applying each t test.
Results
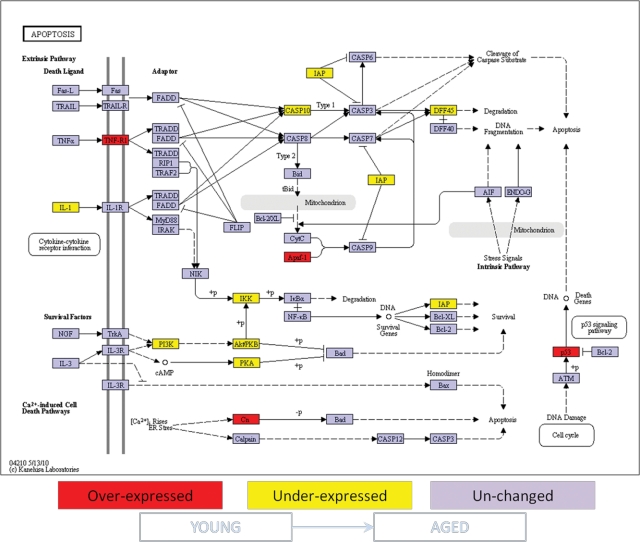
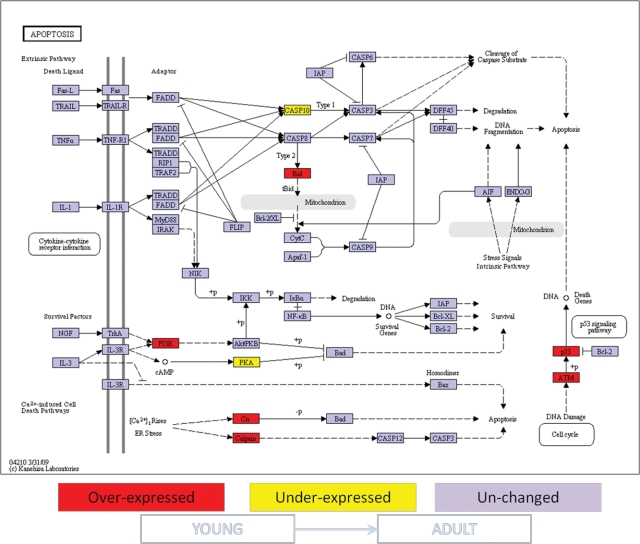
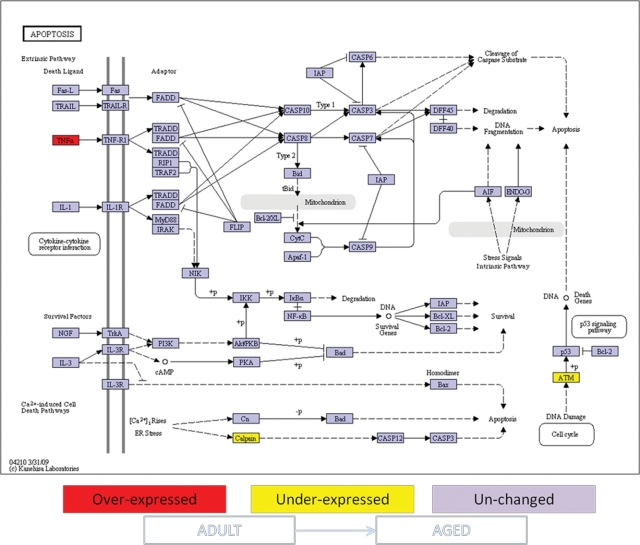
The expression of 88 genes related to apoptosis pathways in healthy gingival tissue was compared among samples from young, adult, and aged animals. Significant differences in gene expression were found between young and either adult (10 genes) or aged (14 genes) gingival tissues. Ontology analysis showed that the differentially expressed genes in gingival tissue with aging involved both pro- and anti-apoptotic molecules. Specifically, the gingival tissue from young animals exhibited higher expression of pro-apoptotic genes such as tumor necrosis factor receptor 1 (TNFR1), the apoptotic peptidase activating factor 1 (APAF-1), p53, calcium-binding protein P22 (Cn), and Calpain and BH3 interacting domain death agonist (BID), and lower expression of negative regulators of apoptosis, e.g., survival genes, including baculoviral IAP repeat-containing 2 (IAP), phosphatidylinositol 3-kinase (PI3K), protein kinase A (PKA). and the IkappaB kinase (IKK), when compared with aged and adult gingival tissues (Figs. 1, 2). The PI3K intracellular signal transducer group of biomolecules was expressed at higher levels in young vs. adult gingival tissues (Fig. 2), although their expression levels were significantly lower in samples from young animals compared with aged animals (Fig. 1). Lower numbers of statistically differentially expressed genes were found when adult and aged gingival tissues were compared (3 genes) (Fig. 3). Interestingly, the expression of the classic inflammatory molecules TNFR1 and TNFα was up-regulated in gingival tissues from young and adult animals, respectively, compared with aged animals (Figs. 1-3).
Figure 1.
Ontology analysis of apoptotic genes expressed in healthy gingival tissues from young vs. aged non-human primates. Comparison of apoptosis gene expression was done between healthy gingival tissues from young vs. aged animals as described in MATERIALS & METHODS. Genes shown in red are over-expressed, and genes shown in yellow are under-expressed in young gingival tissue compared with aged gingival tissue. Expression of genes shown in purple was unchanged at p ≤ 0.05 significance. +p = phosphorylation event; -p = dephosphorylation event.
Figure 2.
Ontology analysis of apoptotic genes expressed in healthy gingival tissues from young vs. adult non-human primates. Comparison of apoptosis gene expression was done between healthy gingival tissues from young vs. aged animals as described in MATERIALS & METHODS. Genes shown in red are over-expressed, and genes shown in yellow are under-expressed in young gingival tissue compared with adult gingival tissue. Expression of genes shown in purple was unchanged at p ≤ 0.05 significance. +p = phosphorylation event; -p = dephosphorylation event.
Figure 3.
Ontology analysis of apoptotic genes expressed in healthy gingival tissues from adult vs. aged non-human primates. Comparison of apoptosis gene expression was done between healthy gingival tissues from young vs. aged animals as described in MATERIALS & METHODS. Genes shown in red are over-expressed, and genes shown in yellow are under-expressed in adult gingival tissue compared with aged gingival tissue. Expression of genes shown in purple was unchanged at p ≤ 0.05 significance. +p = phosphorylation event; -p = dephosphorylation event.
Statistically significant differential expression of 12 genes (adult) and 16 genes (aged) was found when healthy gingival tissues were compared with periodontitis tissues (Table). Strikingly, the genes in the apoptotic pathways either significantly up- or down-regulated in gingival tissue during periodontal disease were unique subsets in samples from adults vs. those obtained from aged animals.
Table.
Genes are Differentially Expressed in Periodontitis vs. Healthy Gingival Tissues from Adult and Aged Non-human Primates
| Adult |
Aged |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe # | Healthy | Periodontitis | Fold-change | P Value | Healthy | Periodontitis | Fold-change | P Value | |
| Pro-apoptosis | |||||||||
| BCL2L10 | MmugDNA.5988.1.S1_at | 159.9 | 236.4 | 1.5 | 0.031 | 175.2 | 255.1 | 1.5 | 0.123 |
| CAPN1 | MmugDNA.2588.1.S1_at | 2694.5 | 3556.5 | 1.3 | 0.038 | 3286.7 | 3488.1 | 1.1 | 0.757 |
| NOD1 | Mmu.9460.1.S1_s_at | 3240.7 | 2909.8 | 0.9 | 0.029 | 3031.3 | 2808.9 | 0.9 | 0.226 |
| PPP3CB | Mmu.9989.1.S1_s_at | 773.5 | 718.3 | 0.9 | 0.004 | 821.2 | 689.9 | 0.8 | 0.257 |
| CHP | MmugDNA.2197.1.S1_at | 315.1 | 224.8 | 0.7 | 0.017 | 276.8 | 318.4 | 1.2 | 0.477 |
| TNFRS11B | MmugDNA.5860.1.S1_at | 244.3 | 152.3 | 0.6 | 0.044 | 182.3 | 188.8 | 1.0 | 0.904 |
| APAF1 | MmugDNA.37857.1.S1_at | 42.6 | 30.8 | 0.7 | 0.617 | 19.6 | 45.6 | 2.3 | 0.006 |
| CSF2RB | MmugDNA.35543.1.S1_at | 3102.3 | 4439.2 | 1.4 | 0.276 | 2848 | 4555.4 | 1.6 | 0.016 |
| CASP3 | MmugDNA.6342.1.S1_at | 1008.3 | 874.5 | 0.9 | 0.575 | 731.2 | 1132.6 | 1.5 | 0.018 |
| DAPK1 | MmugDNA.31762.1.S1_at | 139.1 | 170.3 | 1.2 | 0.151 | 140.7 | 184.3 | 1.3 | 0.009 |
| TRAF 3 | MmugDNA.10177.1.S1_at | 381.0 | 329.8 | 0.9 | 0.405 | 293.0 | 388.2 | 1.3 | 0.005 |
| CASP1 | MmugDNA.31375.1.S1_s_at | 563.7 | 713.4 | 1.3 | 0.172 | 820.8 | 600.3 | 0.7 | 0.002 |
| CASP4 | MmugDNA.24199.1.S1_at | 2352.5 | 2854.4 | 1.2 | 0.396 | 3140.9 | 1997.9 | 0.6 | 0.042 |
| CD40 (TNFRSF5) | MmugDNA.20032.1.S1_at | 27.5 | 24.4 | 0.9 | 0.849 | 14.2 | 5.8 | 0.4 | 0.040 |
| Anti-apoptosis | |||||||||
| IGF1R | MmugDNA.12698.1.S1_at | 18.1 | 46.8 | 2.6 | 0.047 | 23.3 | 10.8 | 0.5 | 0.265 |
| PRKX | MmugDNA.3488.1.S1_at | 25.7 | 41.7 | 1.6 | 0.048 | 41.2 | 42.9 | 1.0 | 0.919 |
| NFKB1 | MmugDNA.25322.1.S1_at | 652.0 | 799.1 | 1.2 | 0.017 | 760.0 | 737.2 | 1.0 | 0.688 |
| PRKAR1A | MmuSTS.3698.1.S1_at | 12146.3 | 10773.6 | 0.9 | 0.036 | 11708.5 | 10383.9 | 0.9 | 0.213 |
| NOL3 | MmugDNA.27369.1.S1_at | 401.6 | 301.6 | 0.8 | 0.044 | 339.1 | 310.4 | 0.9 | 0.481 |
| BNIP2 (II) | MmugDNA.2960.1.S1_at | 43.0 | 18.1 | 0.4 | 0.047 | 14.0 | 27.1 | 1.9 | 0.293 |
| NTRK1 | MmugDNA.14382.1.S1_at | 23.9 | 43.3 | 1.8 | 0.077 | 17.2 | 62.3 | 3.6 | 0.027 |
| BNIP3L | MmugDNA.32220.1.S1_at | 36.7 | 50.0 | 1.4 | 0.339 | 40.1 | 77.3 | 1.9 | 0.026 |
| PRKACA | MmuSTS.3696.1.S1_at | 590.6 | 631.1 | 1.1 | 0.617 | 567.4 | 693.2 | 1.2 | 0.014 |
| RELA | MmuSTS.3377.1.S1_at | 1360.6 | 1312.5 | 1.0 | 0.694 | 1295.9 | 1434.4 | 1.1 | 0.032 |
| BNIP2 (I) | MmugDNA.31249.1.S1_at | 2217.0 | 2374.6 | 1.1 | 0.537 | 2347.5 | 2031.5 | 0.9 | 0.022 |
| BCL2L2 | MmugDNA.17683.1.S1_at | 348.8 | 319.4 | 0.9 | 0.458 | 353.0 | 300.5 | 0.9 | 0.013 |
| CD2 (cytoplasmic tail) | MmugDNA.32305.1.S1_at | 323.6 | 405.6 | 1.3 | 0.213 | 479.8 | 372.7 | 0.8 | 0.025 |
| PIK3CA | MmuSTS.1135.1.S1_at | 1665.3 | 1696.2 | 1.0 | 0.899 | 1749.8 | 1395.0 | 0.8 | 0.021 |
The average expression and fold-change for 28 genes related to apoptotic pathways that showed significant differences (p ≤ 0.05) between healthy and periodontitis gingival tissue from either adult or aged animals is shown. Genes were divided as positive (pro-apoptosis) or negative (anti-apoptosis) regulators of programmed cell death. The corresponding probe number IDs for the listed genes were obtained from the database of the Affymetrix Web site with the NetAffx Query software (www.affymetrix.com).
Discussion
In this study, we sought to determine the potential age-related changes in the expression of genes involved in apoptosis pathways in healthy and diseased gingival tissue from young, adult, and aged non-human primates (M. mulatta). We demonstrated a clear age-related change in the expression of genes in apoptotic pathways in healthy tissues. Additionally, unique subsets of apoptotic genes showed altered expression in health and disease in the adult animals compared with aged animals. Historically, periodontitis in adults and the elderly has been generally considered a similarly chronic immune-inflammatory condition, with the increased risk associated with aging as simply a reflection of the longer-term insult to the periodontium from accumulated oral bacterial biofilms (Schlegel-Bregenzer et al., 1998; Nonnenmacher et al., 2001). However, since we now recognize that the molecular inflammatory fingerprint of gingivitis differs from that of periodontitis (Honda et al., 2006), these findings suggest that tissue destruction in periodontitis may actually have some fundamental differences in mechanism in aged individuals.
Microarray analyses showed that healthy tissues from aged and adult animals appear to exhibit a lower pro-apoptotic and higher anti-apoptotic net gene expression compared with healthy gingival tissues from young animals. These results suggest that apoptotic events that normally occur in the gingival tissue could be reduced in adult/aged individuals compared with young animals. Hypothetically, these physiological changes could involve an adaptation process of the gingival tissue, where the reduced proliferative capacity that is classically observed in the elderly could be compensated for with higher resistance to cell death under stressful environmental conditions associated with the chronic microbial challenge. Similarly, environmental factors as well as aging itself have been associated with the accumulation of gene mutations that could potentially change the ability of the tissues to properly activate specific cell responses as apoptosis (Lee et al., 2010).
The relationship of pro- or anti-apoptotic events with susceptibility for periodontal disease remains unclear. Evidence suggests that the majority of apoptotic events appear to be related to immune cells (i.e., neutrophils, monocytes/macrophages, and lymphocytic cells), rather than epithelial cells or fibroblasts, the major structural cells of the periodontium (Sawa et al., 1999; Gamonal et al., 2003). Hence, with this evidence, along with additional studies that confer an immunomodulatory role on apoptosis (Torchinsky et al., 2010), one could hypothesize that the reduced apoptotic responses in aged gingival tissues could involve failure to control the inflammatory response against the chronic bacterial challenge, resulting in dysregulated clearance of the inflammatory infiltrate. In agreement with this hypothesis, Bodineau et al. recently showed an increased number of mature dendritic cells in elderly patients with chronic periodontitis (Bodineau et al., 2009). Thus, it would be expected that re-establishing the physiological pro-apoptotic mechanisms in aging, specifically within the inflammatory cell population, could reduce the inflammatory response classically observed in periodontal disease, as has been proposed for other chronic inflammatory disorders (Lugering et al., 2006). The susceptibility to undergo apoptosis could vary among different cell types (Higami and Shimokawa, 2000). Therefore, the selection of apoptotic markers in future studies of periodontal disease should include both enhancers and inhibitors of apoptosis, using experimental strategies that facilitate discrimination of the net cell types within the gingival tissue that express these apoptotic changes, and more attention to discrimination of patient populations based upon age.
An additional and startling observation was that the genes differentially expressed in periodontitis vs. healthy gingival tissues from adult animals were mutually exclusive to those genes expressed in tissues from aged animals. These results are consistent with a potential role of apoptosis in the pathogenesis of periodontal disease, as has been previously suggested (Gamonal et al., 2001; Lucas et al., 2010), although further functional studies involving these genes will be required for full evaluation of their role in the complex processes needed to maintain homeostasis in the periodontium. Variations in gene expression associated with periodontitis included both over-expression- and down-regulation of genes that are in pathways to enhance or inhibit apoptosis. Although these findings clearly show gene expression profiles related to regulation of apoptotic events during periodontitis, the net results of altered expression of specific enhancers or inhibitors of apoptosis may change throughout the disease process, emphasizing the need for further prospective controlled studies of the initiation and progression of periodontal lesions.
Changes in the expression of classic inflammatory molecules such as TNFR1 that was observed in gingival tissues from adult but not aged animals, and the significantly increased expression of APAF1 in diseased gingival tissue from aged but not adult animals, are examples reinforcing the fact that the apoptotic mechanisms contributing to periodontitis in aged individuals differ from those in adults. Similarly, there was an age-related expression of the critical enzyme PI3K with fluctuating variations among young, adult, and aged gingival tissues. Importantly, PI3K-dependent resistance to apoptosis during Porphyromonas gingivalis infection has been shown in oral epithelial cells as a tissue invasion mechanism (Yilmaz et al., 2004). Thus, higher apoptosis resistance in aged gingival tissue, related to increased levels of PI3K, may increase the likelihood for periodontopathogenic tissue infection and further onset of periodontitis. Additional results indicated that, among the genes highly expressed in diseased gingival tissues, there were molecules that have not been classically associated with periodontitis, such as IGF1R (insulin growth factor type 1 receptor) in adult gingival tissue, and APAF1 and NTRK1 (neurotrophic tyrosine kinase receptor type 1) in the aged gingival tissues. Future studies addressing the potential role of these molecules in periodontitis appear warranted.
Age-associated diseases (e.g., cardiovascular disease, cancer, arthritis, osteoporosis, type 2 diabetes, Alzheimer’s disease) increase rapidly with aging and are distinguished from the aging process, since all humans age, but not all experience all age-associated diseases. Periodontitis has often been considered a disease of aging, rather than an age-associated disease, being expressed as more continuous undermining of the integrity of the periodontium that accumulates in response to the noxious microbial challenge. Our findings suggest that periodontitis may more accurately reflect an age-associated disease, with somewhat unique/distinct molecular characteristics, as reflected by the gingival apoptotic events inferred by this study.
In general, we have shown that: (i) the net expression of apoptotic genes involved in the homeostasis of the healthy gingival tissue appears to change with age toward a more apoptosis-resistant phenotype, and (ii) the apoptotic pathway gene profile expressed in periodontitis tissues differs in aged when compared with adult gingival tissues. Besides the expression of apoptosis-associated genes, post-transcriptional and post-translational events (e.g., sumoylation, microRNAs, phosphorylation, and ubiquitination) also regulate apoptotic events (Huen and Chen, 2008). Therefore, further investigations to confirm the gingival apoptotic outcomes during health and periodontitis related to age are necessary. These observations will need to be replicated in clinical studies with humans, and could help to clarify inconsistent findings concerning apoptosis in periodontal disease in humans, which normally included populations with a broad range of ages. A better understanding of the age-associated molecular changes of the periodontium and how those variations correlate with oral health or disease is required, since differences in underlying molecular mechanisms of this biologic process in aged individuals could have an effect on more specific therapies to benefit this rapidly expanding subgroup of the US population.
Footnotes
This work was supported by grant 2P20RR020145-06 from the NCRR (National Institutes of Health). We express our gratitude to the Caribbean Primate Research Center (CPRC) for invaluable technical support, especially Edmundo Kraiselburd, for providing guidance and support for the conduct of this study.
References
- Agrawal A, Agrawal S, Cao JN, Su H, Osann K, Gupta S. (2007). Altered innate immune functioning of dendritic cells in elderly humans: arole of phosphoinositide 3-kinase-signaling pathway. J Immunol 178:6912-6922 [DOI] [PubMed] [Google Scholar]
- Albandar JM, Tinoco EM. (2002). Global epidemiology of periodontal diseases in children and young persons. Periodontol 2000 29:153-176 [DOI] [PubMed] [Google Scholar]
- Back M, Hlawaty H, Labat C, Michel JB, Brink C. (2007). The oral cavity and age: a site of chronic inflammation? PLoS One 2:e1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodineau A, Coulomb B, Tedesco AC, Seguier S. (2009). Increase of gingival matured dendritic cells number in elderly patients with chronic periodontitis. Arch Oral Biol 54:12-16 [DOI] [PubMed] [Google Scholar]
- Das P, Chopra M, Sun Y, Kerns DG, Vastardis S, Sharma AC. (2009). Age-dependent differential expression of apoptosis markers in the gingival tissue. Arch Oral Biol 54:329-336 [DOI] [PubMed] [Google Scholar]
- Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. (2008). Transcriptomes in healthy and diseased gingival tissues. J Periodontol 79:2112-2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. (2008). Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol 15:1067-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. (1998). Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 101:890-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamonal J, Bascones A, Acevedo A, Blanco E, Silva A. (2001). Apoptosis in chronic adult periodontitis analyzed by in situ DNA breaks, electron microscopy, and immunohistochemistry. J Periodontol 72:517-525 [DOI] [PubMed] [Google Scholar]
- Gamonal J, Sanz M, O’Connor A, Acevedo A, Suarez I, Sanz A, et al. (2003). Delayed neutrophil apoptosis in chronic periodontitis patients. J Clin Periodontol 30:616-623 [DOI] [PubMed] [Google Scholar]
- Higami Y, Shimokawa I. (2000). Apoptosis in the aging process. Cell Tissue Res 301:125-132 [DOI] [PubMed] [Google Scholar]
- Holt PR, Moss SF, Heydari AR, Richardson A. (1998). Diet restriction increases apoptosis in the gut of aging rats. J Gerontol A Biol Sci Med Sci 53:B168-B172 [DOI] [PubMed] [Google Scholar]
- Honda T, Domon H, Okui T, Kajita K, Amanuma R, Yamazaki K. (2006). Balance of inflammatory response in stable gingivitis and progressive periodontitis lesions. Clin Exp Immunol 144:35-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Chen J. (2008). The DNA damage response pathways: at the crossroad of protein modifications. Cell Res 18:8-16 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. (2010). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38(Database issue):D355-D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Elkon KB, Ma X. (2004). Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity 21:643-653 [DOI] [PubMed] [Google Scholar]
- Koulouri O, Lappin DF, Radvar M, Kinane DF. (1999). Cell division,synthetic capacity and apoptosis in periodontal lesions analysed by in situ hybridisation and immunohistochemistry. J Clin Periodontol 26:552-559 [DOI] [PubMed] [Google Scholar]
- Lee HC, Chang CM, Chi CW. (2010). Somatic mutations of mitochondrial DNA in aging and cancer progression. Ageing Res Rev 9(Suppl 1):47-58 [DOI] [PubMed] [Google Scholar]
- Lucas H, Bartold PM, Dharmapatni AA, Holding CA, Haynes DR. (2010). Inhibition of apoptosis in periodontitis. J Dent Res 89:29-33 [DOI] [PubMed] [Google Scholar]
- Lugering A, Lebiedz P, Koch S, Kucharzik T. (2006). Apoptosis as a therapeutic tool in IBD? Ann NY Acad Sci 1072:62-77 [DOI] [PubMed] [Google Scholar]
- Marzetti E, Leeuwenburgh C. (2006). Skeletal muscle apoptosis, sarcopenia and frailty at old age. Exp Gerontol 41:1234-1238 [DOI] [PubMed] [Google Scholar]
- Meka A, Bakthavatchalu V, Sathishkumar S, Lopez MC, Verma RK, Wallet SM, et al. (2010). Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol 25:61-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher C, Mutters R, de Jacoby LF. (2001). Microbiological characteristics of subgingival microbiota in adult periodontitis, localized juvenile periodontitis and rapidly progressive periodontitis subjects. Clin Microbiol Infect 7:213-217 [DOI] [PubMed] [Google Scholar]
- Sawa T, Nishimura F, Ohyama H, Takahashi K, Takashiba S, Murayama Y. (1999). In vitro induction of activation-induced cell death in lymphocytes from chronic periodontal lesions by exogenous Fas ligand. Infect Immun 67:1450-1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel-Bregenzer B, Persson RE, Lukehart S, Braham P, Oswald T, Persson GR. (1998). Clinical and microbiological findings in elderly subjects with gingivitis or periodontitis. J Clin Periodontol 25(11 Pt 1):897-907 [DOI] [PubMed] [Google Scholar]
- Streckfus CF, Parsell DE, Streckfus JE, Pennington W, Johnson RB. (1999). Relationship between oral alveolar bone loss and aging among African-American and Caucasian individuals. Gerontology 45:110-114 [DOI] [PubMed] [Google Scholar]
- Struillou X, Boutigny H, Soueidan A, Layrolle P. (2010). Experimental animal models in periodontology: a review. Open Dent J 4:37-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchinsky MB, Garaude J, Blander JM. (2010). Infection and apoptosis as a combined inflammatory trigger. Curr Opin Immunol 22:55-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. (1999). Cell death in development. Cell 96:245-254 [DOI] [PubMed] [Google Scholar]
- Yilmaz O, Jungas T, Verbeke P, Ojcius DM. (2004). Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primaryepithelial cells infected with the periodontal pathogen. Porphyromonas gingivalis. Infect Immun 72:3743-3751 [DOI] [PMC free article] [PubMed] [Google Scholar]