Abstract
Accumulating evidence points to a major role for chronic stress of cell renewal systems in the pathogenesis of important human diseases, including cancer, atherosclerosis and diabetes. Here we discuss emerging evidence that epigenetic abnormalities may make substantial contributions to these stress-induced pathologies. Although the mechanisms remain to be fully elucidated, we suggest that chronic stress can elicit heritable changes in the chromatin landscape that ‘lock’ cells in abnormal states, which then lead to disease. We emphasize the need to investigate epigenetic states in disease and links to stress and to consider how the knowledge gained through these studies may foster new means of disease prevention and management.
Over several years, evidence has grown that links chronic cell stress to diseases, which include cancer, cardiovascular disease, diabetes and neurodegenerative disorders1–4. There are many possible explanations for how cell stress promotes disease; for example, direct cytotoxicity, mutagenesis or disruption of intracellular signalling cascades. During development and in adults, chronic stress can also alter the normal balance of cell maturation and division in cell renewal systems and there is emerging evidence that such alterations might be linked to disease.
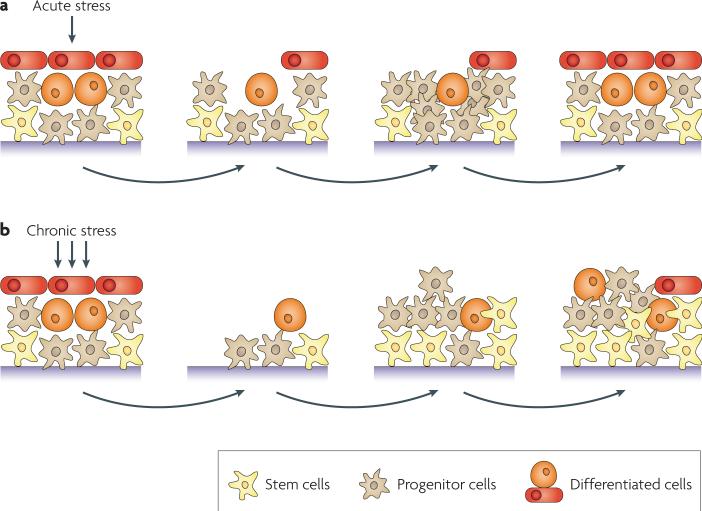
Cell renewal systems, broadly defined, consist of self-renewing stem cells and more committed progenitor cells that give rise to mature cells. Depending on the system, in their basal state, the self-renewing cells may be rapidly dividing (as in the intestinal epithelium) or rarely dividing (as in muscle and neurons). Stress can trigger a change in the stem cell numbers and rate of division5–7. In acute stress, such as transient wounding with rapid repair, increased division of progenitor cells and the subsequent differentiation of their progeny are often required for tissue homeostasis (FIG. 1a). By contrast, prolonged exposure to stress presents a much more severe challenge to cells in self-renewing systems and, to maintain tissue homeostasis, renewal systems are likely to be altered8 (FIG. 1b). Both acute and chronic stress can induce genetic and epigenetic changes although, mathematically, they are more likely to occur during chronic stress. Thus, adaptive pressure due to chronic stress could lead to the evolution of abnormal cell states that contribute to disease.
Figure 1. The effect of stress on cell renewal systems.
a | In an acute stress setting, transient stress might spare tissue stem cells but injure other cell types. This could lead to transient mobilization of progenitor cells (beige) to replenish differentiated cells (red and orange) and achieve tissue homeostasis. b | In a chronic stress setting, there is a more pronounced and ongoing effect on cell renewal systems with injury of all cell types, including stem cells (yellow). The tissue response involves mobilization and continued renewal of both stem cells and progenitor cells. Genetic and epigenetic changes can lead to altered states of these cells with resultant abnormal tissue homeostasis and predisposition to diseases such as cancer.
One outcome of chronic stress might be to establish abnormal cell states that could persist even if the exposure is removed or diminished. A combination of genetic and/or epigenetic changes engendered during the period of stress could result in an altered cellular ‘memory’ that helps to drive disease pathology. epigenetic mechanisms are known to be important for normal cellular memory9. Chromatin compaction and organization — which can be influenced by DNA methylation, histone variants, histone post-translational modifications and nucleosome remodelling — help to determine the gene expression profiles that define and maintain cell identity10–14. Therefore, chromatin structure must be carefully controlled in self-renewing and differentiated cells in cell renewal systems. Indeed, it has been shown that particular cell types have specific epigenetic characteristics, and stem cells differ substantially from terminally differentiated cells13,14.
Importantly, there is now extensive evidence that the cells involved in several major human pathologies have aberrant patterns of epigenetic modifications and, in many cases, these epigenetic patterns may contribute substantially to the disease-causing cellular phenotypes15. In this article, we bring together several examples of diseases in which both chronic stress and epigenetic changes have been implicated. We consider three distinct but potentially coincident consequences of epigenetic alterations that are implicated in disease: abnormal cell proliferation, cell loss and/or dysfunction and alteration of cell state (FIG. 2). These examples lead us to suggest that chronic stress can cause alterations in the chromatin landscape that can drive these three cellular outcomes. Finally, we discuss the future research that is needed to improve our understanding of the role of epigenetics in disease, and how insights into links between stress and epigenetics could have important implications for disease management.
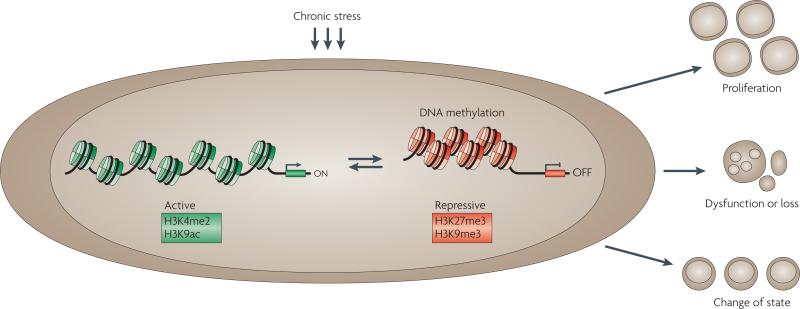
Figure 2. Perturbations caused by chronic stress.
Chronic stress may result in epigenetic changes. For example, alterations in DNA methylation, histone modifications and nucleosome positioning might occur. In the example shown, the changes result in gene silencing. The epigenetic changes may result in heritable patterns of altered gene expression that result in abnormal cell states: proliferation, dysfunction or loss, or a change of state. Such changes can contribute to disease, as discussed in the text. The histone modifications shown here associated with active transcription are histone H3 lysine 4 dimethylation (H3K4me2) and H3 lysine 9 acetylation (H3K9ac); the modifications associated with transcriptional repression are H3 lysine 27 trimethylation (H3K27me3) and H3 lysine 9 trimethylation (H3K9me3).
Abnormal cell proliferation
Cancer
Cancer is currently the most studied disease in terms of understanding how aberrant cell expansion might result from chronic cell stress and the consequent epigenetic alterations. The leading risk states for cancers — including breast, colon, prostate, and lung malignancies — are inflammation and ageing, both of which are states that result in chronic oxidative exposure and increased DNA damage5,8,16–19. These stresses put pressure on stem and/or progenitor cells to increase their numbers and to adapt their survival mechanisms as they attempt to maintain tissue homeostasis5–7,20. For example, in several cell systems, stem cell numbers increase with age but become less efficient in long-term self-renewal and in their capacity to commit to normal cell fates5–7. Such survival adaptations could lead to abnormal cellular changes, which could be sustained by epigenetic alterations that can allow abnormal clones of expanding cells to arise and survive outside their normal context. In tumorigenesis, these changes in epithelial cells may even elicit abnormalities in their cellular environment to support such abnormal growth — that is, evolution of the so-called tumour microenvironment21,22. This scenario constitutes the pre-malignant stages of cancer in which, over time, there is a risk of the expanding cell clones progressing to invasive disease23.
Stress-induced loss of control of cellular proliferation in the early stages of neoplasia can certainly be driven by genetic alterations24, but it is becoming clear that epigenetic processes may influence how genetic abnormalities exert their tumorigenic function25,26. When introduced directly into non-transformed cells, mutations in several key oncogenes or tumour suppressors have different effects depending on the stage of cell differentiation. For example, MYC (also known as c-myc) and KRAS mutations cause apoptosis and/or senescence of mature cells27. loss of adenomatous polyposis coli (APC) function, which leads to increased activity of the Wnt pathway and is a key inducing factor for colon cancer, does not cause chronic expansion of non-stem cells in the intestine28. By contrast, in cells with more primitive or stem cell-like phenotypes, such loss of Apc leads to prolonged proliferation28. The pre-invasive stages of common human cancers can show abnormal promoter DNA hypermethylation and epigenetic silencing of tumour suppressor genes such as APC as an alternative to genetic mutations. Such tumours simultaneously harbour promoter hypermethylation of other genes that play key parts in pathways that regulate stem or progenitor cells25,29.
The loss of function of any of the genes discussed above removes a normal block to expansion of stem-like cell populations and/or prevents normal maturation of these cells25,29. These epigenetic changes associated with cancer are a hallmark of ageing, especially in organs such as the intestine, in which the increased frequency of epigenetic changes parallels the age frequency for colon cancer30,31. epigenetic changes could precede oncogene or tumour suppressor gene mutations or be triggered by them, as suggested for increased KrAS and MYC signalling, which lead to repressive chromatin and gene silencing32,33. Therefore, these changes could create an epigenetic landscape that contributes to the cell becoming chronically dependent on the oncogenic or tumour suppressive pathways that result from a genetic mutation (‘pathway addiction’)34. The stress-induced cell expansion scenarios and epigenetic changes in cancer genes described above can be linked to epigenetic control of development. Many of these cancer genes, including classic tumour suppressors that have abnormal DNA methylation at their promoters in several types of cancer, are marked by repressive polycomb group (PcG) protein complexes during embryonic development35–38. PcG-controlled chromatin maintains these genes in a state of low-level expression, which facilitates retention of pluripotency and self-renewal39,40. These genes are said to be in a ‘poised’ state that allows their transcriptional activation to facilitate cell commitment or differentiation39,40. Control of genes by PcG is not normally associated with promoter DNA methylation in development but PcG complexes can associate with DNA methyltransferases (DNMTs)41 and so they have the potential, under some circumstances, to induce DNA methylation. DNA methylation-mediated gene silencing is less readily reversible than PcG-mediated repression and so the addition of DNA methylation during tumorigenesis would abnormally tighten silencing. This type of event has been observed experimentally by comparing a gene marked by PcG in teratocarcinoma cells with the same gene abnormally DNA hypermethylated in colon cancer cells42. Induction of DNA damage can also recruit PcG proteins to sites of a double-strand DNA break in a gene promoter43. Although this repressive chromatin state usually resolves with repair, clones of cells can evolve in which the promoter retains a memory of the silenced state and progressively accrues DNA methylation43. During tumorigenesis, abnormally tight epigenetic silencing of genes that are normally repressed by PcG in early embryogenesis could lock the cells in a primitive state, which better allows genetic lesions to cause uncontrolled proliferation. Indeed, it is now increasingly recognized that the most aggressive forms of human cancer harbour a genome-wide embryonic stem cell-like gene expression signature, which includes the overexpression of genes encoding many of the key proteins in the PcG complex and the enzymes that catalyse DNA methylation44.
In summary, we hypothesize that the cancer-predisposing states of ageing and chronic inflammation put pressure on stem and progenitor cells in cell renewal systems to expand in hostile environments. Such expansion may involve normal stem and progenitor cell mechanisms, such as poising of key genes in low-level transcription states by PcG occupancy. However, inherent to this scenario is the risk that the PcG complexes may recruit abnormal promoter DNA methylation and histone modification patterns that convert gene poising to stable silencing. Abnormal cell expansion may result in the creation of an epigenetic landscape that fosters the oncogenicity of key gene mutations and/or signalling from such mutations, which can help induce epigenetic abnormalities. The result is an increased risk of development of early cancer stages and progression of tumorigenesis.
Cardiovascular disease
Cardiovascular diseases are one of the leading causes of death in developed countries, and there is mounting evidence of a relationship between stress and altered epigenetic states in these disorders45. As in cancer, some aspects of cardiovascular diseases such as cardiac hypertrophy and atherosclerosis might be understood in terms of abnormal cell expansion states driven by chronic stress.
Contraction of the heart against chronically increased peripheral resistance due to prolonged arterial hypertension can lead to hypertrophy and heart failure46. Hang et al. have recently shown how sustained peripheral resistance induces cardiac hypertrophy through alterations in chromatin structure46. In their study, inducing abnormal pressure on the mouse heart results in cardiomyocyte reversion to a transcription state found during the embryonic stages of cardiac development. This change is mediated by increased expression of the chromatin-remodelling protein BrG1 (also known as SMCA4). During embryogenesis, BrG1 complexes with other chromatin-modifying proteins to repress the expression of adult α-myosin heavy chain and activates the fetal β-myosin heavy chain gene. BrG1 also activates bone morphogenetic protein 10 and inhibits cyclin-dependent kinase inhibitor 1C (also known as p57KIP2) to promote myocyte proliferation. Therefore, the stress-induced expression of BrG1 in the adult reverts cardiomyocytes to an embryonic-like, proliferative state, leading to cardiac hypertrophy. Intriguingly, in a rare human disease, hypertrophic cardiomyopathy of unknown aetiology, increased BrG1 keeps adult cardiomyocytes in an embryonic state through an epigenetic mechanism involving a repressive BrG1-containing complex46.
Atherosclerosis is another case in which stress might lead to aberrant cell expansion. Chronic stress due to inflammation in reaction to oxidized low density lipids (oxlDls), homocysteine and glucose-derived products and shear stress is known to underlie thickening of arterial walls and progressive plaque deposition in coronary arteries and peripheral vessels45,47,48. epigenetic changes are increasingly being associated with the proliferation of vascular smooth muscle cells (vSMCs) in blood vessel walls49,50. In this context, there are similarities to the abnormal perpetuation of more primitive cell states in early cancer evolution and heart hypertrophy. Increased levels of oxlDls have been shown to alter histone acetylation patterns in murine vSMCs by inducing the embryonic transcription factor Krüppel-like factor 4 (KlF4), which collaborates with histone deacetylase 2 (HDAC2) and HDAC5 to hypoacetylate the muscle differentiation gene ACTA2 (REF. 51). loss of muscle-specific gene expression programmes is permissive for vSMC proliferation52, suggesting that this can be an epigenetically mediated process. Further evidence that epigenetic mechanisms may mediate vascular proliferation in atherosclerosis is derived from studies of DNA methylation changes in human vSMCs from atherosclerotic aortas. Abnormal increases in gene promoter DNA methylation for genes such as oestrogen receptor-α and -β have been reported in vSMCs from atheromas, and have been correlated with increased vSMC proliferative capacity50,53,54.
The possibilities outlined above for epigenetic changes in atherosclerosis may also be considered for vessel disease in diabetes, in which abnormal vessel proliferation in the retina contributes to blindness and vessel thickening, and increased extracellular matrix and stromal responses in the kidneys play crucial parts in renal failure55. exposure to increased levels of glucose may foster vSMC proliferative responses that persist even when such cells are returned to a normoglycaemic environment55.
Cellular dysfunction or loss
Cellular dysfunction or loss of functional cell populations is a second potential consequence of stress-induced epigenetic changes and may have a role in multiple diseases55. A role for cellular dysfunction and loss has been emerging for many neurological diseases, in which pathogenesis involves a loss of synaptic firing plasticity and neuronal death56,57. Importantly, some of the same gene changes that predispose to abnormal cell expansion and lead to cancer can result in neuronal cell loss58. Cell dysfunction is also well characterized in diabetes, in which pancreatic islet dysfunction leads to insufficient insulin production. For both neurological diseases and diabetes, evidence is accumulating that cell stress can induce epigenetic changes that give rise to disease states owing to cell dysfunction.
Neurological disorders
Before considering the direct role of stress in neurological disease, it is important to consider that there are a series of genetic disorders that provide firm evidence for the role of epigenetic mechanisms in neuronal dysfunction59. These diseases are caused by mutations in genes that encode regulators of epigenetic processes or by mutations that disrupt epigenetic marks60,61. For example, in fragile X syndrome — the most common heritable mental retardation disorder — expansion of a CGG repeat in the promoter region of the fragile X mental retardation 1 (FMR1) gene leads to abnormal promoter DNA methylation and silencing of FMR1 (REF. 62). Some mental retardation disorders are caused by mutations in proteins involved in modulation of chromatin structure. For example, rett syndrome is caused by mutations in the X-linked gene MECP2, which encodes a protein that binds methylated CpGs and links DNA methylation to chromatin complexes that alter gene expression63–65.
These findings from mental retardation syndromes with defined genetic alterations have helped fuel studies of possible roles for epigenetic changes in several other neurological disorders with as yet undefined causes, and in which chronic stress has been implicated in disease causality and loss of normal neuronal plasticity60. A key example is autism, for which there are accruing data that genetic and epigenetic mechanisms could play a part61,62,66. Autism spectrum disorders (ASDs) include the symptoms of autism in disorders with a broad range of phenotypes. rett syndrome is one of the best understood ASDs and has an epigenetic mechanism for cognitive dysfunction66. More studies are linking ASDs to stress and consequent epigenetic alterations. To date, these studies have looked at DNA methylation or histone modifications at specific genes. For example, increased DNA methylation and the subsequent silencing of genes that could influence brain development — such as for a retinoic acid receptor and the cell survival gene BCL2 — have recently been reported in cerebral tissues and lymphoid-derived cell lines from patients with autism67.
There also is evidence that systemic inflammation and increased oxidative stress contribute to the pathobiology of Alzheimer's disease (AD)68. Oxidative stress has been linked to pathological features of AD, including progressive accumulation of neurofibrillary tangles and amyloid plaques, which are associated with neuronal dysfunction and dementia69. There is also some preliminary evidence of epigenetic changes in AD. For example, the amyloid precursor protein (APP) gene is thought to lose DNA methylation with ageing, and overexpression of this gene can lead to overproduction of APP, which is a hallmark of patients with AD and involved in plaque formation4. More global views of epigenetic changes in brains of patients with AD are currently being generated70. A recent intriguing finding links loss of function of the histone and protein deacetylase SIrT1 to increased amyloid plaques and an Alzheimer-like phenotype in a mouse model71. In this work, SIrT1 is shown to directly activate transcription of the α-secretase ADAM10 gene through deacetylation and co-activation of the retinoic acid receptor71. In this scenario, SIrT1 also induces the Notch pathway, which has been linked to activating the repair of neuronal damage71.
Diabetes
Both type 1 and type 2 diabetes (T1D and T2D) involve loss or dysfunction of pancreatic islet cells. T1D is a disorder characterized by the early loss of insulin-secreting pancreatic islet cells during adolescence. T2D is a progressive disease, which is usually initiated in the setting of obesity by peripheral insulin resistance in fat and muscle tissues, and is later characterized by loss of islet cell function55. Therefore, to understand the basis of diabetes, the control of cell fate in fat, muscle and pancreatic islet cells must be considered in development and adulthood, and these cell fate issues are discussed for T2D later. In terms of the loss of pancreatic islet β-cells as a primary feature of T1D, this disease is thought to be an autoimmune disease that involves T cell-mediated destruction in the setting of chronic β-cell inflammation66. There is now emerging evidence that epigenetic factors may contribute to this stress-induced disease dynamic55,66. For example, genome-wide chromatin immunoprecipitation followed by microarray (ChIP–chip) studies revealed that blood lymphocytes from patients with T1D have an increase in the repressive mark histone H3 lysine 9 dimethylation (H3K9me2) at the promoters of several genes72 — including cytotoxic T-lymphocyte-associated 4, a T1D susceptibility gene — when compared with healthy control individuals. Pathway analyses identified that the functional networks most enriched for altered H3K9me2 were autoimmune- and inflammation-related networks that involved transforming growth factor-β, nuclear factor-κB, p38 mitogen-activated protein kinase, Toll-like receptor and interleukin 6. These findings suggest that chromatin-mediated decreases in the expression of immune response genes may contribute to the pathology of T1D.
Changes in cell state
A third cellular response to stress that could be mediated by changes in epigenetic states is altered cell states in the context of cell renewal. epigenetic control is the key factor that normally regulates changes in cell fate in development and in adult cell renewal systems. For example, DNA methylation changes can be important, as indicated by recent reports that depleting the DNA methylation-catalysing enzyme DNMT1 in normal mouse haematopoietic and human epithelial cells blocks self-renewal and proper cell maturation, leading to depletion of progenitor cells73–75. Such alterations may occur in cells that are normally renewing or through reprogramming of non-renewing cells. We suggest that chronic stress to cell renewal systems leads to the switching of cell fates, and that this process is epigenetically mediated and may play a profound part in the pathology of multiple diseases.
Pre-malignant conditions
The overlap between abnormal cell expansion and altered cell fate can be seen in the evolution of cancer. For example, in the pre-malignant condition Barrett's oesophagus, increased exposure to gastric acid is associated with chronic inflammation and increased oxidative stress of the oesophageal epithelium76. Cells in the exposed region can show increased clonal expansion and harbour abnormal promoter DNA methylation and silencing of tumour suppressor genes77,78. In addition, there is a progressive change of the normal squamous epithelia of the oesophagus to columnar epithelia76. Thus, this epithelial system undergoes a heritable change in cell state due to exposure to a chemical stress.
In the lung, a phenotypic switch of a similar type can occur — from normal columnar epithelia to abnormal squamous epithelium. This is a common pre-cancerous change that is observed as a consequence of exposure to cigarette smoke. Multiple epigenetic gene silencing and promoter DNA hypermethylation events are seen in this pre-cancerous state79. Given the known role of epigenetic mechanisms in maintaining cell fate and the epigenetic abnormalities detected in stress-exposed cells, it seems likely that changes to the epigenetic landscape maintain the altered cell fates in these pre-malignant states.
Diabetes
In addition to cell expansion and dysfunction, altered cell state is involved in the pathogenesis of diabetes. robust experimental observations are allowing us to think about roles for altered epigenetic states and abnormal cell fates induced by stress in diabetes80. There is substantial evidence that the pathobiology of T2D can begin in utero as a function of maternal habits55. In this scenario, increased risk for the offspring might be mediated by epigenetic changes55 arising in utero that later predispose to T2D81. One study showed that in a rodent model of growth restriction there is downregulation of pancreatic and duodenal homeobox1 (PDX1), a crucial β-cell transcription factor82. After birth, Pdx1 was associated with increased H3K9 methylation and decreased H3K4 methylation, modifications which are correlated with transcriptional repression and activation, respectively. On development of diabetes, Pdx1 had promoter-proximal DNA methylation. A particularly exciting observation from this study was that HDAC inhibition reversed these changes and led to expression of Pdx1. This example shows that stress during early development can alter the transcriptional state of genes that are key for cell identity and eventually lead to altered cell state, driving diabetes pathology.
Insulin resistance in T2D is closely linked to glucose metabolism in peripheral mesenchymal tissues, such as adipose tissue and muscle55. Intriguing recent experimental data link insulin resistance to the influence of the stresses of excess glucose and fat consumption on the epigenetic control of fat cell maturation during development and adulthood. Adipocyte differentiation and function are influenced by glucose and the insulin receptor83,84, and differentiation normally proceeds through a series of cell states. Peroxisome proliferator-activated receptor γ (PPARG) activation is an early event that is triggered by transient glucocorticoid receptor occupancy at the PPARG promoter, and memory of this activation persists in the later stages of adipogenesis83. This memory is important for subsequent programmes of gene expression that involve further activation of PPARG and its target genes, which facilitate adipogenic differentiation83,84. In a mouse model of obesity and diabetes, the Pparg promoter becomes abnormally methylated and downregulated in visceral adipose tissue85. Thus, the stress of excess adiposity on this system might induce, through an epigenetic mechanism, more juvenile states of adipocyte renewal that could lead to insulin resistance and diabetes.
This scenario is supported by histology studies of pancreatic islets from patients with T2D, which suggest that, as the disease progresses, stress on the insulin-producing pancreatic β-cells leads to their loss55. recent elegant studies in mice on the epigenetic regulation of cell fate in pancreatic islets help to explain these observations86. In this work, virtually all pancreatic β-cells were experimentally ablated in mature mice, and it was found that through a transdifferentiation event these cells could be replaced by α-cells, which normally secrete the insulin antagonist glucagon. This conversion shows that, in addition to the known plasticity of cells during islet development, there is unanticipated plasticity in adults86. Given previous data that showed that cell state is defined by the epigenetic landscape of the cell, it is likely that as the islet cells respond to toxic stress and β-cell ablation, changes in chromatin mediate transdifferentiation.
Future directions and implications
The examples discussed in this article show links between chronic stress and epigenetic changes that are associated with abnormal cell expansion, cell dysfunctional states, switches in cell fates or combinations of these events in renewing cell systems (FIG. 2). In many of these cases, direct relationships remain to be established. However, based on accumulating evidence we propose that if stress causes epigenetic changes, the heritable nature of these changes lock in the early stages of pathology and so can drive the appearance and progression of disease. We make the important distinction between transient stress to cell renewal systems, such as acute wounding and successful repair, and chronic stress which repeatedly challenges the cell renewal systems that maintain homeostasis and survival (FIG. 1).
The well-documented involvement of epigenetic abnormalities in tumorigenesis and the mounting evidence for these epigenetic changes in other common diseases emphasizes the need for more widespread characterization of normal and disease epigenomes. Projects such as the Cancer Genome Atlas project, the National Institutes of Health roadmap projects and international collaborations87 are timely and illustrate the recognition of this need. A growing armamentarium of technologies based on next-generation sequencing for detailed mapping of DNA methylation and histone modifications are advancing such studies88. Future results from these studies are likely to correct a bias inherent to our present article — namely, we have focused solely on the proximal promoter regions of genes. The above technologies allow visualization of chromatin and DNA methylation patterns at regions more distal to those emphasized here13,14, and will likely prove immensely important for elucidating the epigenetic responses to stress and disease13,14.
Ultimately, no matter what technologies are used, full elucidation of epigenetic abnormalities in diseases, such as cancer, cardiovascular disease, diabetes and neurodegenerative diseases, will only occur when epigenomes in these settings are compared to epigenomes of the cell types in which the diseases arise. These studies will have to include the full range of developmental and disease progression stages. There are great challenges in terms of obtaining high-quality, staged disease samples and the correct cell types from normal cell renewal systems. Improved methods for the isolation of the key cell populations and adaptation of assay platforms for analysis of chromatin from very small cell numbers will be required89. Judicious use of mouse and cell culture models will be helpful towards this end.
The concepts presented here that link stress through epigenetic changes to disease pathogenesis, and the investigative routes discussed to develop these further, could have profound importance for the prevention and treatment of many human diseases. For cancer, there are already several ‘epigenetic therapies’ in the clinic, such as the DNA-demethylating agents 5-azacitidine and deoxy-aza-citidine and various HDAC inhibitors, which are gaining therapeutic traction in neoplastic disorders90,91. The evidence that we have discussed for atherosclerotic vascular disease and diabetes suggests that the reversal of epigenetic abnormalities could be preventative or therapeutic in these diseases. Several of the drugs that are beneficial for T2D work as antagonists to PPArG, which we discussed in terms of epigenetic regulation of adipocyte maturation and insulin resistance. Further understanding of this therapeutic link should improve treatment strategies in diabetes. The observation that replacement of wild-type function of MeCP2 in a mouse model of rett syndrome92 can reverse already established cognitive dysfunctions is a stunning example of how epigenetic therapies might benefit neurological disorders. In this regard, we have cited recent elegant evidence that the deacetylase SIrT1 can reactivate expression of α-secretase and prevent cognitive decline in a mouse model of AD71. SIRT1 has previously been observed to have a protective role against cell stress in vitro and an ability to contribute to resistance to multiple types of stress through protein deacetylation93,94. Most recently, inhibitors of other HDACs have had beneficial effects for a cognitive deficit phenotype in ageing mice95. Therefore, in addition to exploring how epigenetic mechanisms mediate the link between stress and disease, it will be important to explore how epigenetics can be exploited to develop resistance to stress and/or for disease therapies. The possibilities for reversal of epigenetic abnormalities in disease are just beginning to be explored; the research directions we have suggested here may reveal how far such therapies can reach.
“it will be important to explore how epigenetics can be exploited to develop resistance to stress and/or for disease therapies.”
Acknowledgements
The authors thank members of the Baylin laboratory for helpful suggestions and reading of the manuscript and K. Bender for help with manuscript preparation. Portions of the authors’ work cited have been supported by the National Cancer Institute grant CA043318, the National Institute of Environmental Health Sciences grant ES011858 and the National Institutes of Health grant CA116160.
Glossary
- Amyloid plaque
A focal extracellular collection of amyloid protein that surrounds dystrophic neurites in the hippocampus, amygdala and neocortex.
- Committed progenitor cell
A multipotent cell that is committed to a particular lineage and incapable of self-renewal.
- Histone post-translational modification
A covalent alteration of a histone tail residue that alters chromatin structure. Modifications include phosphorylation, methylation, acetylation, sumoylation and ubiquitylation.
- Histone variant
A histone that differs from canonical histones in terms of its structure and regulation.
- Neoplasia
The abnormal proliferation of cells. A neoplasm can be further characterized as benign or malignant based on its ability to invade other tissues.
- Neurofibrillary tangle
A filamentous bundle of tau protein that aggregates in the cytoplasm of neurons and is a histological hallmark of Alzheimer's disease.
- Nucleosome remodelling
An enzymatic process that alters the position of nucleosomes and can influence chromatin condensation.
- Peripheral resistance
The resistance of the peripheral vasculature to blood flow.
- Polycomb group
A family of proteins that are involved in gene silencing during development through methylation of histone H3 lysine 27.
- Pre-malignant
A state in which a cell is benign but poised to become malignant.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visconti R, Grieco D. New insights on oxidative stress in cancer. Curr. Opin. Drug Discov. Devel. 2009;12:240–245. [PubMed] [Google Scholar]
- 4.Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009;46:1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Silva H, Conboy I. Aging and stem cell renewal. StemBook. 2008:1–13. [PubMed] [Google Scholar]
- 7.Chambers SM, et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M. Epigenetics in evolution and disease. Lancet. 2008;372:S90–S96. [Google Scholar]
- 16.Nelson WG, De Marzo AM, DeWeese TL, Isaacs WB. The role of inflammation in the pathogenesis of prostate cancer. J. Urol. 2004;172:S6–S12. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- 17.Pani G, Galeotti T, Chiarugi P. Metastasis: cancer cell's escape from oxidative stress. Cancer Metastasis Rev. 2010;29:351–378. doi: 10.1007/s10555-010-9225-4. [DOI] [PubMed] [Google Scholar]
- 18.Meng X, Riordan NH. Cancer is a functional repair tissue. Med. Hypotheses. 2006;66:486–490. doi: 10.1016/j.mehy.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 19.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 20.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu. Rev. Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 22.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 24.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer — a mechanism for early oncogenic pathway addiction? Nature Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nature Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 27.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 28.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 29.Baylin SB. Stem cells, cancer, and epigenetics. StemBook. 2009:1–14. [PubMed] [Google Scholar]
- 30.Issa JP. CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 31.Issa JP, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nature Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 32.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstein IB. Cancer. Addiction to oncogenes — the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 35.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nature Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 36.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 37.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 40.Chi AS, Bernstein BE. Developmental biology. Pluripotent chromatin state. Science. 2009;323:220–221. doi: 10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 42.Tiwari VK, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J. Cell. Mol. Med. 2010;14:1225–1240. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Victor VM, et al. Oxidative stress, endothelial dysfunction and atherosclerosis. Curr. Pharm. Des. 2009;15:2988–3002. doi: 10.2174/138161209789058093. [DOI] [PubMed] [Google Scholar]
- 48.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 49.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim. Biophys. Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, et al. Epigenetic changes in estrogen receptor β gene in atherosclerotic cardiovascular tissues and in vitro vascular senescence. Biochim. Biophys. Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am. J. Physiol. Cell Physiol. 2008;295:C1175–C1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 53.Ying AK, et al. Methylation of the estrogen receptor-α gene promoter is selectively increased in proliferating human aortic smooth muscle cells. Cardiovasc. Res. 2000;46:172–179. doi: 10.1016/s0008-6363(00)00004-3. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, et al. Epigenetic changes in estrogen receptor β gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim. Biophys. Acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am. J. Physiol. Renal Physiol. 2010;299:F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chauhan V, Chauhan A. Oxidative stress in Alzheimer's disease. Pathophysiology. 2006;13:195–208. doi: 10.1016/j.pathophys.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Morris LG, Veeriah S, Chan TA. Genetic determinants at the interface of cancer and neurodegenerative disease. Oncogene. 2010;29:3453–3464. doi: 10.1038/onc.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 62.Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010;7:264–274. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 64.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl Acad. Sci. USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schanen NC. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 2006;15:R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonda DJ, et al. Oxidative stress in Alzheimer disease: a possibility for prevention. Neuropharmacology. 2010;59:290–294. doi: 10.1016/j.neuropharm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 69.Guglielmotto M, Giliberto L, Tamagno E, Tabaton M. Oxidative stress mediates the pathogenic effect of different Alzheimer's disease risk factors. Front. Aging Neurosci. 2010;2:3. doi: 10.3389/neuro.24.003.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marques SC, Oliveira CR, Outeiro TF, Pereira CM. Alzheimer's disease: the quest to understand complexity. J. Alzheimers Dis. 2010;21:373–383. doi: 10.3233/JAD-2010-100303. [DOI] [PubMed] [Google Scholar]
- 71.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses β-amyloid production by activating the α-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Miao F, et al. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57:3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nature Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 74.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jankowski JA, et al. Molecular evolution of the metaplasia–dysplasia–adenocarcinoma sequence in the esophagus. Am. J. Pathol. 1999;154:965–973. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bian YS, Osterheld MC, Fontolliet C, Bosman FT, Benhattar J. p16 inactivation by methylation of the CDKN2A promoter occurs early during neoplastic progression in Barrett's esophagus. Gastroenterology. 2002;122:1113–11121. doi: 10.1053/gast.2002.32370. [DOI] [PubMed] [Google Scholar]
- 78.Eads CA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 79.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nature Rev. Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 80.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woo M, Patti ME. Diabetes risk begins in utero. Cell Metab. 2008;8:5–7. doi: 10.1016/j.cmet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 82.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J. Clin. Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steger DJ, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lefterova MI, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thorel F, et al. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones PA, et al. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nature Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 90.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 92.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin W, et al. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J. Biol. Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 94.Guarente L. Cell biology. Hypoxic hookup. Science. 2009;324:1281–1282. doi: 10.1126/science.1175679. [DOI] [PubMed] [Google Scholar]
- 95.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]