Abstract
The rapid and extensive spread of the human immunodeficiency virus (HIV) epidemic provides a rare opportunity to witness host–pathogen co-evolution involving humans. A focal point is the interaction between genes encoding human leukocyte antigen (HLA) and those encoding HIV proteins. HLA molecules present fragments (epitopes) of HIV proteins on the surface of infected cells to enable immune recognition and killing by CD8+ T cells; particular HLA molecules, such as HLA-B*57, HLA-B*27 and HLA-B*51, are more likely to mediate successful control of HIV infection1. Mutation within these epitopes can allow viral escape from CD8+ T-cell recognition. Here we analysed viral sequences and HLA alleles from >2,800 subjects, drawn from 9 distinct study cohorts spanning 5 continents. Initial analysis of the HLA-B*51-restricted epitope, TAFTIPSI (reverse transcriptase residues 128–135), showed a strong correlation between the frequency of the escape mutation I135X and HLA-B*51 prevalence in the 9 study cohorts (P = 0.0001). Extending these analyses to incorporate other well-defined CD8+ T-cell epitopes, including those restricted by HLA-B*57 and HLA-B*27, showed that the frequency of these epitope variants (n = 14) was consistently correlated with the prevalence of the restricting HLA allele in the different cohorts (together, P < 0.0001), demonstrating strong evidence of HIV adaptation to HLA at a population level. This process of viral adaptation may dismantle the well-established HLA associations with control of HIV infection that are linked to the availability of key epitopes, and highlights the challenge for a vaccine to keep pace with the changing immunological landscape presented by HIV.
The extent to which HIV is evolving at the population level in response to immune selection pressure is under debate2–6. Resolving the impact of HLA class I alleles on viral evolution is problematic because it can be obscured by other influences, such as founder effect6 (polymorphisms present within the early strains establishing the epidemic in a group). In addition, most HLA alleles do not drive significant selection pressure on HIV, a proportion of escape mutations revert to wild type after transmission, and different HLA alleles may drive the identical escape mutation7.
To test the hypothesis that the frequency of escape mutations in a given population is correlated with the prevalence of the relevant HLA allele in that population, we studied nine distinct cohorts from North America, the Caribbean, Europe, sub-Saharan Africa, Australia and Japan, in which we performed HLA typing, and defined the viral mutations arising within CD8+ T-cell epitopes. We focused initially on a well-characterized mutation, I135X, within the HLA-B*51-restricted epitope, TAFTIPSI (RT 128–135)8, because it arises in acute infection, non-HLA-B*51 alleles do not also select this mutation7,9, and it does not revert to Ile 135 after transmission to HLA-B*51-negative subjects9. Thus, if highly prevalent HLA alleles drive a high frequency of escape mutations in the population, this would be most obvious in relation to HLA-B*51 and the escape mutant I135X. We then considered an additional 13 well-defined escape mutations, including those known to reduce viral fitness and therefore liable to revert after transmission.
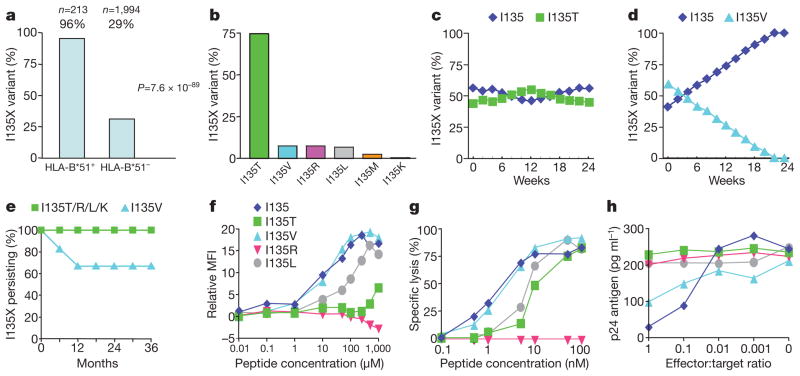
I135X was selected in 205 of 213 (96%) HLA-B*51-positive individuals analysed (Figs 1 and 2, and Supplementary Fig. 1). The I135X variants do not significantly affect viral replicative capacity in vitro, other than the rare I135V mutation. This was the only variant observed to revert to wild-type in vivo during a 3-year follow-up of 38 HLA-B*51-negative subjects identified during acute HIV infection who carried I135X mutant viruses at transmission (Fig. 1e). The I135X mutants substantially affect HLA binding, and therefore also recognition by CD8+ T cells (Fig. 1f–h). Thus, HIV transmission from HLA-B*51-positive subjects would probably involve transmission of I135X, which would persist in the new host. Newly infected HLA-B*51-positive subjects receiving an I135X mutant would be unable to generate an HLA-B*51-TAFTIPSI-specific response.
Figure 1. Selection and fitness cost of I135X escape variants and recognition by the HLA-B*51–TAFTIPSI (RT 128–135)-specific CD8+ T cells.
a, Association between I135X and HLA-B*51 in all study cohorts. b, Ile 135 variation in HLA-B*51-positive subjects. c, d, In vitro competition assays between NL4-3 wild-type virus and I135X viral variants (I135T (c) and I135V (d)). I135R and I135L showed no fitness cost (not shown). e, Persistence of I135X mutants in 38 HLA-B*51-negative subjects followed from acute infection. f, TAFTIPSI variant binding to HLA-B*51 (see Methods). MFI, mean fluorescence intensity. g, h, Recognition of peptide-pulsed HLA-B*51-matched targets and viral variants by representative TAFTIPSI-specific CD8+ T-cell clones
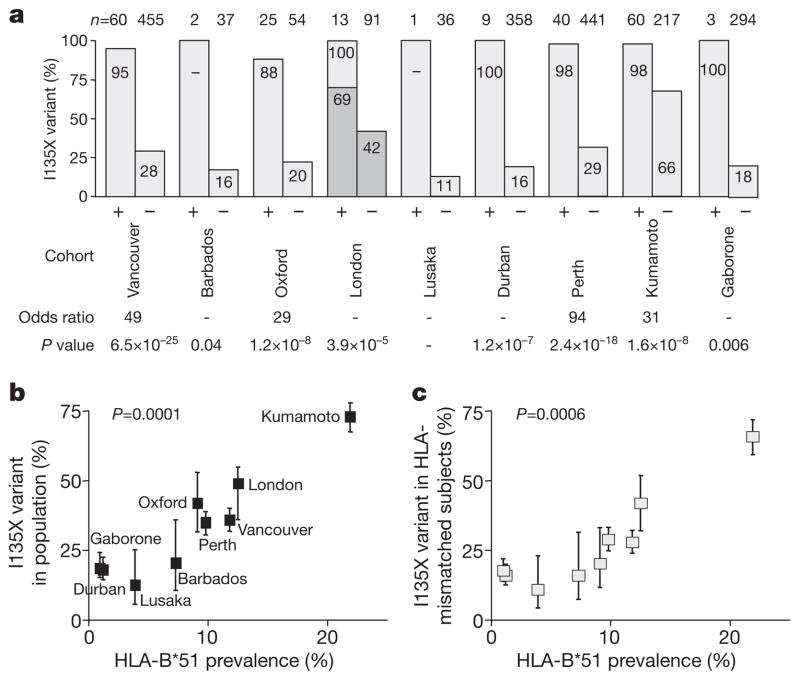
Figure 2. Correlation between frequency of HLA-B*51-associated escape mutations and HLA-B*51 prevalence in study cohorts.
a, Frequency of I135X mutations within TAFTIPSI (RT 128–135) in HLA-B*51-positive (+) and -negative (−) subjects within nine study cohorts. In the acute cohort (London) 69% of HLA-B*51-positive subjects expressed I135X mutant at enrolment, 100% within 2 years of baseline (Supplementary Fig. 1). b, Correlation between frequency of I135X mutation and HLA-B*51 prevalence in the nine study populations. Logistic regression P = 0.0001 (Supplementary Table 1). c, Correlation between I135X frequency in HLA-B*51-negative subjects and HLA-B*51 prevalence in nine study populations. Error bars represent 95% confidence limits, obtained using a binomial error distribution.
To test the hypothesis that the population frequency of I135X is correlated with HLA-B*51 prevalence, HIV sequence and HLA data were collated from the nine study cohorts. One cohort comprised subjects with acute/early HIV infection; the remaining cohorts comprised chronically infected subjects. In all cohorts the odds ratio strongly favoured I135X in the HLA-B*51-positive subjects, even in the acute cohort where I135X was selected sufficiently early to be already over-represented in HLA-B*51-positive subjects (odds ratio 1.65, P = 0.07, Fig. 2a). In Japan, where HLA-B*51 is highly. prevalent10 (21.9% of the study cohort), the frequency of I135X was >50%, and overall across all cohorts the I135X frequency was strongly correlated with HLA-B*51 prevalence (P = 0.0001, Fig. 2b). To control for the possibility that disproportionately more virus sequences from HLA-B*51-positive subjects were analysed, the same analysis comparing I135X frequency in HLA-B*51-negative subjects only was undertaken, with similar findings (Fig. 2c, P = 0.0006). These data suggest that HIV may be adapting to HLA-B*51 with respect to the HLA-B*51–TAFTIPSI response in localities where HLA-B*51 is at high prevalence.
Additional evidence that I135X is accumulating in Japan comes from the observation that only 3 of 14 (21%) HLA-B*51-negative Japanese haemophiliacs infected in 1983 carried I135X, compared with 30 of 43 (70%) HLA-B*51-negative subjects infected between 1997 and 2008 (P = 0.002). Furthermore, HLA-B*51 does not protect against disease progression in Japanese subjects infected between 1997 and 2008, whereas HLA-B*51-positive haemophiliacs infected in 1983 had lower viraemia levels and higher CD4 counts than HLA-B*51-negative haemophiliacs (Supplementary Fig. 2). These data are consistent with fewer HLA-B*51-positive subjects targeting TAFTIPSI during 1997–2008, owing to a population-level increase in the HLA-B*51 I135X escape mutation over this 14–25-year period.
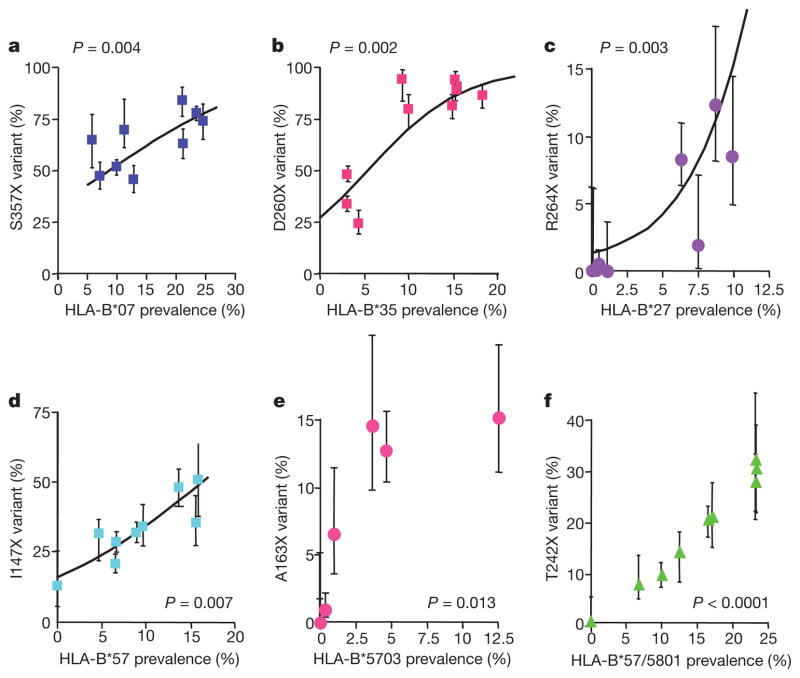
To investigate HIV adaptation to other HLA alleles, we initially examined other escape mutations shown previously to persist stably after transmission5,7. We selected the three non-reverting Gag polymorphisms that, from analysis of 673 study subjects in Durban, South Africa7, were most strongly associated with the relevant restricting allele (P < 10−6 after phylogenetic correction), namely, S357X, D260X and D312X within epitopes restricted, respectively, by HLA-B*07 (GPSHKARVL, Gag 355–363), HLA-B*35 (PPIPVGDIY, Gag 254–262) and HLA-B*44 (AEQATQDVKNW, Gag, 306–316). In addition, we analysed a non-reverting I31V variant (LPPIVAKEI, Int 28–36) previously hypothesized to increase in relation to population HLA-B*51 prevalence5. These additional polymorphisms show a similar relationship to that between I135X and HLA-B*51, overall showing a strongly significant correlation between variant frequency and prevalence of the restricting HLA allele (Figs 3 and 4a, and Supplementary Fig. 3).
Figure 3. Correlation between frequency of HIV sequence variant and HLA prevalence for six additional well-characterized epitopes.
P values calculated after logistic regression analysis as shown (calculations after linear regression analysis are shown in Supplementary Table 1). a, Frequency of the S357X mutation within the HLA-B*07-restricted epitope GPSHKARVL (Gag 355–363). b, Frequency of the D260X mutation within the HLA-B*35-restricted epitope PPIPVGDIY (Gag 254–262). c, Frequency of the R264X mutation within the HLA-B*27-restricted epitope KRWIILGLNK (Gag 263–272). d, Frequency of the I147X mutation within the HLA-B*57-restricted epitope ISPRTLNAW (Gag 147–155). e, Frequency of the A163X mutation associated with the HLA-B*5703-restricted epitope KAFSPEVIPMF (Gag 162–172). f, Frequency of the T242X mutation within the B*57/5801-restricted epitope TSTLQEQIAW (Gag 240–249). Error bars represent 95% confidence limits, obtained using a binomial error distribution.
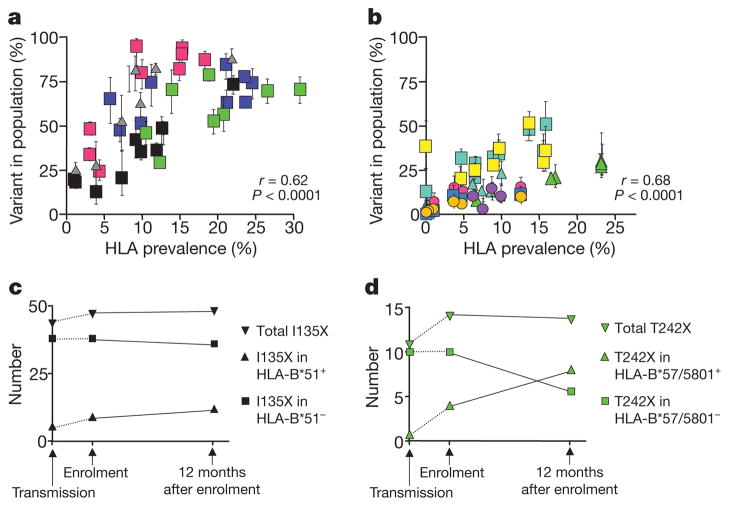
Figure 4. Correlation between HIV variant frequency and HLA prevalence for all epitopes studied.
a, Correlation between HLA prevalence and the five stable, non-reverting variants (symbols in Figs 2 and 3, and Supplementary Fig. 3; grey triangles, I31V; green squares, D312X). b, Eight variants demonstrated to reduce viral fitness (see text, Fig. 3 and Supplementary Fig. 3; turquoise triangles, L268X; yellow squares. A146X; sky-blue squares, V168I; yellow circles, I247X). c, d, Data from acute London cohort. c, Number of HLA-B*51-positive and HLA-B*51-negative subjects carrying the non-reverting I135X variant. The percentage of I135X in HLA-B*51-negative subjects at enrolment (42%) assumed the percentage of I135X in all subjects at transmission (I135X frequency in HLA-B*51-positive subjects at enrolment was 69%, P = 0.07). d, The reverting HLA-B*57/5801-restricted T242X mutation. T242X frequency in HLA-B*57/5801-negative subjects at enrolment was 7%, versus 33% in HLA-B*57/5801-positive subjects (P = 0.01). Error bars represent 95% confidence limits, obtained using a binomial error distribution.
The spectrum of HLA-associated polymorphisms also includes mutations reducing viral fitness1. These either revert to wild type after transmission, or persist in the presence of compensatory mutations. We extended these analyses to include epitopes restricted by HLA-B*27 and HLA-B*57, alleles strongly associated with successful immune control of HIV11,12. The mutations analysed themselves are associated with precipitating loss of immune control13–16 and all inflict a documented viral fitness cost, either demonstrated by in vitro fitness studies and/or in vivo reversion7,14,17–21 (data not shown for V168I).
Again, a strong correlation between escape mutant frequency and prevalence of the restricting HLA allele was observed (Figs 3c–f and 4b, and Supplementary Fig. 3; overall, for these nine variants affecting viral fitness, r = 0.69, P < 0.0001). Unexpectedly, this correlation remained significant even when comparing HLA prevalence with variant frequency in the HLA-mismatched population (r = 0.40, P = 0.0004). As anticipated, non-reverting variants such as I135X accumulate at the population level, but even rapidly reverting18,20 mutations such as T242N can accumulate, if the selection rate exceeds the reversion rate (Fig. 4c, d).
Although frequency of the analysed HIV polymorphisms and HLA prevalence were strongly correlated overall, some anomalies were observed. For example, despite a 0% prevalence of HLA-B*57 in Japan10, 38% of the Japanese cohort had the HLA-B*57-associated A146X variant. One potential explanation might be A146X selection by non-HLA-B*57 Japanese alleles. Analysing Gag sequences from Japanese study subjects, we observed a strong association between A146P and HLA-B*4801 (P = 0.00035), and then that A146P is indeed selected in HLA-B*4801-positive subjects (Supplementary Fig. 4a, b). We defined a novel HLA-B*4801-restricted epitope (Gag 138–147), showing also that A146P is an escape mutant (Supplementary Fig. 4c–f). These data illustrate that more than one HLA allele can drive the selection of a particular escape mutant (Supplementary Fig. 5). Also, in populations where HIV-specific CD8+T-cell responses are incompletely characterized, the influences of locally prevalent HLA alleles on HIV sequence variation are unknown.
These data show a strong correlation between HLA-associated HIV sequence variation and HLA prevalence in the population (r = 0.69, P < 0.0001, Supplementary Fig. 6), suggesting that the frequency of the studied variants is substantially driven by the HLA-restricted CD8+ T-cell responses. Non-reverting variants5,7, as well as those previously shown to arise at a fitness cost7,14,16–21, were studied. The latter constitute approximately 55–65% of HLA-associated polymorphisms7,20. This current analysis included epitopes whose role in HIV immune control is unknown, as well as those believed to contribute significantly to containment of HIV4,7,13–19. Analysis of well-characterized epitopes only also served to limit potential confounding influences of epitope clustering (selection of the same variant by different HLA alleles) and of founder effect. Either would be capable of obscuring a true HLA effect on population variant frequency.
The HLA-B*57-associated A146X mutation illustrates the complexity that may result from epitope clustering. A146X is selected by at least six distinct HLA alleles (Supplementary Fig. 5). A true correlation existing between mutation frequency and individual HLA allele prevalence might thus be obscured by selection of the same mutation by other alleles.
Founder effect also has an undoubted influence on population frequencies of particular polymorphisms6. Phylogenetic correction of sequence data excludes founder effect as a confounder6,7,9, and the highly significant associations between the presence of particular HLA alleles and all 14 HIV polymorphisms studied, persisting after phylogenetic correction (Supplementary Table 3), provide compelling evidence that the effects observed here are substantially HLA-driven. The large numbers of study subjects in these current studies reduce the likelihood of genuine HLA associations with HIV amino acid polymorphisms being obscured by founder effects. The relative impact of HLA and founder effect on variant frequency is harder to quantify, and is likely to differ substantially between particular populations.
The consequence of HIV adapting to certain CD8+ T-cell responses is unknown. For non-reverting polymorphisms such as HLA-B*35-associated D260E, the variant approaches fixation, because even at population frequencies of 90%, D260E is still significantly selected in HLA-B*35-positive subjects (Supplementary Fig. 7b). Important questions relevant to vaccine design include the extent and rate of sequence change in populations. Relevant factors include the selection rate in subjects expressing the HLA allele, the reversion rate in HLA-mismatched subjects, the population HIV transmission rate, and HLA allele prevalence. Models would need to include factors such as the selection of compensatory mutations to slow reversion rates, and antiretroviral therapy access that would slow transmission rates.
HLA adaptation to certain CD8+ T-cell responses may also alter currently established HLA associations with slow disease progression. Data here suggest that, whereas 25 years ago HLA-B*51 was protective in Japan11,12, this is no longer the case (Supplementary Fig. 2). The apparent increase in I135X frequency in Japan over this time supports the notion that HLA-B*51 protection against HIV disease progression hinges on availability of the HLA-B*51-restricted TAFTIPSI response. However, whether this is the case remains unknown.
For HLA-B*27 and HLA-B*57, there is more clear-cut evidence that their association with HIV control depends on the Gag-specific epitopes presented and analysed here4,7,13–15,18,19. For each of the HLA-B*27- and HLA-B*57-associated Gag mutations studied, an in vitro fitness cost or in vivo reversion has been observed. A strong correlation between variant frequency and HLA prevalence even for rapidly reverting variants can be explained, either by mutant acquisition exceeding reversion rate (Fig. 4D), or by selection of compensatory mutations slowing or halting reversion altogether. The clearest example of the latter is the HLA-B*27-associated R264K mutation, ‘corrected’ by S173A19. Compensatory mutations are also well described for the HLA-B*57-associated Gag mutations14,18. These data suggest that the escape mutations in these HLA-B*27- and HLA-B*57-restricted epitopes are accumulating over time. Several studies have now demonstrated that transmission of viruses encoding escape mutants in the critical Gag epitopes to individuals expressing the relevant MHC class results in failure to control viraemia2,21,22. The accumulation at the population level of these escape mutations in HLA-B*27 and HLA-B*57 Gag epitopes is therefore likely to reduce the facility of these alleles to slow HIV disease progression.
The longer-term consequences of this process for immune control of HIV are unknown. Loss of currently immunodominant epitopes would promote subdominant CD8+ T-cell responses, which can be more effective23,24. Also, the adapted virus provides new epitopes that can be presented, potentially with beneficial effects. In hepatitis C virus, for example, HLA-A*0301 holds a particular advantage, but only against the specific strain of virus responsible for the Irish outbreak25. In HIV, HLA-B*1801 is associated with high viraemia in C clade but not in B clade infection10,11,26; the opposite applies to HLA-B*5301.
Thus, the data presented here, showing evidence that the virus is adapting to CD8+ T-cell responses, some of which may mediate the well-established associations (HLA-B*57, HLA-B*27 and HLA-B*51) with immune control of HIV, highlight the dynamic nature of the challenge for an HIV vaccine. Important questions to be addressed include the speed and extent of sequence change, particularly in Gag, the most effective target for CD8+ T-cell responses1,7,13,21. The induction of broad Gag-specific CD8+ T-cell responses may be a successful vaccine strategy, but such a vaccine will be most effective if tailored to the viral sequences prevailing, and thus may need to be modified periodically to keep pace with the evolving virus. Moreover, the strong associations between certain HLA class molecules, such as HLA-B*57, HLA-B*27 and HLA-B*51, and slow disease progression may decline as the epidemic continues, particularly where these HLA alleles are highly prevalent, and where HIV transmission rates are high.
METHODS SUMMARY
Overall 2,875 subjects were studied, from 9 previously established study cohorts. These cohorts comprised subjects from North America, the Caribbean, Europe, sub-Saharan Africa, Australasia and Asia. All subjects were antiretroviral-therapy-naive. Apart from the London acute cohort (n = 142), all cohorts comprised chronically infected subjects. The 14 variants studied are well-defined escape mutations within well-characterized CD8+ T-cell epitopes, and included those persisting after transmission and likely to have little effect on viral fitness (n = 5), as well as those shown previously to reduce viral fitness (n = 9). Autologous HIV-1 sequences, and HLA class I types, were determined for all study subjects. The replicative capacity of I135X variants selected within the HLA-B*51-restricted epitope TAFTIPSI (RT 128–135) was assessed via in vitro competition assays and also via longitudinal follow-up of HLA-B*51-negative subjects infected acutely with I135X variants. Polymorphism frequency in the study cohorts was compared with prevalence of the relevant HLA molecule in the study cohort using a logistic regression model taking into account the different numbers of study subjects in each cohort. Demonstration of an HLA allele driving escape at Gag 146 in the Japanese cohort was undertaken first by identification of an association between HLA-B*4801 and A146P, subsequent definition of an HLA-B*4801-restricted CD8+ T-cell response to a novel epitope Gag 138–147 (LI10), and finally demonstration that A146P reduced viral recognition by LI10-specific CD8+ T cells.
Supplementary Material
Acknowledgments
This work is funded by grants from the National Institutes of Health (RO1AI46995 (P.G.), 1 R01 AI067073 (B.D.W.), R01AI64060 (E.H.)), the Wellcome Trust (P.G., P.K.), the UK Medical Research Council (J.F., A.P. and P.M.), and the Mark and Lisa Schwartz Foundation, the Ministry of Health, Labour and Welfare (Health and Labour HIV/AIDS Research Grants 012), the NIHR Biomedical Research Centre Programme and the Ministry of Education, Science, Sports and Culture (number 18390141), Japan (M.T.). P.G. is an Elizabeth Glaser Pediatric AIDS Foundation Scientist; J.G.P. is a Marie Curie Fellow (contract number IEF-041811). The authors are also grateful to A. McLean and H. Fryer for discussions of the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Y.K., K.P., J.F. and P. M. undertook much of the experimental work and data analysis, and contributed equally. M.T. and P.G. undertook much of the project conception, planning, supervision, analysis and writing of the manuscript, and contributed equally.
Author Information Accession numbers for newly determined viral sequences are included in Supplementary Information. Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to P.G. (philip.goulder@paediatrics.ox.ac.uk).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Goulder PJR, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nature Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulder PJR, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 3.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 4.Draenert R, et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leslie AJ, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya T, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 7.Matthews P, et al. Central role of reverting mutations in HLA associations with viral setpoint. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomiyama H, et al. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum Immunol. 1999;60:177–186. doi: 10.1016/s0198-8859(98)00113-x. [DOI] [PubMed] [Google Scholar]
- 9.Brumme Z, et al. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- 10.Itoh Y, et al. High throughput DNA tying of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 11.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nature Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien SJ, Gao X, Carrington M. HLA and AIDS: a cautionary tale. Trends Mol Med. 2001;7:379–381. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- 13.Kiepiela P, et al. CD8+T-cell responses to different HIV proteins have discordant associations with viral load. Nature Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 14.Leslie AJ, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nature Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 15.Goulder PJR, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nature Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 16.Feeney ME, et al. Immune escape precedes breakthrough HIV-1 viremia and broadening of the CTL response in a HLA-B27-positive long-term nonprogressing child. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Picado J, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford H, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneidewind A, et al. Escape from a dominant Gag-specific CTL response in HLA-B27+subjects is associated with a dramatic reduction in HIV-1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brumme Z, et al. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goepfert P, et al. Transmission of Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki S, et al. Transmission of SIV carrying multiple cytotoxic T lymphocyte escape mutations with diminished replicative capacity can result in AIDS progression in Rhesus macaques. J Virol. 2008;82:5093–5098. doi: 10.1128/JVI.02607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallimore A, Dumrese T, Hengartner H, Zinkernagel RM, Rammensee HG. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtappels R, et al. Subdominant CD8 T-cell epitopes account for protection against cytomegalovirus independent of immunodomination. J Virol. 2008;82:5781–5796. doi: 10.1128/JVI.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKiernan SM, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 26.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.