Abstract
Storage conditions are considered to be a critical component of DNA-based microbial community analysis methods. However, whether differences in short-term sample storage conditions impact the assessment of bacterial community composition and diversity demands systematic and quantitative assessment. Therefore, we used barcoded pyrosequencing of bacterial 16S rRNA genes to survey communities, harvested from a variety of habitats (soil, human gut (feces) and human skin) and subsequently stored at 20°, 4°, −20°, and −80°C for 3 and 14 days. Our results indicate that the phylogenetic structure and diversity of communities in individual samples was not significantly influenced by storage temperature or duration of storage. Likewise, the relative abundances of most taxa were largely unaffected by temperature even after 14 days of storage. Our results indicate that environmental factors and biases in molecular techniques likely impart greater amounts of variation to microbial communities than do differences in short-term storage conditions, including storage for up to two weeks at room temperature. These results suggest that many samples collected and stored under field conditions without refrigeration may be useful for microbial community analyses.
Keywords: microbial community storage conditions, environmental and human metagenomic studies, barcoded bacterial 16S rRNA gene pyrosequencing, phylogenetic- and taxonomic-based community analyses, soil, human fecal and human skin microbiota
Introduction
The treatment and handling of samples after collection is a critical aspect of study design when using DNA-based methods to compare the composition and diversity of microbial communities from environmental samples. It is widely assumed that microbial DNA must be extracted from the samples immediately after collection or, if this is not possible, that samples must be frozen (Rochelle et al., 1994). Samples stored at room temperature even for a short period before DNA is extracted are often considered unfit for downstream analyses because of changes to the microbial community. Although these assumptions are widespread, few and conflicting studies have directly tested the influence of storage conditions on DNA-based bacterial community analyses. For example, Dolfing et al. (2004) and Klammer et al. (2005) used DNA fingerprinting methods to show that the overall structure of soil bacterial communities was not strongly affected by storage conditions. Likewise, Roesch et al. (2009) reported only modest shifts in bacterial diversity in only one of four human gut samples after 72 hours of storage at room temperature. In contrast, both Tzeneva et al. (2009) and Ott et al. (2004) observed significant effects of storage conditions on the composition and diversity of microbial communities in soil and human gut samples, respectively. Nechvatal et al. (Nechvatal et al., 2008) tested a series of room-temperature methods for preserving stool for up to 5 days and found considerable variability among methods, but tested each sample with a different stool specimen, limiting their ability to draw general conclusions because of the large degree of variability among stool specimens from different human subjects.
Because of the diversity of samples, conditions tested and analytic methods employed, we still lack a comprehensive understanding of how and whether storage of samples prior to DNA extraction impacts bacterial community analyses and the magnitude of these potential storage effects. In particular, we do not know whether variation in storage conditions (temperature and length of storage) influences our ability to resolve differences in bacterial community composition and diversity between samples. To address these knowledge gaps, we analyzed bacterial communities in soil, human skin, and human fecal samples stored for different amounts of time and at varying temperatures using a barcoded pyrosequencing procedure with the sequence data from each sample analyzed using both phylogenetic and taxonomic community analysis procedures.
Methods
Sample collection and storage conditions
Microbial communities were sampled from three distinct habitats: surface soils, human skin, and human feces. Fecal samples (Fecal 1 and 2; c. 100 g each) were donated by two anonymous male participants. Immediately after collection, each sample was homogenized by stirring with a sterile spatula without added buffer in a sterile container. Replicate sub-samples (n=24) of each homogenized fecal sample were obtained by inserting sterile cotton swabs into each sample, and then placing the swab into its own separate dry, sterile 15 ml conical tube. Soil was collected (3- 2.5 × 10 cm cores) from two locations on the campus of the University of Colorado (40° 0′ N, 105° 16′ W) in June 2009. One set of cores was collected from underneath a Pinus ponderosa tree (Soil 1) while the other was collected from an irrigated lawn (Soil 2). Replicate cores were composited and sieved through 2 mm mesh and thoroughly homogenized by hand. From these two soil samples, forty-eight 1g sub-samples (n=24 per sample) were each placed in 1.5 ml centrifuge tubes. Skin samples were taken from the axillae (armpits) of one male and one female volunteer using sterile cotton swabs that had been pre-moistened in a sterile solution of 0.15 M NaCl and 0.01% TWEEN 20 (Paulino et al., 2006; Fierer et al., 2008). The axillary surface was swabbed for 30 s with all 24 swabs per individual at one time. The swabs were then placed in sterile 15 ml conical tubes for storage.
Replicate sub-samples of each community type (n=3) were subsequently stored at 20°, 4°, −20° and −80°C for either 3 or 14 days before DNA extraction. All sample-treatment combinations (4 storage temperatures; 2 storage times; 6 unique samples) were analyzed in triplicate as described in the next paragraph. Participants in the study gave informed consent under the sampling protocol approved by the University of Colorado Human Research Committee (protocol 1007.39).
DNA extraction, PCR , amplicon pooling and pyrosequencing
Genomic DNA was extracted from all samples with the MoBio Power Soil DNA extraction kit (MoBio, Carlsbad, CA, USA) as described previously (Fierer et al., 2008; Lauber et al., 2009). Note that samples were placed in bead tubes containing solution C1 and incubated at 65°C for 10 min followed by 2 min bead beating with the MoBio vortex adapter; the remaining steps of the extraction were performed as directed by the manufacturer. PCR amplification of bacterial 16S rRNA genes using primers directed at variable regions V1 and V2 (positions 27 to 338 according to E. coli 16S rRNA gene numbering scheme) was achieved following the protocol described in our earlier publications (Fierer et al., 2008; Lauber et al., 2009). Briefly, amplicons generated from three PCR reactions per sample were pooled to reduce per-PCR variability and purified with the MoBio Ultra Clean PCR clean up kit according to the manufacturer’s instructions and quantified (PicoGreen; Invitrogen, Carlsbad, CA, USA). No-template PCR controls were also performed. PCR products generated from each sub-sample contained a sub-sample specific, error correcting barcode, which allowed us to assemble a single composite sample for pyrosequencing by combining equal amounts of amplicon from each sub-sample. The composite sample was then gel purified (Qiaquick gel Clean up kit, Qiagen, Valencia, CA, USA) and precipitated with ethanol to remove any remaining contaminants. DNA was sequenced using a Roche 454 FLX pyrosequencer.
Sequence analysis
16S rRNA sequences were processed according to methods described in our previous publications (Fierer et al., 2008; Hamady et al., 2008). Briefly, sequences less than 200 nt or greater than 300 nt or with average quality scores of < 25 were removed from the dataset, as were those with uncorrectable barcodes, ambiguous bases or if the bacterial 16S rRNA gene-specific primer was absent. Sequences were then assigned to the specific sub-samples based on their unique 12 nt barcode and then grouped into phylotypes at the 97% level of sequence identity using cd-hit (Li & Godzik, 2006) with minimum coverage of 97%. We chose to group the phylotypes at 97% identity because this matches the limits of resolution of pyrosequencing (Kunin et al., 2010) and because the branch length so omitted contributes little to the tree, and therefore to phylogenetic estimates of beta diversity (Hamady et al., 2009b). A representative for each phylotype was chosen by selecting the most abundant sequence in the phylotype with ties being broken by chosing the longest sequence. A phylogenetic tree of the representative sequences was constructed using the Kimura 2-parameter model in Fast Tree (Price et al., 2009) after sequences were aligned with NAST (minimum 150 nt at 75% minimum identity) (DeSantis et al., 2006a) against the GreenGenes database (DeSantis et al., 2006b). Hypervariable regions were screened out of the alignment using PH Lane mask (http://greengenes.lbl.gov/). Differences in community composition for each pair of samples were determined from the phylogenetic tree using the weighted and unweighted UniFrac algorithms (Lozupone & Knight, 2005; Lozupone et al., 2006). UniFrac is a tree-based metric that measures the distance between two communities as the fraction of branch length in a phylogenetic tree that is unique to one of the communities (as opposed to being shared by both). This method of community comparison accounts for the relative similarities and differences among phylotypes (or higher taxa) rather than treating all taxa at a given level of divergence as equal (Lozupone & Knight, 2008). Although UniFrac depends on a phylogenetic tree, it is relatively robust to differences in tree reconstruction method or to the approximation of using phylotypes to represent groups of very similar sequences (Hamady et al., 2009b).
UniFrac calculates the unique fraction of branch length for a sample from a phylogenetic tree constructed from each pair of samples in a data set. Because the UniFrac metric is a phylogenetic estimate of community similarity, it avoids some of the problems associated with analyses that compare communities at arbitrarily defined levels of sequence similarity (Lozupone & Knight, 2008; Hamady & Knight, 2009a). The phylogenetic diversity of each sample was determined from 1000 randomly selected sequences per sample using Faith’s phylogenetic diversity metric (Faith’s PD; Faith, 1992), which calculates the amount of branch length for each sample within the relaxed-neighbor-joining tree. The taxonomic identity of each phylotype was determined using the RDPII taxonomy (60% minimum threshold) (Cole et al., 2005). All sequences have been deposited in the GenBank short read archive (accession number SRA012078.1).
Statistical analyses
The effect of temperature and length of storage on relative taxon abundance (minimum 1% abundance per sample-treatmet combination) was assessed using the Kruskal-Wallis test in systat 11.0 for sequences classified to the level of order (fecal and skin) or family (soil). Statistical differences in overall community composition (UniFrac distances) were performed within each sample type with the permanova package in primer v6 using Sample, Day, Temperature and Day × Temperature as main factors. Pairwise UniFrac distances were visualized by non-metric multidimensional scaling in primer v6 (Clarke & Warwick, 2001). Differences in Faith’s PD due to temperature and length of storage were assessed using the Kruskal-Wallis test.
Results and discussion
Sequence analysis and overall results
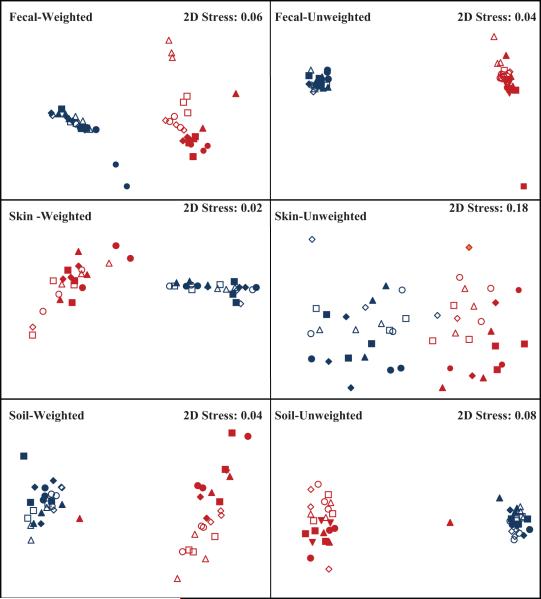
After eliminating low quality sequences, the number of reads ranged from 1304 to 3022 per sub-sample with an average of 2019 sequences per sub-sample and a total of 290,696 sequences for the data set. One sub-sample was omitted from the data set (Fecal 1 Day 14, 20°C replicate 2) due to visible fungal growth prior to DNA extraction. Each sample type yielded a similar total number of bacterial 16S rRNA sequences (97,943 for feces, 97,527 for skin and, 95,226 for soil). These distinct sample types harbored communities that were distinct with respect to their composition and diversity (Figs. 1, 2 and Tables 1, 2). The human-derived samples tended to have few dominant phyla (3 per sample type) accounting for 95 to 98% of the sequences: in contrast, in the soils, the six most abundant phyla accounted for only c. 83% of the sequences. Fecal samples were dominated by members of the phylum Bacteroidetes (62%), and the Firmicutes (35%), while skin samples had a relatively even distribution of Firmicutes (39%), Bacteroidetes (31%) and Actinobacteria (25%). Soil samples contained many phyla including the Bacteroidetes (32%), Acidobacteria (27%), and Proteobacteria [Alphaproteobacteria (10%), Betaproteobacteria (6.5%), Gammproteobacteria (5.2%) and the Deltaproteobacteria (2.7%)]. The unique distribution of phyla was also seen in overall community composition as NMDS visualization of pairwise UniFrac distances showed clustering by individual sample rather than temperature or length of storage (Fig. 1). Sample types also differed with respect to community diversity levels with soil bacterial communities harboring the highest levels, followed by fecal and skin samples (Faith’s PD = 40, 11 and 10, respectively). As noted below, each pair of sub-samples within a given sample type had bacterial communities that differed with respect to their composition and diversity and these differences were irrespective of the storage conditions (see Table 1 and below).
Fig.1.
Non-metric Multidimensional Scaling (NMDS) plots of UniFrac weighted and unweighted pairwise distances. Overall community composition was not affected by temperature or duration of storage for weighted UniFrac distances (P> 0.1 in all cases). Length of storage significantly affected the skin communities for the unweighted UniFrac metric (P = 0.02). The remaining unweighted UniFrac distances were not significantly different by day or temperature.
Blue=sample 1, red = sample 2. Open symbols = Day 3, closed symbols = Day 14. ▲= 20°C, ■ = 4°C, ● = −20°C, ◆ = −80°C.
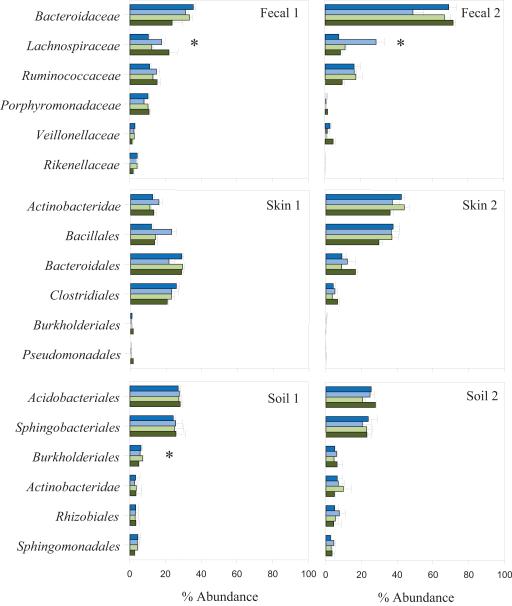
Fig. 2.
The relative abundance of various bacterial taxa in fecal, skin and soil samples after 14 days of storage. Bars represent the mean abundance of each group at each temperature in descending order (e.g. 20, 4, −20, and −80°C) with 1 standard error of the mean. Abundances of bacterial taxa were classified to family for the fecal samples and to order for the skin and soil samples using the RDPII nomenclature. Differences in relative abundances due to storage temperature were assessed within individual samples using the Kruskall-Wallis test. Asterisk (*) indicates P- values < = 0.05.
Table 1.
permanova results of Sample, Day, Temperature and Day × Temperature effects on 387 pairwise weighted and unweighted UniFrac distances. Bold text indicates tests where P< 0.05.
| dfa | SSa | MSa | Pseudo-F | P | |
|---|---|---|---|---|---|
| Unweighted UniFrac | |||||
| All Samples | |||||
| Sample | 5 | 26 | 5.1 | 25 | 0.001 |
| Day | 1 | 0.27 | 0.27 | 0.73 | 0.75 |
| Temperature | 3 | 0.64 | 0.21 | 0.56 | 1.0 |
| Day × Temperature | 3 | 0.66 | 0.22 | 0.57 | 1.0 |
| Fecal | |||||
| Sample | 1 | 3.3 | 3.3 | 19 | 0.001 |
| Day | 1 | 0.26 | 0.26 | 1.09 | 0.24 |
| Temperature | 3 | 0.58 | 0.19 | 0.79 | 0.83 |
| Day × Temperature | 3 | 0.57 | 0.19 | 0.77 | 0.86 |
| Soil | |||||
| Sample | 1 | 1.4 | 1.4 | 6.2 | 0.001 |
| Day | 1 | 0.27 | 0.27 | 1.11 | 0.15 |
| Temperature | 3 | 0.68 | 0.23 | 0.92 | 0.87 |
| Day × Temperature | 3 | 0.66 | 0.22 | 0.89 | 0.98 |
| Skin | |||||
| Sample | 1 | 1.0 | 1.0 | 4.8 | 0.001 |
| Day | 1 | 0.32 | 0.32 | 1.4 | 0.02 |
| Temperature | 3 | 0.67 | 0.22 | 0.97 | 0.61 |
| Day × Temperature | 3 | 0.72 | 0.24 | 1.05 | 0.22 |
| Weighted UniFrac | |||||
| All Samples | |||||
| Sample | 5 | 14 | 2.8 | 301 | 0.001 |
| Day | 1 | 0.04 | 0.04 | 0.41 | 0.76 |
| Temperature | 3 | 0.05 | 0.02 | 0.15 | 1.0 |
| Day × Temperature | 3 | 0.04 | 0.01 | 0.12 | 1.0 |
| Fecal | |||||
| Sample | 1 | 0.83 | 0.83 | 101 | 0.001 |
| Day | 1 | 0.06 | 0.06 | 2.28 | 0.107 |
| Temperature | 3 | 0.07 | 0.02 | 0.93 | 0.475 |
| Day × Temperature | 3 | 0.03 | 0.01 | 0.38 | 0.894 |
| Soil | |||||
| Sample | 1 | 0.46 | 0.46 | 56 | 0.001 |
| Day | 1 | 0.04 | 0.04 | 2.07 | 0.11 |
| Temperature | 3 | 0.04 | 0.01 | 0.67 | 0.70 |
| Day × Temperature | 3 | 0.02 | 0.01 | 0.38 | 0.99 |
| Skin | |||||
| Sample | 1 | 0.87 | 0.87 | 79 | 0.001 |
| Day | 1 | 0.02 | 0.02 | 0.51 | 0.53 |
| Temperature | 3 | 0.03 | 0.01 | 0.28 | 0.93 |
| Day × Temperature | 3 | 0.05 | 0.02 | 0.53 | 0.73 |
df=degrees of freedom: SS= sum of squares; MS= mean squares
Table 2.
Faith’s phylogenetic diversity (Faith’s PD) analysis of samples. Mean Faith’s PD and (SEM) are shown for individual sub-samples, and each temperature on both days. Temperature had a significant effect on Faith’s PD for Fecal sample 1 on Day 3 and was significantly greater on Day 14 for Fecal 1(P< 0.05 in both cases). The phylogenetic diversity of the remaining samples was unaffected by temperature or length of storage.
| Day 3 | Day 14 | |
|---|---|---|
| Fecal 1 | 12 (0.9) | 15 (0.7) |
| 20°C | 9.2 (0.4) | 15 (0.4) |
| 4°C | 12 (0.1) | 15 (0.8) |
| −20°C | 13 (0.3) | 15 (0.3) |
| −80°C | 13 (0.3) | 14 (0.4) |
| Fecal 2 | 7.7 (0.2) | 7.9 (0.3) |
| 20°C | 7.6 (0.3) | 8.1 (0.4) |
| 4°C | 7.7 (0.1) | 7.6 (0.3) |
| −20°C | 7.6 (0.2) | 8.2 (0.4) |
| −80°C | 7.8 (0.1) | 7.8 (0.1) |
| Skin 1 | 13 (1.5) | 13 (0.7) |
| 20°C | 12 (0.2) | 13 (0.7) |
| 4°C | 12 (0.2) | 14 (0.04) |
| −20°C | 13 (0.4) | 13 (1.3) |
| −80°C | 16 (2.3) | 12 (0.4) |
| Skin 2 | 8.2 (1.0) | 7.6 (0.7) |
| 20°C | 7.0 (0.1) | 8.7 (0.8) |
| 4°C | 8.1 (0.9) | 7.0 (0.1) |
| −20°C | 7.8 (1.2) | 7.8 (0.9) |
| −80°C | 9.7 (0.9) | 6.9 (0.2) |
| Soil 1 | 38 (1.0) | 39 (0.9) |
| 20°C | 37 (0.9) | 39 (1.2) |
| 4°C | 36 (1.3) | 39 (1.3) |
| −20°C | 39 (0.1) | 40 (0.8) |
| −80°C | 40 (0.5) | 40 (0.6) |
| Soil 2 | 43 (1.3) | 40 (0.5) |
| 20°C | 40 (0.6) | 40 (0.1) |
| 4°C | 43 (0.3) | 39 (0.6) |
| −20°C | 44 (1.4) | 39 (0.2) |
| −80°C | 45 (0.7) | 40 (0.8) |
Storage effects: fecal samples
Bacterial communities in the fecal samples did not change appreciably with different storage conditions and retained their unique composition even after 14 days of storage (Fig. 1 and Table 1). Fecal 2 was dominated by the Bacteroidaceae (c. 75%), while Fecal 1 had a more even distribution of the six most abundant taxa across all temperatures (Fig. 2). Although the relative abundances of a few individual taxa were affected by temperature (Fig. 2), this did not have a significant effect on overall community composition. The UniFrac distance between bacterial communities from the two hosts was significantly greater (permanova P = < 0.001) for both weighted and unweighted UniFrac than the distance between samples stored at different temperatures and durations (P > 0.1, Table 1). This minimal effect of storage on the overall structure of the communities is evident from Fig. 1 which shows that replicate samples tended to cluster by host. Likewise, the phylogenetic diversity of the fecal samples remained consistent across the temperatures (Table 2). Our results extend those reported by Roesch et al. (2009) who found minimal differences in community composition and relative taxon abundances after 72 h of storage at the one temperature tested (room temperature) for 3 of the 4 samples in their study.
In summary, even though we did observe shifts in the abundance of some taxa in our small sample set under different storage conditions, this did not mask interpersonal differences in overall fecal bacterial community composition, and did not affect our ability to differentiate the host origin of the two fecal samples.
Skin samples
As with the fecal communities, each skin sample was distinctive in terms of dominant taxa and overall community composition. Skin 1 had relatively more Bacteroidales and Clostridiales (c. 20-30%) while Skin 2 had greater abundances of the Actinobacteridae and Bacillales (c. 35-55%) (Fig. 2). These person-to-person differences in taxon abundance were also evident in the UniFrac analyses, as each sample clustered by host rather by temperature or length of storage (Fig. 1). Weighted pairwise UniFrac distances were significantly greater between the samples from the two individuals (P < 0.001) than between replicate sub-samples stored at different temperatures (P = 0.93) or durations (P = 0.53). Similarly, we saw no significant effect on the phylogenetic diversity between replicate sub-samples analyzed after 3 days of storage versus 14 days of storage, irrespective of the storage temperature (P > 0.05 in all cases). The fact that the highly personalized nature of skin-associated bacterial communities (Gao et al., 2007; Grice et al., 2009; Costello et al., 2009) were still apparent after 14 days at a range of temperatures, with storage conditions having relatively little impact on community composition or diversity, has important implications for mass sampling efforts sponsored by various international human microbiome projects, which aim to relate microbial community structure and function to physiologic and pathophysiologic features in people experiencing a range of lifestyles in a variety geographic locations, some remote (Turnbaugh et al., 2007).
Soil samples
The two soils harbored unique bacterial communities, with temperature and length of storage having little effect on overall community composition (Figs. 1 and 2, Table 1). Soils retained similar abundances of the six most numerous taxa across the range of storage temperatures tested, except for the Burkholderiales, which were marginally affected by temperature in Soil 1 (P = 0.05, Fig. 2). Although each sample had similar abundances of most taxa, the two soil communities were clearly distinct regardless of storage conditions (P < 0.001, Fig. 1 and Table 1). Analysis of UniFrac pairwise distances showed no significant effect of Day, Temperature or Day × Temperature on overall community composition for sub-samples immediately frozen at −80°C and those stored at 20°C for 14 days (P > 0.05 in all cases). Likewise, phylogenetic diversity was unaffected by temperature or length of storage (P > 0.05 in all cases, Table 2).
Conclusion
We surveyed microbial communities from multiple environments under a broad range of storage conditions, and demonstrated that bacterial community composition in the samples was largely unaffected by differences in short-term storage conditions. Although it is not currently possible to resolve changes in bacteria at the species or strain level using pyrosequencing given the limitations of read length and error rate (Kunin et al., 2010), our results are consistent with other studies and indicate that the provenance of samples has a greater effect on microbial community structure than do the conditions under which samples are stored prior to conducting DNA extractions. Critically, these differences persist both at a broad level (e.g. between soil and skin) and at the more subtle level of specific samples (e.g. different soils, or skin from different people). Sub-samples stored under different conditions did not have identical bacterial communities, perhaps due to insufficient sample homogenization or the inherent variability in DNA extractions and PCR amplification between sub-samples. Importantly, these other potential sources of variability were more important than the variability introduced by differences in storage temperature and duration between sub-samples even after 14 days of storage at room temperature. Although specific taxa may change in relative abundance with different storage conditions, our data suggest that the types of samples in this study can be stored and shipped at room temperature without having a significant impact on the assessment of overall community composition or the relative abundances of most major bacterial taxa.
Acknowledgements
We thank Donna Berg-Lyons for her help with the sample processing, Jill Manchester for her help with DNA sequencing, plus Micah Hamady and Elizabeth Costello for assistance with the bioinformatics analyses. We would also like to thank members of the Fierer lab group for help on previous drafts of this manuscript. This work was supported by grants from the National Science Foundation (EAR 0724960), the U.S. Department of Agriculture (2008-04346) (N.F.), and the Howard Hughes Medical Institute (R.K.), the Bill and Melinda Gates Foundation, and the Crohn’s and Colitis Foundation of America, and NIH (R01 HG004872) (R.K. and J.I.G.)
References
- Clarke KR, Warwick RM. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Marine Ecology-Progress Series. 2001;216:265–278. [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Knight R, Fierer N. Bacterial Variation in Human Body Habitats Across Space and Time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006a;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb. 2006b;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfing J, Vos A, Bloem J, Ehlert PAI, Naumova NB, Kuikman PJ. Microbial Diversity in Archived Soils. Science. 2004;306:813a. doi: 10.1126/science.306.5697.813a. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation Evaluation and Phylogenetic Diversity. Biological Conserv. 1992;61:1–10. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. P Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei ZH, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. P Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. NatMethods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009a;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone CA, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009b;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klammer S, Mondini C, Insam H. Microbial community fingerprints of composts stored under different conditions. Ann Microbiol. 2005;55:299–305. [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl Environ Microb. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac - An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Micrbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar APN, Kato I. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Meth. 2008;72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Timmis KN, Hampe J, Wenderoth DF, Schreiber S. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Micro Infec Dis. 2004;50:237–245. doi: 10.1016/j.diagmicrobio.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LFW, Casella G, Simell O, Krischer J, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. Influence of Fecal Sample Storage on Bacterial Community Diversity. The Open Microbiology Journal. 2009;3:40–46. doi: 10.2174/1874285800903010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochelle PA, Cragg BA, Fry JC, Parkes RJ, Weightman AJ. Effect of sample handling on estimation of bacterial diversity in marine sediments by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. 1994;15:215–225. [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The Human Microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeneva VA, Salles JF, Naumova N, de Vos WA, Kuikman PJ, Dolfing J, Smidt H. Effect of soil sample preservation, compared to the effect of other environmental variables, on bacterial and eukaryotic diversity. Res Microbiol. 2009;160:89–98. doi: 10.1016/j.resmic.2008.12.001. [DOI] [PubMed] [Google Scholar]