Abstract
When it comes to microtubules, kinetochores leave nothing to chance. Recent studies have provided insight into an amazingly choreographed dance between the proteins in the kinetochore and their substrate and power source the microtubule.
Kinetochores are complex protein machines assembled on the centromeres of segregating chromatids that use microtubules of the mitotic spindle to perform the work of moving chromosomes. To do this they must coordinate at least three activities: they ensure that the two sister chromatids bind microtubules from opposite poles of the spindle (biorientation) [1]. Second, they generate spindle checkpoint signals that prevent cell cycle progression until each chromosome is bioriented [2]. Third, kinetochores use the energy stored in the microtubule to perform the work of moving chromosomes [3]. The complexity of vertebrate kinetochores is boggling, as they contain over 80 different proteins each at hundreds of copies per kinetochore [4]. Each human kinetochore will bind approximately 20 microtubules kinetochores are built upon repetitive elements where each tightly controls a single microtubule. However recent work has started to bring order from this chaos; as webs of protein interactions and maps of dependencies for assembly have melded with groupings of proteins by function to produce working models for kinetochore structure and function (reviewed in [5] [4]). A microtubule that is captured is put through a series of steps that are accomplished by dramatic reorganizations of the kinetochore. Our goal is to highlight the series of events from the initial binding of a vertebrate kinetochore to using it to move chromosomes. We will correlate these events to activities and structural changes in the kinetochore and highlight recent papers that have provided new insights in this rapidly developing field.
Initial Attachments and Checkpoint coordination
In many cells with open mitoses the initial attachments of kinetochores to microtubules are through the minus end directed motor cytoplasmic dynein. Although we have known this for over 20 years [6], it is still unclear why dynein binds first, however in figure 2A we present a model that could explain these initial attachments. Mitotic spindles are highly polarized structures with two poles and microtubules plus ends pointing towards the center. It is a reasonable assumption that after nuclear envelope breakdown, chromosomes have random orientations in relationship to the poles of the spindle prior to microtubules being encountered. Each kinetochore binds multiple microtubules and therefore any kinetochore that is not facing directly towards a pole has a chance to bind microtubules from both poles [1]. Thus it is believed that the first event of the dance is for kinetochores to spin chromosomes so that the two kinetochores are facing opposite poles. This is accomplished by having the minus-end directed motor dynein and its regulators associated with the kinetochore where they are positioned to make the initial microtubule attachments and pull the kinetochore towards the poles [6]. Chromosome arms contain multiple plus-end directed kinesins that also bind microtubules and push the arms toward the center [7]. The combined actions of these opposing forces may spin chromosomes so that the two kinetochores face opposite poles.
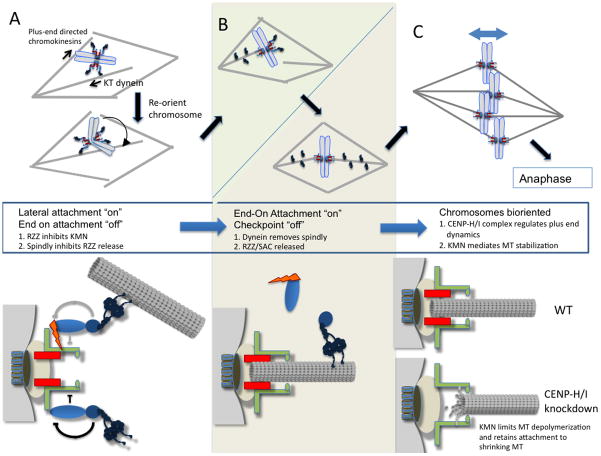
Figure 2.
The progression of chromosome microtubule interactions from initial attachment to final end-on attachment. (A) The plus end directed activity of chromokinesins, bound to chromosome arms, combined with the kinetochore bound minus directed motor activity of the dynein complex may serve to rotate the chromosomes so that their KT face opposing sides of the microtubule spindle and increase the likelihood that sister KT will attach to opposite poles. While KT lack end-on attachment, they produce a mitotic checkpoint signal possibility through the recruitment of checkpoint proteins by RZZ. During this time, RZZ also acts to inhibit the Ndc80 complex from producing end-on MT attachment. Spindly in turn inhibits RZZ. Grey arrows indicate that it is unclear if RZZ still inhibits KMN after dynein binds a microtubule (B) As end-on MT attachment is achieved through the KMN, spindly is removed from kinetochores by dynein mediated stripping. This removal relieves the inhibition of RZZ by spindly. RZZ can then leave the kinetochore in the absence of spindly and the mitotic checkpoint is silenced. (C) KMN binds the lateral surface and CENP H/I regulates the plus ends of microtubules that are “end-on” attached. CENP-H/I depletion results in less stable MT plus ends that can results in MT depolymerization, which is rescued by the KMN.
Ultimately chromosomes must align on the metaphase plate and thus move toward the plus-end of the microtubule, opposite to the action of dynein. To accomplish this dynein transport toward the minus end is transient and the microtubule is handed to a second kinetochore-microtubule binding complex composed of KNL-1, Mis12 and Ndc80 subcomplexes and referred to as “KMN”; believed to be the site where plus-end of microtubules are inserted “end-on” into the kinetochore [5]. While molecular details of this hand-off of microtubules from dynein to KMN is mysterious, it is clear that dynein and many of its associated proteins disengage from the microtubule after the event. A member of RZZ (Zw10) has been visualized moving down the microtubule in a process known as “streaming” [8] [9]. Thus there is a large change in the kinetochore architecture that accompanies the conversion from initial to “end-on’ attachments (figure 1). This change in structure not only turns “off” dynein dependent initial attachments but it also underlies much of the temporal regulation of the kinetochore. A complex that streams with dynein is called RZZ (figure 1), which is essential to generate the spindle checkpoint signal that inhibits anaphase onset in the absence of end-on microtubule attachments [10]. Thus the physical removal of RZZ from kinetochores after attachments is a mechanism to silence the checkpoint signal. Recent work has identified additional roles for RZZ that suggest that it plays an active role to ensure that dynein attachments precede end-on attachments. Cells lacking a protein known as “spindly” have RZZ at kinetochores but dynein and its regulators are missing [11]. Surprisingly Gassmann et al. showed that knockdown of the spindly proteins in C. elegans produced phenotypes reminiscent of knockdown of the KMN complex (very poor congression of chromosomes to the metaphase plate), while knockdown of RZZ gave significantly weaker phenotypes. Knockdown of RZZ and spindly rescued the stronger phenotype [12] [13]. The simplest interpretation is that RZZ inhibits the end-on binding of KMN until RZZ is cycled off the kinetochore (figure 2), which argues that the kinetochore is actively suppressing end-on attachments until dynein makes initial attachments. The position of dynein in the kinetochore as the protein furthest from the centromere would also contribute to its binding microtubules first.
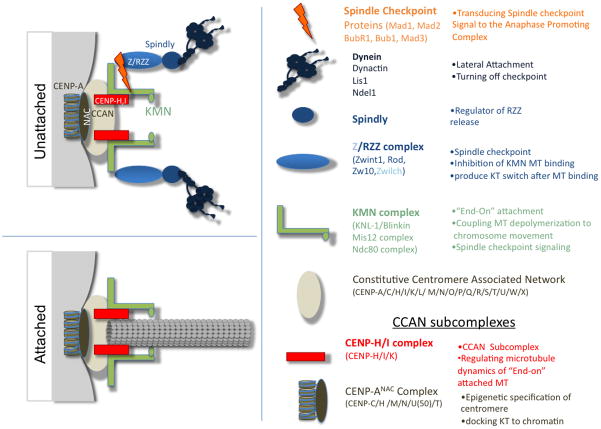
Figure 1.
Kinetochores have two structures depending upon whether they are attached to microtubules. The centromere/kinetochore can be subdivided into functional groups and biochemical subcomplexes, which are summarized according to the proteins in the subcomplexes and their major functions. Only the complexes discussed in this review are depicted. Unattached kinetochores (KT), which are assembled upon the constitutive centromere, recruit complexes with MT binding functions (Dynein and KMN) and the components of the mitotic checkpoint, which are thought to sense both occupancy and tensions across the centromere as a measure of proper microtubule attachment of the kinetochore. End-on microtubule attachment results in changes in the protein complexes present at the kinetochores. The constitutive centromere and components of the KMN network are retained; however, the RZZ complex, spindly, dynein and the mitotic checkpoint proteins are removed from the kinetochores upon end-on attachments (for a complete review see [5] [4]).
The simplest model for turning off the checkpoint would have been that dynein physically pulls RZZ off the kinetochore by walking down the microtubule. However two other recent studies on spindly suggests that this model is too simple. Cells lacking spindly take a long time to align chromosomes, but they are able to turn off the checkpoint and release RZZ efficiently even though dynein is not recruited ([13] and New Gassmann et al 2010). In contrast, mutants of spindly that don’t bind dynein prevent RZZ release and maintain checkpoint signaling. Thus spindly appears to be an inhibitor of RZZ release suggesting that the dynein removal of spindly is the requisite event for the changes that accompany initial to end-on attachment and turning off the checkpoint. However, spindly removal is not sufficient for checkpoint satisfaction, which is consistent with models that there are redundant mechanisms of checkpoint signaling from kinetochores [14].
RZZ shares its ZW10 subunit with another complex called NRZ (NAG/Zw10/RINT), which is involved in ER vesicle trafficking (reviewed in [15]). During membrane trafficking the NRZ proteins serve as tethering factors that recruit coated vesicles to ER membranes, for subsequent undocking by the SNARE (SYNTAXIN 18 SNARE) proteins. Civril et al. show that Rod (R of RZZ) and NAG are structurally related proteins containing three common motifs a β-propeller, NRH domain and then a Sec39 related region (New Civril et al. 2010). The RINT and Zwilch components of these two complexes do not show any recognized structural similarity, suggesting how a common motif is adapted to very different processes. What are the similarities of kinetochore streaming and ER vesicle docking? At this point it is very unclear and many interesting possibilities were outlined in a recent review[15]. However, future experiments from both the ER transport and kinetochore fields will need to identify the common mechanism used by this structural motif as well as how they are adapted to perform unique functions.
End-On attached kinetochores
Kinetochores mature from dynein association to KMN binding, which holds the plus ends of microtubules and is thought to play an essential role in moving chromosomes. KMN is part of a coupler that remains bound to a depolymerizing microtubule by sliding down the lateral surface [5]. Microtubule depolymerization is unidirectional and can generate force that, in association with the coupler, powers the movements of chromosomes.
In a bioriented chromosome, as long as most of the microtubules attached to one side of a kinetochore are depolymerizing there will be net movement of the chromosome to that side. Since there are numerous end-on attached microtubules at each kinetochore, there must be some coordination of microtubule polymerization and depolymerization within a kinetochore and perhaps even across sisters. In an exciting recent paper Amaro et al. suggest that a group of constitutive centromere proteins are regulators of plus-end MT dynamics and thus provide the first real insight into this classic problem [16]. It has been known for many years that the half-life of microtubules embedded in kinetochores (K-fibers) is much longer than other microtubules of the spindle [17] and these fibers are stabilized by the KMN [18]. By following the dynamics of photo-activatable GFP-α-tubulin at the plus-end of the MT embedded in the metaphase plate, Amaro et al. showed that in cells lacking the CENP H/I complex the K-fiber microtubules at the metaphase plate were less stable, and exhibited similar dynamics to non-kinetochore spindle microtubules. Outside the metaphase plate, K-fibers had substantially slowed dynamics. The interesting answer to this apparent paradox is that CENP-H/I proteins are required to suppress the plus end dynamics of the very plus end of k-fibers but another complex, likely KMN, has an activity to prevent complete depolymerization of an end-on attached microtubule (figure 2C). Prior to this work, it was tempting to consider the CCAN as a mere foundation for the assembly of the transient kinetochore, based on its position within the centromere close to the CENP-A containing chromatin and its presence at the centromere throughout the cell cycle. However, in actuality, the physical location of the CENP-H/I proteins internal to the KMN microtubule attachment point places them in the ideal location to regulate plus ends [19]. The authors go on to show that CENP-H/I complex also regulates the oscillatory movements of metaphase aligned chromosomes, consistent with a role is coordination of groups of end-on attached microtubules. These data redefine how we think about the constitutive centromere and open up numerous studies to explore how the KMN coupler and CENP H/I regulator of microtubule dynamics regulators coordinate chromosome movements.
Contributor Information
P. Todd Stukenberg, Email: Pts7h@virginia.edu.
Daniel R. Foltz, Email: dfoltz@virginia.edu.
Cited work
- 1.Tanaka TU. Bi-orienting chromosomes: acrobatics on the mitotic spindle. Chromosoma. 2008;117(6):521–33. doi: 10.1007/s00412-008-0173-5. [DOI] [PubMed] [Google Scholar]
- 2.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9(1):33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 5.Welburn JP, I, Cheeseman M. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15(5):645–55. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113(4):805–15. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154(6):1135–46. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155(7):1159–72. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basto R, et al. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr Biol. 2004;14(1):56–61. doi: 10.1016/j.cub.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 10.Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15(7):386–92. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177(6):1005–15. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22(17):2385–99. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barisic M, et al. Spindly/CCDC99 Is Required for Efficient Chromosome Congression and Mitotic Checkpoint Regulation. Mol Biol Cell. 2010 doi: 10.1091/mbc.E09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoufias DA, et al. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA. 2001;98(8):4492–7. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt HD. Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends Cell Biol. 2010;20(5):257–68. doi: 10.1016/j.tcb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Amaro AC, et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 12(4):319–29. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131(3):721–34. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127(5):969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 19.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137(4):672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]