Synopsis
Mutations in MECP2 (methyl CpG binding protein 2) are linked to the severe postnatal neurodevelopmental disorder Rett Syndrome (RTT). MeCP2 was originally characterized as a transcriptional repressor that preferentially bound methylated DNA, however, recent data indicates MeCP2 is a multifunctional protein. MeCP2 binding is now associated with certain expressed genes and involved in nuclear organization as well, indicating its gene regulatory function is context dependent. In addition, MeCP2 is proposed to regulate mRNA splicing and a mouse model for RTT shows aberrant mRNA splicing. To further understand MeCP2 and potential roles in RTT pathogenesis, we have employed a biochemical approach to identify the MeCP2 protein complexes present in the mammalian brain. We show that MeCP2 exists in at least four biochemically distinct pools in the brain and characterize one novel brain-derived MeCP2 complex that contains the splicing factor Prpf3. MeCP2 directly interacts with Prpf3 in vitro and in vivo and many MECP2 RTT truncations disrupt the MeCP2-Prpf3 complex. In addition, MeCP2 and Prpf3 associate in vivo with mRNAs from genes known to be expressed when their promoters are associated with MeCP2. This data supports a role for MeCP2 in mRNA biogenesis and suggests an additional mechanism for RTT pathophysiology.
Keywords: MeCP2, Prpf3, Rett Syndrome, RNA
Introduction
MeCP2 was originally identified by its ability to preferentially bind double stranded DNA containing symmetrically methylated CpG dinucleotides and is the founding member of the methyl-CpG binding domain (MBD) family of proteins [1,2]. The first biological role for MeCP2 was illustrated by showing the protein interacts with methylated DNA in vivo and could repress transcription by association with a transcriptional co-repressor complex containing Sin3A and histone deacetylase [3–5]. In 1999 a genetic analysis identified mutations in MECP2 as causal for Rett Syndrome (RTT), providing the first direct link between an epigenetic regulator and a human disease [6]. RTT is a severe postnatal neurodevelopmental disorder and one of the most common causes of mental retardation in females [7]. First described in 1966 by Andreas Rett [8], RTT is characterized by a period of apparently normal development from birth to 6–18 months followed by a regression of obtained language and motor skills [7]. RTT patients usually exhibit a deceleration of head growth, respiratory dysfunction, scoliosis, cognitive impairment, seizures, and social withdraw [8,9]. In addition to RTT, numerous MECP2 mutations have now been linked to a variety of additional disorders, including autism, Angelman syndrome, learning disabilities, and mental retardation syndromes [7,10–14].
MeCP2 has been reported to associate with myriad protein partners including Sin3A [3,5], c-REST and Suv39h1 [15], c-Ski and N-CoR [16], Brm [17], and HP1 [18], all supporting a model of MeCP2 interacting with or being a stable component of transcriptional co-repressor complexes, resulting in targeted transcriptional repression of methylated DNA through modification of the chromatin state or chromatin associated proteins. However, the biological relevance and implications towards RTT for these numerous documented MeCP2 interactions is not clear due in part to the particular methods utilized and non-neuronal choices for initial cellular protein sources. In fact, contradicting these numerous studies, it has been proposed that endogenous MeCP2 does not form any stable protein-protein interactions in vivo [19]. Compounding the issue, recent work has expanded MePC2’s proposed gene regulatory role beyond mere transcriptional repression; MeCP2 is implicated in transcriptional activation, genome-wide transcriptional silencing, mediating chromatin and nuclear architecture, and regulating pre-mRNA splicing as well [20–23]. Thus, the in vivo protein-protein interaction profile of endogenous MeCP2, particularly in the brain, is still an open question and increasingly more important to understand as new functions for MeCP2 are emerging.
Genetic studies in mice suggest that expression of functional MeCP2 in neurons is essential for normal synapse formation and neuronal function during postnatal development and re-expression of MeCP2 in differentiated neurons alone rescues a RTT mouse model [24–29]. However, this idea is being challenged by a recent study that indicates the lack of MeCP2 specifically in glial cells contributes to RTT phenotypic neurons by an unknown secreted glial factor [30]. This discrepancy illustrates the need for more unbiased approaches in determining the molecular roles of MeCP2 in both normal and RTT brain; thus, intact mammalian brain tissue would be the ideal source to study endogenous MeCP2 protein function. Here we use the power of biochemistry to characterize MeCP2 in the mammalian brain and show that native MeCP2 protein purified from adult rat brain exists in multiple biochemically distinct pools/complexes, consistent with MeCP2 working as a multi-functional protein. We further characterize one brain-derived MeCP2-complex that contains Prpf3, a known spliceosome-associated protein [31], as well as the Sdccag1 [32], a mediator of nuclear export [33]. MeCP2 shows specific, direct interactions with Prpf3 and Sdccag1 and these interactions are disrupted by certain RTT mutations. In addition, we show that MeCP2 and Prpf3 co-associate in vivo with mRNAs from genes activated by MeCP2, further supporting the previously identified regulatory role of MeCP2 in mRNA biogenesis [23] and providing another potential mechanism disrupted during RTT pathogenesis.
MATERIALS AND METHODS
MeCP2 Protein Purification
Rat brain nuclei were isolated generally as described [34]. Adult rat brains (n=200 per extract preparation, Pel-Freez Biologicals) were thawed on ice, homogenized, and nuclei were collected. Nuclei were suspended in Buffer A (20mM Hepes pH 7.5, 1.5mM MgCl2, 1 mM EGTA, 10% glycerol, 0.5mM DTT, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml aprotinin) supplemented with 350mM NaCl (A-350) and extracted for 30 min at 4°C. Insoluble material was removed by centrifugation at 200,000g for 20min at 4°C. The supernatant was diluted with Buffer A to <200mM NaCl and used as a soluble protein source for chromatography.
Chromatography was preformed using an AKTA-FPLC (GE Healthcare) at 4°C in Buffer A with indicated concentrations of NaCl. Soluble protein (130 mg per purification) was fractionated over a MonoQ10/10 column with bound protein eluted by a 20 CV linear salt gradient from 100mM to 1000mM NaCl, collecting 0.5 CV fractions. The MeCP2 containing fractions peaking at 230mM NaCl (QB1/2) were pooled, diluted with buffer A and fractionated over MonoS5/5 with bound protein eluted by a 20 CV linear salt gradient from 250mM to 1000mM NaCl, collecting 0.5 CV fractions. The MeCP2 containing fractions peaking at 550 mM NaCl (QB2) were combined and loaded onto a 1ml Heparin FastFlow column and eluted with a 10 CV linear gradient from 450mM to 1000mM, collecting 0.5 CV fractions. Pooled heparin column fractions containing MeCP2 were applied to a 110 ml superose6 column and fractionated in buffer A-150 with 0.1% Trition-X100. Subsequently, 0.5 ml of each fraction was trichloroacetic acid-precipitated, subjected to SDS-PAGE, and used for western blotting or silver staining. Silver-stained polypeptide bands precisely co-fractionating with MeCP2 (by western) were excised and analyzed by mass spectrometry.
For the nuclease treatment experiments, MonoQ eluted MeCP2 was dialyzed against buffer A-100 containing 1mM CaCl2, 10mM MgCl2 and without EGTA then either treated with or without 500 U of benzonase nuclease (Sigma) for 30 min. at 37°C, and re-tested for ability to bind the MonoQ resin.
Antibodies and Western Blot Analysis
Protein samples were separated by SDS-PAGE and transferred to ECL nitrocellulose membrane (GE Healthcare) for western blotting by standard methods. Image manipulation was in accordance with Biochemistry guidelines. For each experiment, the western blotting images presented are from the same exposure on the same piece of film, linearly adjusted for brightness in Adobe Photoshop. The anti-MeCP2 3998 antibody is a rabbit polyclonal derived from bacterially expressed recombinant protein encoding the full-length human MeCP2e2 isoform. The anti-MeCP2 7–18 antibody is a rabbit polyclonal derived from bacterially expressed recombinant protein encoding amino acid residues 310 to 388 of human MeCP2e2 isoform. Antibodies were used at the following concentrations: anti-MeCP2 7–18, 1:2000; anti-MeCP2 (Upstate 07–013) 1:1000; anti-Prp3 (MBL D171-3) 1:2000; and anti-HA High Affinity (clone 3F10) at 1:1000.
Mass spectrometry (ms)
Ms was carried out at the Protein Sciences Facility at the University of Illinois. FPLC purified fractions were separated by SDS-PAGE, visualized by ms compatible silver staining. Polypeptide bands were excised, destained (50% acetonitrile, 25 mM ammonium bicarbonate), and digested in 25 µl of Sequencing Grade Trypsin (12.5 ng/µl in 25 mM ammonium bicarbonate, G-Biosciences St. Louis, MO) using a CEM Discover Microwave Digestor (Mathews, NC) for 15 min at 55°C (60W). Digested peptides were extracted using 50% acetonitrile with 5% formic acid, dried in a Savant SpeedVac and suspended in 13 µl of 5% acetonitrile containing 0.1% formic acid with 10 µl of sample used for ms analysis. The mass spectrometer used was Waters quadrupole time-of-flight mass spectrometer (Q-ToF) connected to a Waters nano-Acquity UPLC. The column used was Waters Atlantis C-18 (0.075 mm × 150 mm) with a flow rate of 250 nl per min. Peptides were eluted using a linear gradient of water/acetonitrile containing 0.1% formic acid (0–60% B) in 60 min. The mass spectrometer was set for data dependent acquisition; ms/ms was performed on the most abundant four peaks at any given time. Data analysis was performed using Waters Protein Lynx Global Server 2.2.5, Mascot (Matrix Sciences) and BLAST against NCBI NR database.
Generation of plasmids
RNA was purified from rat brain tissue or HeLa cells using Trizol Reagent (Invitrogen) per manufacturers instructions. All cDNAs were generated using SuperScript III One-Step RT-PCR with Platinum Taq (Invitrogen). All PCRs were performed with Phusion polymerase (New England Biolabs) and cloned into pGEM-T easy (Promega) for sequencing prior to sub-cloning into pGEX-5X1 (GE Life sciences), pCDNA 3.1 (Invitrogen), or the pCDNA 3.1 HA vector [35]. All primers are listed in Table S1. All human MeCP2 constructs were generated from the human MECP2E2 cDNA (NM_004992). To generate constructs for in-vitro synthesized proteins, cDNAs were PCR amplified using primers listed in Table S1 and sub-cloned into the specified restriction sites of pCDNA 3.1. Rat cDNAs were sub-cloned between NotI and XhoI; the human Prpf3 cDNA clone was sub-cloned between EcoRI and XhoI; the human full-length MeCP2 was amplified from full-length cDNA and sub-cloned between NotI and XhoI. Constructs for bacterially generated GST fusion proteins were PCR amplified from full-length cDNAs, sub-cloned into the specified restriction sites of pGEX5-X1 using primers listed in Table S1. Rat GST-Prpf3 was sub-cloned between BamHI and XhoI. The rat and human GST-MeCP2 full-length, GST-MeCP2 deletion, and human GST-MeCP2 RTT cDNAs were sub-cloned between EcoRI and XhoI. For the pCDNA3PHA-MECP2 vector, full-length human MeCP2 was sub-cloned between NotI and XhoI restriction sites of the pCDNA3.1 HA vector and subsequent digestion with NdeI and XhoI for cloning of the HA-MeCP2 fragment into the pCDNA3.1P puromycin vector.
Cell culture
HT-22 cells were transfected with pCDNA 3.1P HA-MeCP2 using Fugene HD transfection reagent (Roche). Stable integrants were selected as pools in puromycin (1µg/ml), and maintained in Dulbecco’s modified Eagle’s medium (Biowhittaker) supplemented with 10% fetal bovine serum, glutamine, antibiotics and puromycin.
GST Pull-down assay
Assays were preformed essentially as previously described [35], using recombinant GST-fusion proteins and in vitro transcribed/translated proteins radiolabeled with [35S]methionine using the T7 TnT Quick Coupled Transcription/Translation System (Promega) with or without 250 units of Benzonase nuclease.
Co-IP
Co-IPs from fractionated brain extracts were carried out as follows: MonoQ fractionated rat brain nuclear extracts (QB2) were diluted to 100mM NaCl with Buffer A(0). Protein A Dynabeads (Invitrogen Corp) were blocked with BSA and added to the extracts with 10 µl of either anti-MeCP2 3998 or normal rabbit serum IgG for 2 hrs at 4°C with rotation. Co-IPs were washed three times with buffer A225, eluted by boiling in Laemmli buffer, separated by SDS-PAGE and subjected to Western blot analysis. Co-IPs from cell culture were carried out as previously described [23] with modifications. Confluent 10-cm plates of HT-22 cells stably expressing HA-MeCP2 were lysed in 1ml of IPH buffer (50 mM Tris·HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% IGEPAL, 1 mM PMSF) and debris removed by centrifugation. Lysates were pre-cleared with protein A agarose then incubated for 4 hrs with anti-HA (Sigma, E6779) or irrelevant IgG with protein A agarose beads (Santa Cruz Biotechnologies) at 4°C with rotation. Nuclease treatment was preformed by suspension of protein bound beads with 125 units of benzonase in manufactures recommended buffer for 10 min at 37 °C. IPs were washed three times with IPH buffer, eluted with Laemmli buffer, separated by SDS-PAGE and subjected to Western blot analysis.
RIP and Re-RIP
Experiments were performed as previously described with slight modifications [36]. HT-22 cells (1 × 108) stably expressing HA-MeCP2 were collected, washed, and suspended in 4ml 1X PBS. Crosslinking buffer (100mM NaCl, 1mM EDTA, 0.5 mM EGTA, 50mM HEPES, 11% formaldehyde) was added (400 µl), incubated at RT for 30 min with rocking and quenched for 5 min at RT. Cell pellets were washed with 1X PBS and lysed in 1 ml FA buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors) by sonication. The lysed cells were treated with 200 U DNase I (Promega) in 25 mM MgCl2, 5 mM CaCl2, and 6 µl RNasin (Promega) for 30 min at 37 °C, cleared by centrifugation at 4 °C, diluted 1:10 with ChIP dilution buffer, and incubated with either anti-HA antibody or non-specific IgG with 4 µl RNasin, for 12 hrs, rotating at 4 °C. Protein A or G Dynabeads were added for 1 hr rotating at 4 °C and washed 3 × 10 min with wash buffer (50mM Tris pH 7.4, 500mM NaCl, 1% Triton X-100, 0.1% SDS). For Re-RIP experiments, beads were washed and bound immune-RNA complexes were released in 20mM DTT for 30 min at 37°C and resuspended in one volume of ChIP dilution buffer for Re-RIP with antibody for Prpf3 and washed as before. Beads were brought up in 200 µl of elution buffer (200mM NaCl, 50 mM Tris pH 7.4, 20ug Proteinase K) for 1 hr at 42°C, and cross-links reversed at 65°C. Samples were extracted with acid equilibrated (pH 4.8) phenol:chloroform (5:1) and ethanol precipitated. Precipitated material was suspended in 50 µl DEPC-H2O and used for RT-PCR analysis with SuperScript III One-Step RT-PCR with Platinum Taq (Invitrogen) for mouse Cdk10 and Frg1. Reactions were analyzed by electrophoresis on a 2% agarose gel.
RESULTS
MeCP2 exists in at least four distinct protein pools in rat brain nuclei
A large-scale biochemical purification of endogenous MeCP2 protein from rat brain nuclear extract was performed to characterize the native MeCP2 in the mammalian brain. Whole rat brains were homogenized under non-denaturing conditions, the intact nuclei were purified by centrifugation through a sucrose cushion, and nuclear proteins were extracted under mild ionic conditions. Proteins were initially fractionated by strong anion exchange (MonoQ) column chromatography with the MeCP2 protein being tracked by western blotting with multiple MeCP2-specific antibodies (Figures 1A and S1 and data not shown), all providing virtually identical profiles. The majority of soluble MeCP2 protein (~90%) did not associate with the MonoQ resin agreeing with a previous report [19], however, a significant fraction (~10%) of this brain-derived MeCP2 consistently bound to MonoQ resin and eluted in two distinct peaks along the linear salt gradient (Figure 1A) indicating multiple biochemically distinct pools of MeCP2.
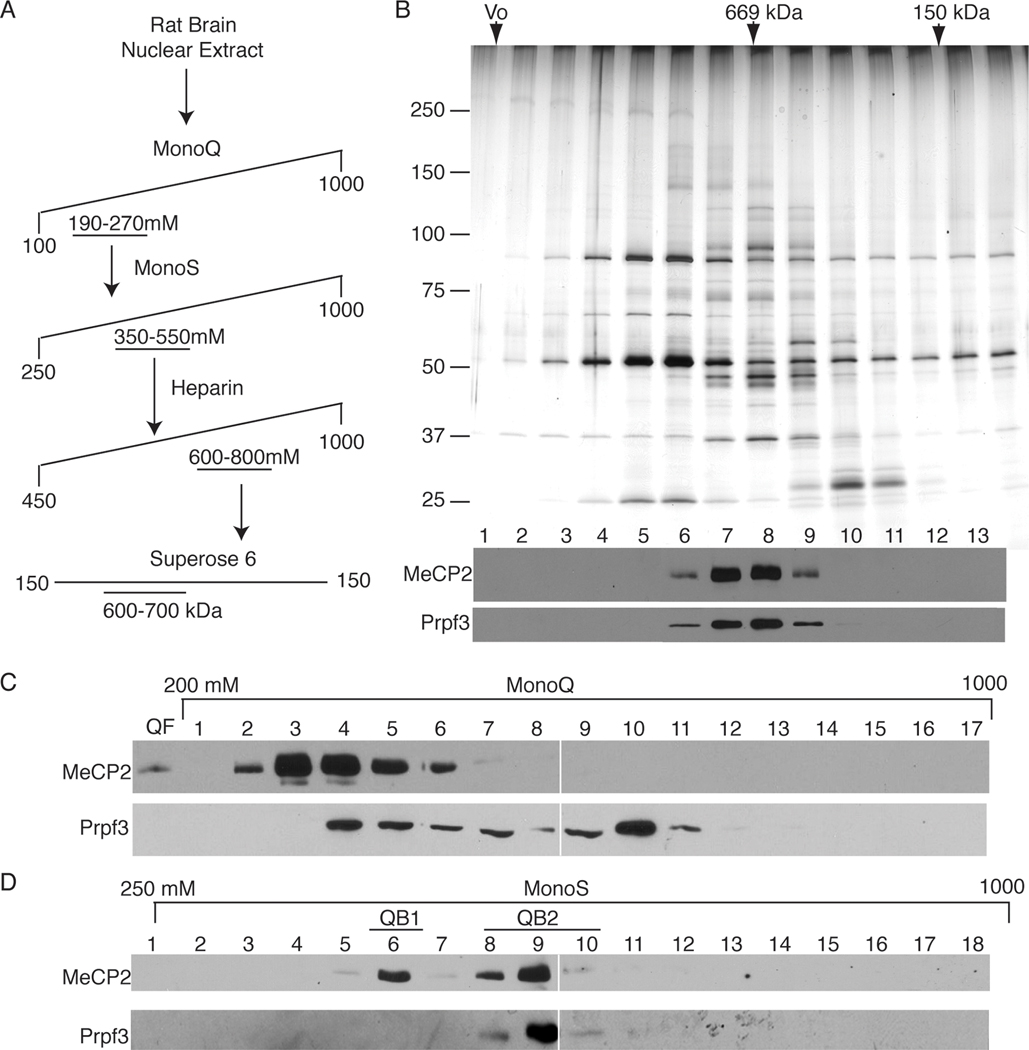
Figure 1. Brain-derived nuclear MeCP2 exists in multiple biochemically distinct pools.
(A) Chromatographic separation of crude rat brain nuclear extract by strong anion exchange (MonoQ resin) results in three distinct pools of MeCP2 as indicated by western blot using the MeCP2 7–18 antibody (Figure S1). The majority of MeCP2 does not bind the column (QF), while the bound MeCP2 elutes in two peaks, at 230mM NaCl (QB1/2) and 450mM NaCl (QB3). (B) MonoQ resin-bound fractions of MeCP2 were treated with (+) or without benzonase nuclease and tested for ability to re-bind the Mono Q. Western blot analysis for MeCP2 of MonoQ flow thru, wash, and 1000mM NaCl step elution shows benzonase treatment does not affect binding of MeCP2 to the Mono Q column. (C) Plasmid spiked MonoQ fractions treated with benzonase (+) or untreated (−) in parallel served as controls for benzonase treatment.
The MonoQ bound (QB) MeCP2 elution profile showed an initial broad peak (QB1/2) concentrated at 230mM NaCl and tailing to 370mM, suggesting multiple MeCP2 complexes, with a minor yet distinct peak (QB3) centered at 450mM NaCl (Figure 1A). Since MeCP2 is a DNA binding protein and also known to interact with RNA, the potential of nucleic acids mediating the anionic association of MeCP2 with the cationic resin was addressed. The QB fractions were treated with benzonase nuclease to remove both DNA and RNA and then MonoQ chromatography was repeated. We found that all of the nuclease-treated QB MeCP2 still bound the MonoQ resin and was released by step elution, indicating the interaction was both RNA and DNA independent (Figure 1B, C). Finally, the presence of MeCP2 in the MonoQ flow through (QF) and each elution peak (QB1/2 and QB3) was subsequently confirmed by mass spectrometry (Figure S2).
The early eluting MonoQ bound pool of MeCP2 (QB1/2) was characterized further for complexity and content (Figure 1A). The QB1/2 peak fractions were pooled and fractionated over the MonoS strong cation exchange resin, resolving into two peaks of MeCP2 protein (Figure 2D, top panel), the first eluting at 400mM NaCl (QB1) and the second eluting at 550mM NaCl (QB2). These results indicate at least three biochemically distinct pools of MeCP2 exist in the initial QB fraction and four MeCP2 pools exist overall in rat brain nuclear extract (QF and QB1, 2, and 3).
Figure 2. The QB2 pool of MeCP2 co-purifies with 6 candidate proteins including Prpf3.
(A) MeCP2 peak fractions were purified using a four-step process including the MonoQ strong anion exchange resin, MonoS strong cation exchange resin, Heparin affinity resin and by Superose6 gel filtration. (B) (Upper panel) Silver-stain analysis of the Superose6 fractionation of the QB2 MeCP2 pool. (Lower panel) Western blot analysis of Superose6 fractions shows MeCP2 protein peaks in fractions 7 and 8, precisely co-fractionating with Prpf3 protein. (C) The MonoQ fractionation of brain-derived nuclear extract reveals an overlap of Prpf3 and MeCP2 yet pools of each protein do not co-fractionate and remain independent of the other. (D) Peak fractions of the QB1/2 fractionated over the MonoS resin show two distinct pools of MeCP2 peaking at 400 mM NaCl (QB1) and 550 mM NaCl (QB2) by western blot analysis. Corresponding MonoS fractions probed for Prpf3 protein shows precise co-fractionation with QB2 Mecp2.
Brain-derived MeCP2 exists in a complex with the splicing factor Prpf3
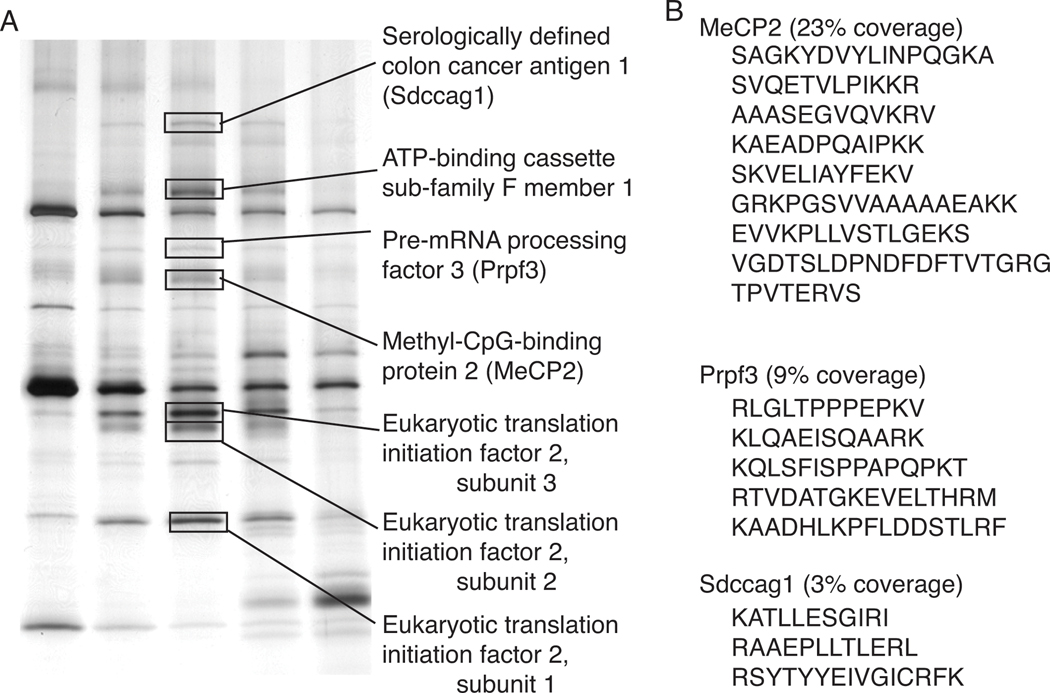
In order to identify potential MeCP2-interacting proteins, the more abundant MonoS bound pool of MeCP2 (QB2) was purified further by fractionation using heparin affinity chromatography and gel filtration through Superose6 (Figure 2A). Western blotting and silver stain analysis of the final fractionation showed MeCP2 peaking with the apparent molecular weight of 600–700kDa and precisely co-purifying with six additional polypeptides (Figures 2B, and 3). Since monomeric MeCP2 has previously been shown to exhibit an unusual Superose6 gel filtration profile migrating equivalent with an apparent mass of approximately 450kDa [19], the apparent mass for QB2 MeCP2 is consistent with being associated with additional proteins. Therefore, all six MeCP2 co-purifying polypeptides were identified by mass spectrometry with significant coverage (Figures 3 and S3) as: Prpf3, Sdccag1, ATP-binding cassette 50, and 3 components of a translation initiation complex (Eif2 subunits 1, 2, and 3). Since a commercial antibody was available against Prpf3, western blot analysis of the size exclusion chromatography fractions were carried out and show MeCP2 and Prpf3 precisely co-fractionate (Figure 2B, bottom) confirming the silver-stain analysis (Figure 2B, top).
Figure 3. Identification of candidate MeCP2 complex proteins.
(A) Polypeptides from silver stained Superose6 fractions were excised and identified by mass spectrometry (tandem LC MS/MS). MeCP2 and two other nuclear proteins, Sdccag1 and Prpf3 were identified as well as components of a translation initiation complex. (B) Mass spectrometry peptides identifying MeCP2, Prpf3, and Sdccag1.
With Prpf3 identified as a putative MeCP2-interacting protein, the MeCP2 fractionation scheme (Figure 2A) was analyzed for Prpf3 by western blotting to determine if Prpf3 was present in all MeCP2 pools or specific to QB2 and similarly to determine if all Prpf3 in brain extracts associated with MeCP2. Not surprisingly, only a fraction of Prpf3 in brain extracts co-fractionated with MeCP2 when assaying the MonoQ separation profile (Figure 2C), however, all of the detectable Prpf3 overlapping with MeCP2 specifically co-fractionated with the QB2 pool of MeCP2 from the MonoS fractionation and not with the QB1 MeCP2 pool (Figure 2D). Thus, Prpf3 distinguished QB2 from other MeCP2 protein pools. Because Prpf3 is a known component of the spliceosome [31] and MeCP2 has been shown to interact with another spliceosome-associated protein, YB-1 [23], the QB2 fractionation was screened for YB-1. However, YB-1 was not found by western blotting and YB-1 was absent from the mass spectrometry analysis indicating YB-1 did not co-purify with QB2 MeCP2 under these conditions (Figure 3, data not shown).
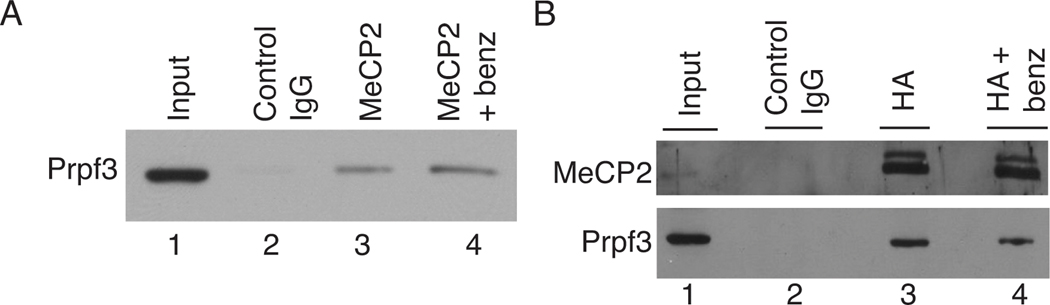
To confirm that native MeCP2 and Prpf3 are in a complex in vivo, co-IP experiments were performed (Figure 4). In brain extracts, as determined by the biochemical fractionations described above, the vast majority of MeCP2 is not associated with Prpf3 and likewise, the majority of Prpf3 in brain is not associated with MeCP2 making it difficult to visualize any interaction by co-IP directly from crude nuclear extracts. Thus, two approaches were used; co-IPs from fractionated brain nuclear extract (Figure 4A) and co-IPs from crude extracts using tissue culture cells overexpressing an epitope-tagged version of MeCP2 (Figure 4B). Rat brain nuclear extract was prepared and fractionated over MonoQ as described above. The QB1/2 MeCP2 fractions were diluted to 100 mM NaCl in buffer A(0) and used for co-IP experiments assayed by western blotting. Using the anti-MeCP2 3998 antibody to IP, Prpf3 was specifically co-IP’ed from these fractions and the interaction between MeCP2 and Prpf3 was not dependent on nucleic acids (Figure 4A, lane 3 and 4). Alternatively, to show co-IP interaction between MeCP2 and Prpf3 from crude cell extracts, the murine hippocampal HT-22 cell line was used to generate a pool of cells stably expressing HA epitope-tagged human MeCP2. Immunofluorescence showed that HA-MeCP2 in these cell lines predominantly concentrated to DAPI-rich heterochromatic foci as expected for endogenous MeCP2 (Figure S4). Anti-HA antibodies were then used to specifically co-IP HA-MeCP2 and MeCP2-associated proteins from these cell lines. Western blotting showed that HA-MeCP2 specifically co-IP’ed a fraction of the endogenous mouse Prpf3 (Figure 4B, lane 3), indicating that Prpf3 and MeCP2 exist in a stable complex within these neuronal cells, and their interaction was not dependent on nucleic acids (Figure 4B, lane 4). We conclude that MeCP2 and Prpf3 associate in vivo.
Figure 4. Co-IPs confirmed MeCP2 associates with Prpf3 in vivo.
(A) Western blotting of a co-IP experiment from fractionated brain extract showing: a peak MeCP2 fraction (lane 1, 5% input), control IgG IP (lane 2), anti-MeCP2 IP (lane 3), and anti-MeCP2 IP treated with benzonase (lane 4), demonstrates that Prpf3 interacts with MeCP2 in the brain independent of nucleic acids. (B) Western blotting of a co-IP experiment from HA-MeCP2 transfected HT22 cells showing: whole cell extract (lane 1, 5% input), IgG IP (lane 2), anti-HA IP (lane 3) and anti-HA IP treated with benzonase (lane 4), demonstrates that Prpf3 interacts with MeCP2 cell culture independent of nucleic acids.
MeCP2 interacts directly with Prpf3 and Sdccag1, independent of nucleic acids
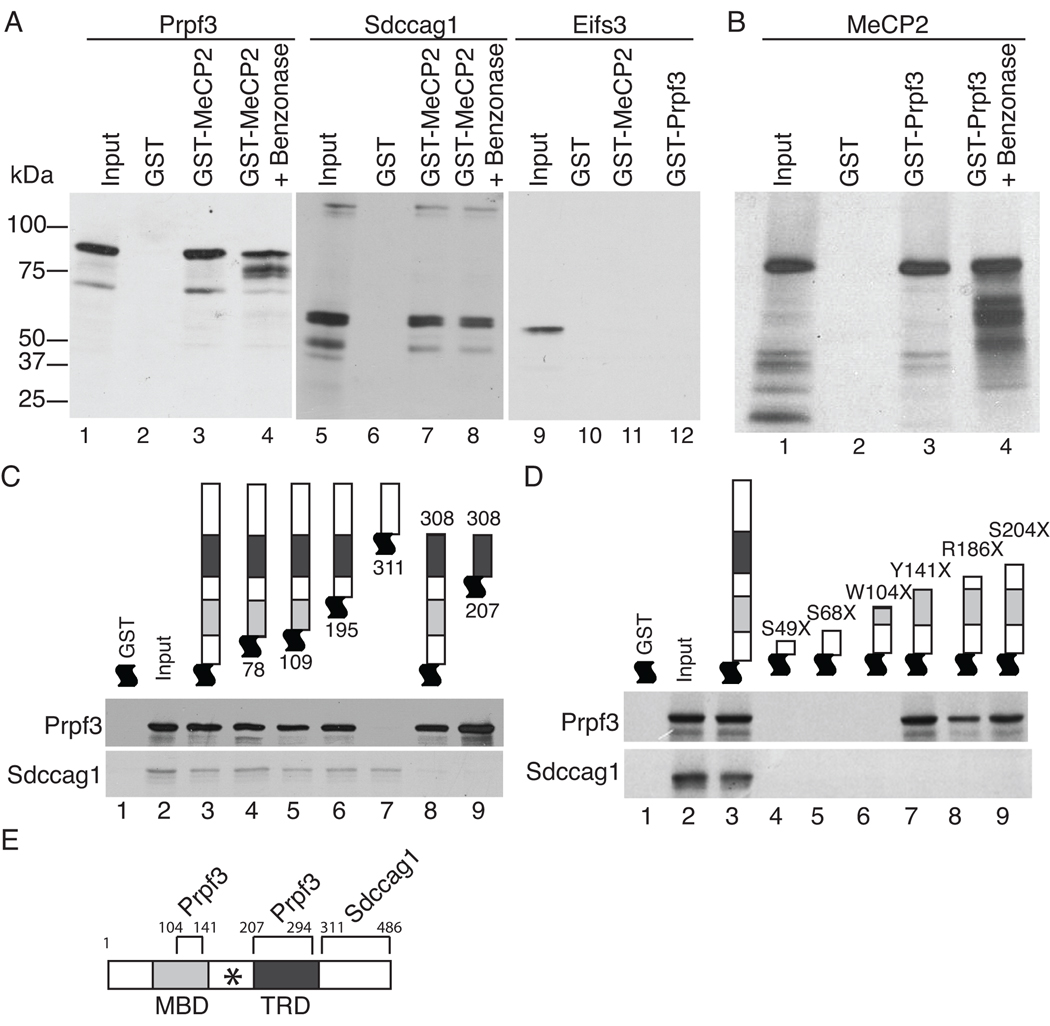
To investigate which of the co-purifying candidate proteins directly interacted with MeCP2, the cDNAs for each identified protein was cloned and the recombinant proteins were tested for the ability to interact with MeCP2 in vitro (Figures 5 and S4). GST pull-down assays using a GST-MeCP2 fusion protein showed that rat Prpf3 specifically interacted with full-length rat MeCP2, but not GST alone (Figure 5A, left panel), and reciprocally, rat GST-Prpf3 interacted with rat MeCP2 (Figure 5B). Furthermore, treatment of the GST-MeCP2/Prpf3 reaction with benzonase nuclease showed the interaction was not dependent on nucleic acids (Figure 5A, B). Similarly, rat Sdccag1 specifically interacted with rat GST-MeCP2 in a nucleic acid independent manner (Figure 5A, middle panel). Interestingly, neither rat GST-MeCP2 nor rat GST-Prpf3 interacted with any of the other four identified polypeptides supporting the specificity of the observed interactions of MeCP2, Prpf3 and Sdccag1 (Figure 5A, right panel and data not shown).
Figure 5. MeCP2 directly interacts with Prpf3 and Sdccag1.
For all experiments, the GST-tagged protein was bacterially generated and purified and the visualized [35S]-methionine labeled interacting proteins were generated by in vitro transcription and translation. Coomassie blue staining of the gels showing input GST fusion proteins are shown in Figure S4. (A) Prpf3 (left) and Sdccag1 (middle) interact directly with GST tagged MeCP2 but not GST alone. Eif2s3 (right) does not interact with either GST MeCP2 or GST Prpf3. Benzonase treatment indicates these interactions are independent of nucleic acids. (B) Reciprocally, MeCP2 interacts with GST tagged Prpf3 independent of nucleic acids. (C) GST-MeCP2 deletion constructs mapped the region of MeCP2 required for the direct interactions with Prpf3 or Sdccag1 in vitro. (D) GST tagged MeCP2 containing the indicated RTT nonsense mutations disrupt Prpf3 binding if truncations are prior to amino acid 104, but identify a second Prpf3 binding site on MeCP2 in the MBD between amino acids 104 and 141. All RTT truncations tested abolished Sdccag1 binding to MeCP2. (E) Map of MeCP2e2 protein showing the Prpf3 and Sdccag1 interaction domains with boundary amino acid numbers. The MBD, TRD, and (*) RNA binding domain [37] are indicated.
Mapping the Prpf3 and Sdccag1 interaction domains of MeCP2
The regions of MeCP2 that directly interact with Prpf3 and Sdccag1 were mapped by GST pull-down analysis. A series of bacterially generated human GST-MeCP2 deletion mutants were tested for their ability to interact with in vitro synthesized human Prpf3 (Figures 5C, D). Prpf3 retained the ability to interact with MeCP2 N-terminal deletions up through amino acid residue 195 as well as a C-terminal deletion lacking amino acid residues 309 – 486. However, Prpf3 did not interact with an MeCP2 deletion lacking amino acid residues 1 – 308, indicating that the TRD region is required for interaction (Figure 5C, upper panel). Therefore, a fragment of MeCP2 that contains only amino acid residues 207–308, corresponding to the TRD domain, was tested and found to interact with Prpf3 with similar apparent affinity as full length MeCP2. Similarly, Sdccag1 was synthesized in vitro and subjected to the same series of GST-MeCP2 deletions as Prpf3 (Figure 5C, lower panel). Sdccag1 was able to interact with all of the N-terminal MeCP2 deletion proteins tested, but failed to interact with a MeCP2 deletion lacking the region C-terminal to the TRD. Thus, the interaction domain with Sdccag1 resides between amino acid residues 309–486, adjacent with the Prpf3 interaction domain (Figure 5E).
Considering most known RTT mutations reside in MECP2 and many are nonsense mutations resulting in a truncated MeCP2 it is likely that some of these mutations would also disrupt the interactions of Prpf3 and/or Sdccag1 with MeCP2. A series of bacterially generated human GST-MeCP2 RTT nonsense mutants were produced and tested for their ability to interact with in vitro synthesized human Prpf3 and rat Sdccag1 (Figure 5D). The series of RTT mutations truncate MeCP2 ranging from amino acids 49 to 204. All of these RTT nonsense mutants lack the C-terminal region of MeCP2 and therefore disrupted its interaction with Sdccag1 as expected (Fig 5D, bottom panel). The Prpf3 interaction was disrupted by RTT truncations at amino acids S49X, S68X and W104X as well. However, the Prpf3 interaction with MeCP2 was maintained with the RTT truncations Y141X, R168X, and S204X. This indicates there are two MeCP2 regions capable of interacting with Prpf3 flanking the inter-domain region between the MBD and TRD of MeCP2. Interestingly, this same inter-domain region of MeCP2 has been previously characterized as the RG domain and is required for MeCP2’s RNA binding activity [37]. We conclude that MeCP2 contains two domains sufficient for interaction with Prpf3, one in the MBD between amino acids 104 and 141, and the second in the TRD between amino acids 207 and 294 and one domain for MeCP2’s interaction with Sdccag1, residing between amino acids 311 and 486 (Figure 5E). Therefore, any RTT mutation truncating MeCP2 at or before amino acid residue 104 will abolish the Prpf3 and Sdccag1 interaction, while any RTT truncation at or before amino acid residue 297 would disrupt the Sdccag1 interaction with MeCP2, all of which could affect MeCP2’s role in RNA biogenesis.
MeCP2 interacts with mRNA in vivo
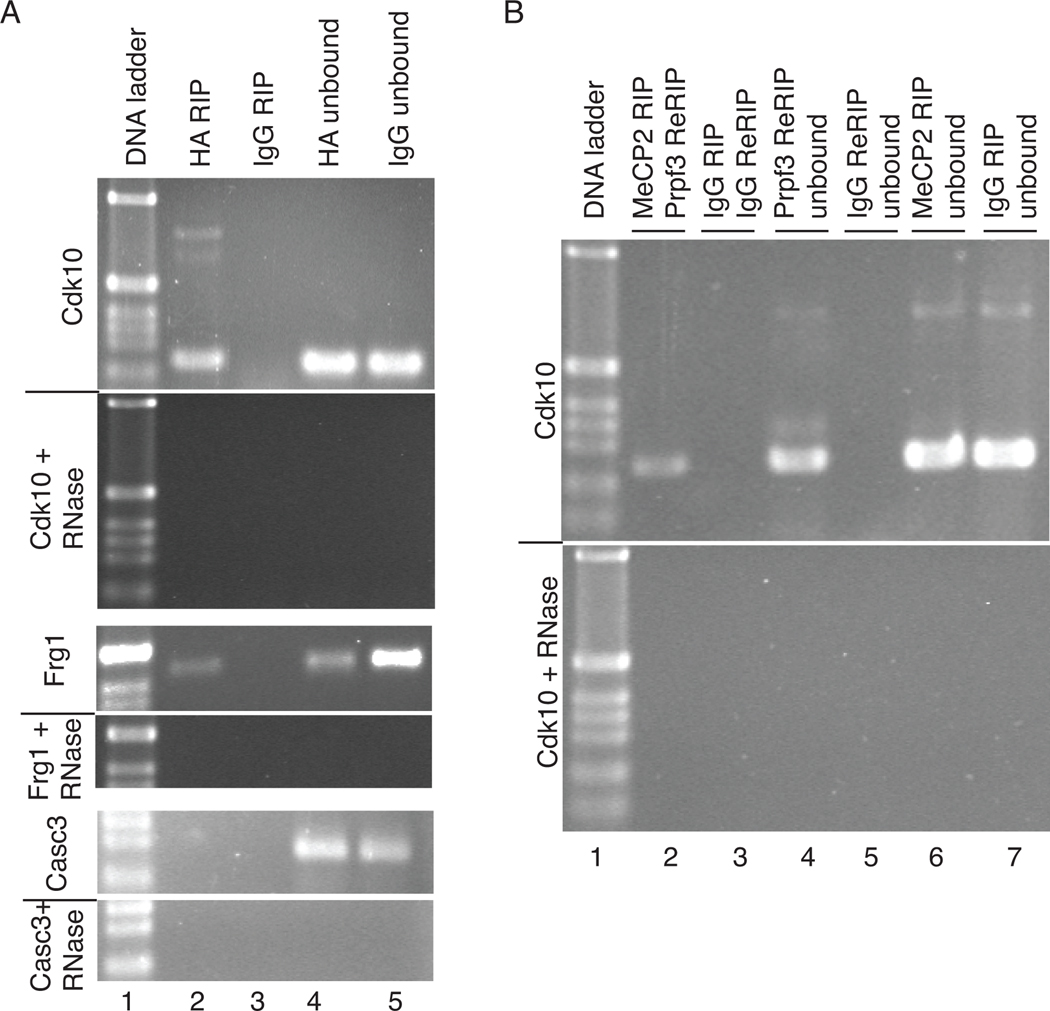
Several lines of evidence implicate that MeCP2 is associated with mRNA biogenesis; MeCP2 binds RNA in vitro [37], is part of a RNP complex with YB-1 in cell culture [23], MeCP2 knock-out mice show misspliced transcripts in the brain [23], it has been identified as a transcriptional activator at the majority of gene promoters it has been shown to regulate [21], and here MeCP2 is shown to interact with the splicing factor Prpf3. To determine if MeCP2 interacts with the RNA transcripts expressed from the genes it regulates in vivo, RNA immunoprecipitations (RIP) were performed. Using anti-HA antibodies on HT-22 (HA-MeCP2) cell lysates, mRNAs for the MeCP2-regulated genes Cdk10 and Frg1 were able to be specifically RIP’ed (Figure 6A). Assaying the IP’s by RT-PCRs using oligonucleotide primers designed to amplify across exon junctions of Cdk10 and Frg1 show MeCP2 is associated with the spliced form of the genes, while MeCP2 is not associated with the Casc3 gene transcript (Figure 6A), a gene not regulated by MeCP2 [21], supporting the specificity of the MeCP2-mRNA interaction. The RIPs were RNase sensitive confirming that RNA, and not DNA, was specifically IP’ed. Interestingly, RT-PCR for RIPs using primers for Cdk10 additionally show what appears to be pre-mRNA by size and RNase sensitivity (Figure 6A top). Although the Cdk10 pre-mRNA product was not observed in 100% of the RT-PCR analyses for Cdk10, it was observed repeatedly and suggests that MeCP2 associates with the Cdk10 RNA at a step prior to the completion of splicing and remains bound after RNA processing is completed.
Figure 6. The MeCP2-Prpf3 complex interacts with mRNA in vivo.
(A) RT-PCR analysis of a RIP from HA-MeCP2 stably transfected HT-22 cells indicates an association of HA-MeCP2 with Cdk10 mRNA (top, lane 2), and FRG1 mRNA (middle, lane 2), but not Casc3 mRNA (bottom, lane 2). Control RIPs using normal rabbit serum (IgG) (lane 3) show no RT-PCR product. RT-PCRs using unbound RNA from RIPs indicate target mRNAs were present in all RIP samples (lanes 4 and 5) and all RT-PCRs were RNase sensitive (+ RNase) confirming RNA and not DNA was being assayed. (B) An anti-HA RIP followed by anti-Prpf3 Re-RIP experiment from HA-MeCP2 HT-22 cells was assayed for Cdk10 mRNA by RT-PCR (lane 2). Control RIP and Re-RIP experiments using normal rabbit serum (IgG) showed no product by RT-PCR (lane 3). Unbound mRNA was assayed by RT-PCR for the RIP (lanes 6 and 7) and Re-RIPs (lanes 4 and 5) to confirm the presence of the Cdk10 mRNA in the reactions. All RT-PCRs were sensitive to RNase treatment (lower panel) confirming the amplifications were from RNA and not DNA templates.
To investigate if Prpf3 was part of the HA-MeCP2/mRNA complex, a RIP and Re-RIP approach was implemented. Anti-HA antibodies were used to RIP HA-MeCP2/mRNA complexes from HT-22 (HA-MeCP2) cell lysates, the bound complexes were eluted from the HA antibodies intact using DTT to disrupt the IgG structure, and then anti-Prpf3 antibodies were used to Re-RIP. Therefore, any RNAs present in the Re-RIP must have been associated with both MeCP2 and Prpf3. RT-PCR analysis showed that RIP for HA-MeCP2 and subsequent Re-RIP for Prpf3 protein IP-ed mRNA for Cdk10, indicating that both proteins are in fact associated with this target mRNA (Figure 6B), further supporting their being in a complex in vivo.
DISCUSSION
MeCP2 has been characterized as a multifunctional protein using a variety of techniques and from numerous cellular contexts, however only the biochemical characteristics of the endogenous MeCP2 protein in the mammalian brain are relevant to RTT. MeCP2 mutations identified from RTT patients are generally point mutations resulting in a single missense or nonsense amino acid change, and have been identified throughout all protein domains of MECP2 (www.rettsyndrome.org/), suggesting that all domains are critical for MeCP2 function [38]. Although all these mutations produce clinical RTT pathology, certain mutations are more strongly associated with particular symptoms and disease severity suggesting that all RTT mutations in MECP2 are not equal, with some potentially being more disruptive towards MeCP2’s many functions [39]. The underlying mechanisms of how these myriad mutations lead to RTT pathophysiology remain unclear due to a lack of understanding toward the complete scope of MeCP2’s normal function in the brain. Here, we identified four distinct brain-derived MeCP2 protein pools, agreeing with the functional data proposing multiple roles for MeCP2 [38]. Characterizing one complex as containing MeCP2, Pprf3, Sdccag1, and mRNA strongly complements previously published data that MeCP2 is an RNA binding protein involved in mRNA splicing [23].
MeCP2’s association with Prpf3, a major component of the spliceosome, supports MeCP2 as having a role in modulating mRNA splicing [23]; however, it is not clear what exactly that role might be. MeCP2 is present at the promoter of many actively transcribed genes [20], and activates a majority of genes it regulates in the mouse hippocampus [21]. Interestingly, a recent study suggests that differential gene body methylation may play a role in RNA transcript splicing [40]. Genome-wide methylation profiling in multiple human cell types found that exons were more highly methylated than introns, and that there were sharp transitions of DNA methylation at exon-intron boundaries [40]. Experimentally, it is known that MeCP2 binds RNA in vitro with a similar affinity as to methylated DNA, and that the two activities are mutually exclusive [37]. Thus, MeCP2 at an active promoter or within the body of a transcribed gene would be well positioned spatially upon its release to interact with the transcripts of the genes it activates and influence splice site selection. Therefore, it is reasonable to propose that MeCP2-activated genes would also be targets for splicing regulation by a MeCP2 containing complex. Supporting this model, RNA transcripts from Cdk10, a gene positively regulated by MeCP2 binding at its promoter [21] and abnormally spliced in Mecp2308/Y mice [23], were associated with both MeCP2 and Prpf3 in vivo (Figure 6).
Based on what is known about MeCP2 and Prpf3 independently, it is possible that the direct interaction of MeCP2 with Prpf3 could function in splice site selection. Prpf3 is one of multiple associated proteins of the U4/U6.U5 tri-snRNP complex that is recruited to the splice site to form and stabilize the functional spliceosome, and is essential for pre-mRNA splicing [31,41]. Importantly, MeCP2 can bind RNA directly [37], and has been implicated as a regulator of alternative splicing in the HeLa and Neuro2A cell lines through an RNA-dependent interaction with YB-1 [23], a protein known to participate in splicing of mRNAs [42]. While we were unable to detect YB-1 as a component of the MeCP2/Prpf3/Sdccag1 complex in brain, it is inefficient to inhibit the highly active and stable RNases that would abolish any YB-1/RNA/MeCP2 interaction during purification so we cannot rule out that YB-1 could be part of a larger RNA-dependent alternative splicing complex. Nevertheless, a direct interaction of MeCP2 and Prpf3 further supports a role for MeCP2 in mRNA splicing regulation. Notably, Mecp2308/Y mice, which produce a truncated form of MeCP2 and reproduce many of the classical features of RTT [43], have been shown to have multiple genes that are abnormally spliced in the brain [23]. This suggests the C-terminal portion of MeCP2, which we have identified as the putative Sdccag1 interaction domain, plays a critical role in regulating alternative splicing.
Sdccag1 was originally identified from colon cancer patients by a serological analysis of recombinant cDNA expression libraries (SEREX) [32] and later identified as a tumor suppressor by its ability to cause cell cycle arrest in a non-small-cell lung cancer cell line [44]. The significance of finding Sdccag1 as part of the MeCP2-Prpf3 complex can only be implied due to the limited information on its function in vertebrates and Drosophila. Bioinformatically the Sdccag1 protein contains a predicted RNA-binding domain homologous to a eukaryotic small nuclear RNP [45], suggesting the capacity to function in mRNA splicing. Functionally, the Drosophila Sdccag1 homolog Caliban has been shown to interact with and mediate the nuclear export of the Prospero homeodomain transcription factor (Prox in mammals) and this interaction and function is conserved in mammalian cells [33]. This raises the possibility that the MeCP2/Prpf3/Sdccag1 complex may not only be involved in splicing mRNA, but also transporting mRNAs. Consistent with this model, MeCP2 has been shown to have both a nuclear and cytoplasmic localization in neuronal cell lines [46].
Considering the range of mutations throughout MECP2 in RTT, it is not surprising that many RTT mutations are within, or predicted to affect both the Prpf3 and Sdccag1 binding domains, disrupting the MeCP2/Prpf3/Sdccag1 complex and presumably MeCP2-mediated splicing regulation and mRNA transport. With MeCP2 emerging as a multifunctional protein and its biological role in RTT unclear, identifying biochemically distinct pools of MeCP2 in the brain containing novel MeCP2 interacting proteins is a valuable tool towards an understanding MeCP2 function and its dysfunction in RTT. This study adds to the mounting evidence indicating that one such critical function of MeCP2 in the brain involves RNA biogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly appreciate the gift of HT-22 cells from Dr. Stephanie Ceman. We thank Dr. Ann Cheever and Dr. Brian Imai for technical support and Dr. Takako I. Jones for help with the manuscript.
FUNDING
This work was supported by the National Institutes of Health, National Institute of Child Health and Human Development [grant number 5K22HD001338 to PLJ]; and start-up funds for PLJ from the School of Molecular and Cellular Biology and Department of Cell and Developmental Biology, University of Illinois at Urbana-Champaign.
Abbreviations used
- CV
column volume
- IP
immunoprecipitation
- MeCP2
methyl CpG binding protein 2
- MBD
methyl cytosine binding domain
- ms
mass spectrometry
- Prpf3
pre-mRNA processing factor 3
- RIP
RNA IP
- RTT
Rett Syndrome
- Sdccag1
serologically defined colon cancer antigen 1
- TRD
transcription repression domain
Footnotes
AUTHOR CONTRIBUTION
SWL performed biochemistry, IPs, Co-IPs, RIPs, transfections and wrote the paper; JO initiated the project, performed biochemistry and the initial MeCP2 fractionation into multiple complexes; PMY performed the mass spectrometry and identifications; PLJ conceived the project, funded the project, performed biochemistry, and wrote the paper.
REFERENCES
- 1.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 4.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 5.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 6.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 7.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Rett A. On a unusual brain atrophy syndrome in hyperammonemia in childhood. Wien Med Wochenschr. 1966;116:723–726. [PubMed] [Google Scholar]
- 9.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 10.Ylisaukko-Oja T, Rehnstrom K, Vanhala R, Kempas E, von Koskull H, Tengstrom C, Mustonen A, Ounap K, Lahdetie J, Jarvela I. MECP2 mutation analysis in patients with mental retardation. Am J Med Genet A. 2005;132A:121–124. doi: 10.1002/ajmg.a.30416. [DOI] [PubMed] [Google Scholar]
- 11.Zoghbi HY. MeCP2 dysfunction in humans and mice. J Child Neurol. 2005;20:736–740. doi: 10.1177/08830738050200090701. [DOI] [PubMed] [Google Scholar]
- 12.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 13.Lam CW, Yeung WL, Ko CH, Poon PM, Tong SF, Chan KY, Lo IF, Chan LY, Hui J, Wong V, Pang CP, Lo YM, Fok TF. Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet. 2000;37:E41. doi: 10.1136/jmg.37.12.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, Clayton-Smith J. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 16.Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 17.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal N, Hardt T, Brero A, Nowak D, Rothbauer U, Becker A, Leonhardt H, Cardoso MC. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- 20.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 25.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 26.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 28.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 29.Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang A, Forman-Kay J, Luo Y, Luo M, Chow YH, Plumb J, Friesen JD, Tsui LC, Heng HH, Woolford JL, Jr, Hu J. Identification and characterization of human genes encoding Hprp3p and Hprp4p, interacting components of the spliceosome. Hum Mol Genet. 1997;6:2117–2126. doi: 10.1093/hmg/6.12.2117. [DOI] [PubMed] [Google Scholar]
- 32.Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Bi X, Jones T, Abbasi F, Lee H, Stultz B, Hursh DA, Mortin MA. Drosophila caliban, a nuclear export mediator, can function as a tumor suppressor in human lung cancer cells. Oncogene. 2005;24:8229–8239. doi: 10.1038/sj.onc.1208962. [DOI] [PubMed] [Google Scholar]
- 34.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 35.Matzat LH, Berberoglu S, Levesque L. Formation of a Tap/NXF1 homotypic complex is mediated through the amino-terminal domain of Tap and enhances interaction with nucleoporins. Mol Biol Cell. 2008;19:327–338. doi: 10.1091/mbc.E07-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C, Yang L, Yang JJ, Huang Y, Liu ZR. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 38.Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, Zoghbi H, Percy A, Glaze DG. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauber J, Plessel G, Prehn S, Will CL, Fabrizio P, Groning K, Lane WS, Luhrmann R. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- 42.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–3830. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 44.Carbonnelle D, Jacquot C, Lanco X, Le Dez G, Tomasoni C, Briand G, Tsotinis A, Calogeropoulou T, Roussakis C. Up-regulation of a novel mRNA (NY-CO-1) involved in the methyl 4-methoxy-3-(3-methyl-2-butenoyl) benzoate (VT1)-induced proliferation arrest of a non-small-cell lung carcinoma cell line (NSCLC-N6) Int J Cancer. 2001;92:388–397. doi: 10.1002/ijc.1197. [DOI] [PubMed] [Google Scholar]
- 45.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyake K, Nagai K. Phosphorylation of methyl-CpG binding protein 2 (MeCP2) regulates the intracellular localization during neuronal cell differentiation. Neurochem Int. 2007;50:264–270. doi: 10.1016/j.neuint.2006.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.