Abstract
Embryonic stem (ES) cell technology may serve as a platform for the discovery of drugs to treat diseases such as diabetes. However, because of difficulties in establishing reliable ES cell differentiation methods and in creating cost-effective plating conditions for the high-throughput format, screening for molecules that regulate pancreatic beta cells and their immediate progenitors has been limited. A relatively simple and inexpensive differentiation protocol that allows efficient generation of insulin-expressing cells from murine ES cells was previously established in our laboratories. In this report, this system is characterized in greater detail to map developmental cell stages for future screening experiments. Our results show that sequential activation of multiple gene markers for undifferentiated ES cells, epiblast, definitive endoderm, foregut, and pancreatic lineages was found to follow the sequence of events that mimics pancreatic ontogeny. Cells that expressed enhanced green fluorescent protein, driven by pancreatic and duodenal homeobox 1 or insulin 1 promoter, correctly expressed known beta cell lineage markers. Overexpression of Sox17, an endoderm fate-determining transcription factor, at a very early stage of differentiation (days 2–3) enhanced pancreatic gene expression. Overexpression of neurogenin3, an endocrine progenitor cell marker, induced glucagon expression at stages when pancreatic and duodenal homeobox 1 message was present (days 10–16). Forced expression (between days 16 and 25) of MafA, a pancreatic maturation factor, resulted in enhanced expression of insulin genes, glucose transporter 2 and glucokinase, and glucose-responsive insulin secretion. Day 20 cells implanted in vivo resulted in pancreatic-like cells. Together, our differentiation assay recapitulates the proceedings and behaviors of pancreatic development and will be valuable for future screening of beta cell effectors.
Introduction
Embryonic stem (ES) cell technology is a promising tool for drug discovery, because of the ability of ES cells to indefinitely self-renew and differentiate into many cell lineages.1 Recent studies utilizing ES cell technology as a platform for chemical screening have identified molecules implicated in the maintenance or cytotoxicity of undifferentiated ES cells as well as those that direct the commitment of ES cells toward very early endodermal, mesodermal, or ectodermal progenitor cells.2–5 This progress has occurred because techniques used to maintain ES cells in vitro have become relatively standard and are now widely available to many laboratories. In contrast, screening for molecules that influence more differentiated lineage-specific cell types is limited because of the difficulties in establishing appropriate and reliable lineage-specific cell culture methods for ES cell differentiation and in creating cost-effective plating conditions for the high-throughput screening (HTS) format. The reports on HTS of chemicals administered to early pancreatic progenitor6 and neuronal7 cells are only recently emerging. For example, a prior well-defined human ES cell to early pancreatic endoderm differentiation system8–10 has enabled the success of the chemical screening reported by Chen and coworkers.6 However, challenges remain for screening of effectors that regulate the more mature pancreatic beta-like cells because glucose-responsive insulin-secreting cells cannot yet be robustly generated in vitro.9 In addition, the expansive use of a number of recombinant proteins, such as Activin, wingless-int, fibroblast growth factor (FGF)10, and insulin-like growth factor, may be cost prohibitive for large-scale HTS projects.
The pancreas derives from the definitive endoderm and its development is governed by the sequential activation of a number of cell-autonomous transcription factors and noncell autonomous growth factors.11 For example, Sox17, a high mobility group box-containing transcription factor, is critical for definitive endoderm formation.12 Ectopic expression of XSox17 can force naive Xenopus ectoderm to become endoderm,13 demonstrating a cell fate conversion capability. Pancreatic and duodenal homeobox 1 (Pdx1), a homeodomain-containing transcription factor, is important for both the commitment of early pancreatic progenitors and the maintenance of mature beta cells.14–17 Neurogenin3 (Ngn3), a basic helix-loop-helix–containing transcription factor, was originally discovered in neural crest cells and then found to be present in pancreatic endocrine progenitors.14,18 It is initially expressed at low levels at E9.5 in pancreatic buds, then peaks at E13.5, and is reduced greatly by E17.5.19 Ngn3 is necessary for pancreatic endocrine differentiation, as demonstrated by Ngn3-deficient mice that do not develop pancreatic endocrine cells and die immediately following birth.20 MafA is a member of the basic leucine zipper family of transcription factors and is a direct regulator of insulin production in mature beta cells.21–23 During pancreatic development, MafA is differentially expressed in insulin-expressing cells.21–23 Mice that lack MafA have normal islet architectures with a normal beta/alpha cell ratio at birth but progressively become diabetic at 8 weeks of age,24 which suggests that MafA is required for beta cell maturation. The diabetes phenotype of these MafA-mutant mice is associated with reduced levels of insulin 1, insulin 2, Pdx1, and glucose transporter 2 (Glut2).24 In addition, MafA has a potent direct effect on the transcription of insulin 2 promoter in alpha-like cells, as stable transfection of MafA alone in an alpha cell line, αTC6, induces insulin 2 expression.22 The growth factor FGF10 is also required for the expansion of early Pdx1-expressing progenitors in the developing pancreas.25
Our laboratories are interested in screening of regulators that may affect proliferation and/or maturation of the later pancreatic insulin-producing beta cells and their immediate progenitors. We have previously established a relatively simple and inexpensive differentiation protocol that allows efficient generation of the pancreatic-like, insulin-expressing cells from murine ES cells.26,27 The present study sought to characterize this assay system in greater detail, first, to verify whether pancreatic-lineage cell types properly developed in vitro according to what is expected from current developmental biology literature and, second, to define appropriate target cell stages before our further investment in HTS. Four different experimental strategies were employed. First, detailed time course analysis for endogenous gene expression during culture was analyzed using quantitative reverse transcription (RT)–polymerase chain reaction (PCR) with TaqMan® Probes. Second, cells that expressed enhanced green fluorescent protein (EGFP), driven by Pdx1 or insulin 1 promoter, were sorted and analyzed for their expression of known beta and progenitor cell markers. Third, specific key transcription (such as Sox17, Ngn3, and MafA) or growth (such as FGF10) factors were forced expressed or added exogenously in various stages of differentiation to test whether pancreatic genes were activated accordingly. Finally, differentiated cells were transplanted and graft morphology was evaluated at 5 weeks posttransplantation. In each method, we found gene expression patterns of the in vitro differentiated or induced cells consistent with what has been reported regarding pancreatic developmental biology. Further, grafts in vivo contained cells that displayed characteristics consistent with pancreatic histology. In summary, the data validated our culture system as a suitable cellular tool to target pancreatic beta cells, or their immediate progenitors, for screening of molecules that affect their proliferation and/or maturation.
Materials and Methods
Growth and Differentiation of ES Cells
Maintenance and differentiation of mouse ES lines R1, Pdx1-EGFP,28 mouse insulin promotor (MIP)-EGFP,29 Ngn3-EGFP,27 AinV-Sox17, AinV-Ngn3, and A2lox-MafA were performed as previously described26,27 (summarized in Fig. 1A). Briefly, embryoid bodies (EBs) were grown in the presence of selected batches of fetal calf serum (FCS) (Gemini BioProducts, West Sacramento, CA; Tissue Culture Biologicals, Seal Beach, CA) for 6 days. Serum lots were selected based on their ability to support the differentiation of EBs toward insulin 1, glucagon, and amylase A2 gene expression in the later stage (days 18–20). From day 0 to 2 and 2 to 6 of EB culture, 6 and 0.6 mM of monothioglycerol (Sigma, St. Louis, MO) was added to culture, respectively. Whole day 6 EBs were washed, counted, and plated (75 or 80 EBs/well) in six-well attachment tissue culture plates (Corning Incorporated, Corning, NY). The attachment culture medium contained Dulbecco's modified Eagle's medium/F-12 (1:1) (Mediatech, Herndon, VA) and 15% knockout serum replacement (Invitrogen, Carlsbad, CA). From day 6 to 10 of attachment culture, 5% serum was added to basic media to hasten EB attachment. On day 13 of the culture, 10 mM nicotinamide (N) (Sigma), 10 ng/mL human recombinant activin βB (A) (R&D Systems, Minneapolis, MN), and 0.1 nM exendin-4 (E) (Sigma) were added. The media were changed every 3–4 days. Human recombinant FGF10 (R&D Systems) was used together with 1 unit/mL of heparin sodium (APP Pharmaceuticals, Schaumburg, IL) and was added on day 10 cells.
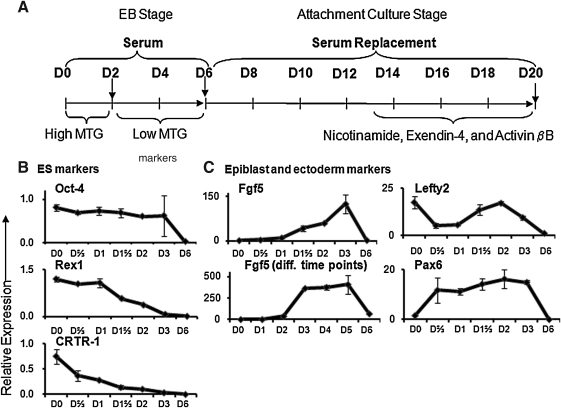
Fig. 1.
Kinetics studies of endogenous gene expression in our ES cell differentiation assay. (A) Protocol for differentiation of murine ES cells to pancreatic-like lineages in vitro. (B–E) R1 ES cells were differentiated and gene expression was analyzed by quantitative RT–PCR with Taqman probes. Data represent the mean and standard deviation from duplicated samples. ES, embryonic stem; PCR, polymerase chain reaction.
RNA Isolation, RT-PCR, and Quantitative PCR Analysis
Procurement of cells, extraction of total RNA, and RT-PCR analysis were preformed as previously described26 with minor modifications. Briefly, undifferentiated ES cells were dissociated into a single-cell suspension by incubation with 0.25% trypsin–ethylenediaminetetraacetic acid (3 min). Cells from attachment culture were detached by incubation with 0.25% trypsin–ethylenediaminetetraacetic acid (3 min, 37°C), washed, and then dissociated with 4 mg/mL collagenase B (Roche Diagnostics, Indianapolis, IN) and 1,000 U/mL deoxyribonuclease I (Calbiochem, Gibbstown, NJ) (30 min, 37°C) to produce a single-cell suspension. Total RNA was extracted from cells using RNeasy Mini Kit (Qiagen, Valencia, CA), and genomic DNA was digested with deoxyribonuclease I (Invitrogen). Equal amounts (1 μg) of RNA were used for reverse transcription (Invitrogen or Qiagen). One-twentieth of the cDNA mixture was used for PCR amplification of each gene, and commercially available predesigned primers and TaqMan Probes (Applied Biosystems, Foster City, CA) were used to analyze gene expression. Either cyclophilin G (Fig. 1B, C) or β-actin (all other figures) was used as the internal control. Data for quantitative PCR analysis were expressed as relative expression to internal control among the samples analyzed on the same run of PCR. Thus, it should be noted that levels of gene expression between the left and right panels shown in Figure 1D cannot be directly compared. TaqMan Probes used were undifferentiated ES cells (Oct-4, Rex1, Cp2-related transcriptional repressor-1 [CRTR-1]), epiblast (Fgf5, Lefty2), ectoderm (Pax6), mesendoderm (Brachyury, Cerberus), definitive endoderm (Sox17, CXCR4, Foxa2, Cerberus), visceral endoderm (Sox7), endothelial progenitors (Flk-1), cardiac progenitors (Nkx2.5), gut tube hepatocyte nuclear factor (HNF1b, HNF4a), early pancreatic progenitors (Pdx1, Sox9, Ngn3, HNF6, Hlxb9, Nkx6.1), and later pancreatic (insulin 1, insulin 2, glucagon, somatostatin, amylase 2A, cytokeratin 7 [CK7]) and hepatic (albumin) cells.
Culture of Dissociated Cells in Semisolid Media
Day 16 culture was dissociated into a single-cell suspension and placed in a semisolid media (5 × 104 cells/0.5 mL/well) as previously described.27 In short, cells were mixed thoroughly with cold culture mixture before subsequent incubation at 37°C in 5% CO2 air. One microliter of culture mixture contained 1% 1,500 centipoise (high-viscosity) methylcellulose (Sinetsu Chemical, Tokyo, Japan), 5% Matrigel™, 50% day 16 conditioned media, 5% FCS, 1 ng/mL vascular endothelial growth factor-A, 10 mmol/L nicotinamide, 0.1 nmol/L exendin-4, and 10 ng/mL human recombinant activin βB. Day 16 conditioned media were obtained by adding the Dulbecco's modified Eagle's medium/F-12 media containing 15% serum replacement, nicotinamide, exendin-4, and activin βB on day 13 culture and collecting the medium on day 16 culture.
Flow Cytometric Analyses and Sorting
On designated days, cultures were dissociated into a single-cell suspension and analyzed or sorted by flow cytometry. Events were acquired and analyzed by either a CyAn™ ADP 9 color (Beckman Coulter, Brea, CA) with FlowJo software (TreeStar, Ashland, OR) or an Accuri Cytometer and software (Accuri Cytometers, Ann Arbor, MI). All events were gated with forward scatter (cell size) and side scatter (cell granularity) profiles. Cells were also stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/mL) to discriminate dead cells. Undifferentiated ES cells (labeled as D0 in Fig. 2) were used for negative control gating. Cell sorting was performed on a MoFlo™ MLS (Beckman Coulter).
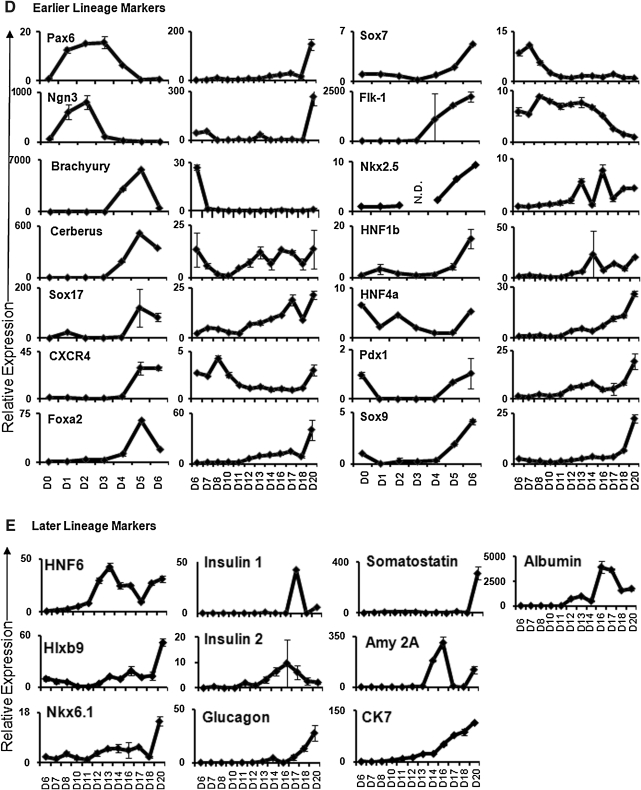
Fig. 2.
Flow cytometric and gene expression analyses of in vitro differentiated Pdx1- or insulin 1-expressing cells. Differentiated EGFP+ and EGFP− cells, derived from Pdx1-EGFP (A) or MIP-EGFP (B) ES line, were analyzed by flow cytometry and subsequently sorted (left panels). Undifferentiated ES cells were used for negative gating of EGFP expression. Gene expression in various populations of cells was analyzed for designated markers by quantitative RT-PCR (right panels). Data represent the mean and standard deviation from duplicated samples. *Expression of the gene is different at P < 0.01, compared with presort control. Pdx1, pancreatic and duodenal homeobox 1; EGFP, enhanced green fluorescent protein. Color images available online at www.liebertonline.com/adt
Generation of Inducible AinV-Sox17, AinV-Ngn3, and A2lox-MafA ES Cells
Using gene-specific PCR primers and the TOPO TA Cloning kit (Invitrogen), full-length murine Sox17 and Ngn3 cDNA was isolated from R1 ES-derived day 526 and 1627 cells, respectively. The fidelity of Sox17 and Ngn3 cDNAs was confirmed by sequencing. Sox17, Ngn3, and mouse MafA30 were cloned into the multicloning site of pLox targeting vectors, which contain a PGK promoter followed by a loxP site for genomic integration into a doxycycline-inducible locus31 or wild-type and mutant loxP sequences for cassette exchange recombination at the same locus.32 The targeting vector with the cDNA of interest and a cyclization recombinase expression vector were electroporated into AinV or A2lox ES cells; clones resistant to G418 selection were hand-picked, expanded, and cryopreserved for further characterization and experimentation.
Glucose-Stimulated C-Peptide Release In Vitro
Thirty day 6 EBs were plated per well in 24-well plates. On day 16, cells received either 0 or 0.2 μg/mL of doxycycline. On day 25, cells were incubated overnight with Krebs-Ringer solution (KRBH; 129 mM NaCl, 4.8 mM KCl, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5 mM NaHCO3, 10 mM HEPES, 0.1% bovine serum albumin) containing 10% FCS and 2.5 mM d-glucose (glucose). The next day, cells were washed thrice with KRBH containing 2% FCS and 2.7 mM d-glucose and then sequentially incubated with 2.7 mM, 16.7 mM, and 2.7 mM d-glucose, followed by 16.7 mM glucose plus 10 mM theophylline (37°C, 0.5 mL/well; 2 h for ES-derived cells, ∼250,000 cells/well; 30 min for murine islets, 15 islets/well). Cells were washed thrice when the solution was changed from 16.7 to 2.5 mM glucose to eliminate carryover. The C-peptide concentration in the buffer was measured using the murine C-peptide enzyme-linked immunosorbent assay kit, U-Type (Shibayaji Co., Gunma, Japan), which has a detection limit of 30 pg/mL. Normal adult islets were used as positive controls. The stimulation was expressed as the fold change of C-peptide concentrations from the second to the fourth treatments, compared with the first treatment of the same well.
Transplantation of ES-Derived Cells
Ten- to 16-week-old SCID-beige male mice (Taconic, Hudson, NY) were used as recipients. At least 5 days prior to transplantation, mice were injected intraperitoneally with 200 mg/kg of freshly dissolved streptozotocin (STZ; Sigma) in citrate buffer (pH 5.5). On the day of transplantation, only those mice with two sequential measurements of daily blood glucose >200 mg/dL (Glucometer Elite XL monitoring system; Bayer Corporation, Pittsburgh, PA) were used as recipients. Day 20 cells (from 180 EBs/mouse) were scraped from the tissue culture wells and placed under the kidney capsule. Five weeks after transplantation, mice were sacrificed and grafts were cut (5 mm), fixed in phosphate-buffered saline containing 4% formalin (pH 7.5, 24 h), embedded in paraffin, sectioned, and processed for hematoxylin and eosin (H&E) and immunohistological staining.
Immunohistology
Immunofluorescent staining was performed as previously described.26 Guinea pig anti-swine insulin serum (1:100) was obtained from Dako (Carpinteria, CA), rabbit anti-human amylase antibody (1:100) from Sigma, rabbit anti-human C-peptide antibody (1:200) from Cell Signaling Technology (Danvers, MA), and mouse anti-porcine glucagon antibody (1:100) from Sigma. When indicated, sections were counterstained with DAPI for nuclei.
Statistical Analysis
Student's t-test was used to determine statistical significance.
Results
Kinetic Studies of ES, Epiblast, Germ Layer, and Early Lineage Cell Markers During EB Differentiation
In previous studies, we established a stepwise culture system that induces murine ES cells to commit to early endocrine pancreas.26,27 In summary (Fig. 1A), ES cells were differentiated in suspension culture (6 days) to form EBs, which were then allowed to attach and further differentiate into cells that express Pdx1, insulin 1, insulin 2, glucagon, and amylase 2A, a set of markers that suggests pancreatic specification. A population of cells that resembles the primitive first wave33,34 of pancreatic endocrine differentiation that simultaneously expressed insulin/C-peptide and glucagon, as detected by double immunofluorescent staining, could be found in day 17 culture.11 Insulin+ cells that do not express glucagon, that is, the definitive second-wave–like cells, were also generated by ∼day 19 in culture, after the addition of a combination of beta cell specification and differentiation factors: nicotinamide, exendin-4, and activin βB.26
To further characterize the developmental kinetics of EBs in detail, quantitative RT-PCR was performed in two ES cell lines, R1 and Pdx1-EGPF, using a panel of TaqMan probes (Fig. 1 and Supplementary Fig. S1, respectively; Supplementary Data are available online at www.liebertonline.com/adt). Within the Oct-4+ pluripotent cell population, Rex1 and CRTR-1 are highly expressed in the inner cell mass (ICM) and downregulated upon differentiation toward epiblast.35,36 During the first 3 days of differentiation, the expression levels of Oct-4 were maintained, whereas the levels of Rex1 and CRTR-1 were significantly downregulated by day 2 or 3 (Fig. 1B), suggesting that ES cell differentiation was occurring in these early EBs.
Markers for epiblast (Fgf5 and Lefty2) were then examined. Fgf5 is undetectable in the ICM until epiblast formation, and its expression is maintained during gastrulation.36–38 Expression of Fgf5 gradually increased during the first 3 days of differentiation, was maintained from days 3 to 5, and was downregulated on day 6 (Fig. 1C). Lefty2 has been reported to be enriched in mouse epiblast-derived stem cells.39 There was a biphasic expression of Lefty2: it was first detected in undifferentiated ES cells, downregulated during 16–24 h of differentiation, upregulated on day 2, and then downregulated on days 3–6 (Fig. 1C). These results suggest that epiblast-like cells were formed at least by days 2–3 of EB culture.
Markers for the three germ layers and lineage-specific progenitors were then examined. The ectoderm marker, Pax6,40 was expressed as early as 2 h (not shown) and until day 4 (Fig. 1C, D). Ngn3, which when expressed at a later stage may represent pancreatic endocrine differentiation,14 was upregulated in days 1–2 EBs (Fig. 1D); its expression may also reflect ectoderm differentiation at this early stage.18 The mesendoderm markers, Brachyury41 and Cerberus,42 were detectable as of day 3 (not shown), and their levels increased dramatically during days 4–5 of EB differentiation (Fig. 1D). Levels of the definitive endoderm markers, Sox17, CXCR4,43 Cerberus,44 and Foxa2,45 increased at day 5 (Fig. 1D). By day 6, an increase in expression of markers for visceral endoderm (Sox7),12 endothelial progenitor (Flk-1),46 cardiac progenitor (Nkx2.5),47 gut tube (HNF1b48 and HNF4a),49 and early pancreatic progenitors (Pdx114–17 and Sox9)50 was observed (Fig. 1D). Protein expression of Sox17 and Foxa2 was further tested by flow cytometric and/or immunohistological analyses. Consistent with quantitative RT-PCR analysis (Fig. 1D), undifferentiated ES cells did not express Sox17, whereas 15%, 9%, and 7% of total cells were Sox17+ in days 5, 6, and 16 cultures, respectively (Supplementary Fig. 2A). Sox17 (Supplementary Fig. 2B, C) or Foxa2 (Supplementary Fig. 2D) staining coincided with the DAPI+ nuclei staining, as anticipated for transcription factors. These results confirmed protein expression of Sox17 or Foxa2 during differentiation. Overall, these results suggest that waves of cells that resembled ectoderm, epiblast, mesendoderm, definitive endoderm, gut tube, early pancreatic endoderm, and other lineage progenitors were formed during EB differentiation.
Kinetic Studies of Later Lineage Cell Markers During Attachment Culture
Kinetic expression of pancreatic-lineage genes during the later attachment culture stage was determined (Fig. 1D, E). Between days 6 and 10, levels of the pancreatic endoderm and progenitor cell markers, HNF1b, HNF4a, Pdx1, and Sox9, were relatively unchanged (Fig. 1D). In contrast, starting from ∼day 11 culture, expression of these genes significantly increased (Fig. 1D). Concurrently, expression of other pancreatic progenitor markers, HNF6,51 Hlxb9,52 and Nkx6.1,53 were also enhanced after day 11 (Fig. 1E). Upregulation of endocrine (insulin 1, insulin 2, glucagon, and somatostatin), exocrine (amylase 2A), and ductal (CK7) genes began later, starting between days 13 and 20, as assessed in two independent cell lines (Fig. 1E and Supplementary Fig. 1B). Overall, the kinetics of genes of the later stage culture appeared to be more variable than the earlier EB culture among the two cell lines. However, expression of most of the late lineage-specific genes had an upward trend toward day 20 culture (Fig. 1E and Supplementary Fig. 1B), suggesting that cells may continue to differentiate or mature beyond this time point. Interestingly, expression of the hepatocyte marker, albumin, was present as of day 12 (Fig. 1E), whereas expression of the visceral endoderm marker, Sox7, and the endothelial cell marker, Flk-1, decreased over time (Fig. 1D), suggesting that our culture system preferentially supports the development of definitive endoderm-like lineages during attachment culture. Taken together, the time course data were consistent with our prior studies26,27 and suggest that our differentiation protocol supports the development of endoderm and pancreatic-lineage–like cells in vitro.
Gene Expression Patterns from Sorted Insulin 1 or Pdx1 Promoter-Driven EGFP+ Cells
Pdx1 is expressed not only in early multipotential pancreatic progenitors but also in terminally differentiated insulin-expressing beta cells.14–17 We investigated whether Pdx1- or insulin 1-expressing cells, isolated from our late stage culture, expressed known pancreatic markers. Two preestablished reporter murine ES cell lines (Pdx1-EGFP28 and MIP-EGFP)29 were differentiated using our protocol. On day 16 or 20, cells were dissociated, sorted, and gene expression of the EGFP+ cells was analyzed by quantitative RT-PCR (Fig. 2). Day 16 Pdx1-EGFP+ cells expressed higher levels of Pdx1, insulin 1, insulin 2, glucagon, Ngn3, Sox9, HNF1b, Foxa2, and CK7, compared with the presort cells, suggesting that this population of cells contained a mixture of committed beta-like cells, pancreatic progenitors, and ductal-like cells. In contrast, sorted day 20 MIP-EGFP+ cells (EGFP driven by murine insulin 1 promoter) expressed higher levels of insulin 1 and insulin 2, but not other markers, compared with the presort cells. These results suggest that insulin 1-EGFP+ cells were enriched for the committed beta-like cells, but not the endocrine progenitors or other lineage cells. Thus, ES-derived Pdx1 or insulin 1 promoter-driven EGFP-expressing cells correctly expressed markers for beta cell lineage.
Effects of Overexpression of Sox17, Ngn3, or MafA at Specific Time During Differentiation
Several transcription factors are shown to affect endoderm or pancreas development when expressed ectopically.11 Endogenous gene expression analyses shown in Figures 1 and 2 do not necessarily predict that our ES-derived cells would behave similarly to their in vivo counterparts. We therefore designed gain-of-function experiments to test the effects of overexpression of key transcription factors in a time- and dose-dependent manner, during differentiation. Inducible AinV-Sox17, AinV-Ngn3, and A2lox-MafA ES lines were generated using Tet-On murine ES cell technology.31,32 Characterization of the inducible AinV-Sox17, AinV-Ngn3, and A2lox-MafA ES lines (undifferentiated clonal cells) is presented in Supplementary Figure 3. All of the selected clonal lines maintained undifferentiated states (unchanged levels of Oct-4 or Rex1 and lack of spontaneous expression of germ layer markers) and expressed high levels of their respective gene of interest after doxycycline (1 μg/mL), the inducer, was added to the culture media for 24 h.
Consistent with the cell fate determination effects demonstrated in Xenopus,13 stable expression of Sox17 in undifferentiated human ES cells leads to a homogenous population of definitive endoderm progenitor-like cells that can be cultured over 50 passages and is capable of further differentiation into endodermal lineages, including hepatic- and pancreatic-like cells in vitro.54 Human ES cells are thought to resemble murine epiblasts.39 We therefore tested the hypothesis that overexpression of Sox17 in epiblasts may result in enhanced pancreatic differentiation in the later culture. Our days 2–3 EBs expressed both ectoderm (Pax6) and epiblast (Fgf5 and Lefty2) markers (Fig. 1C, D). Graded doses of doxycycline (inducer) were therefore added to day 2 EBs for 24 h, and the resulting day 20 cells were analyzed by quantitative RT-PCR for insulin 1, insulin 2, somatostatin, pancreatic polypeptide, glucagon, ghrelin, Pdx1, and amylase 2A (Fig. 3A). Expression of all of these genes was increased, compared with the noninduced controls, when doses of 0.2 or 1 μg/mL doxycycline were used, suggesting that both endocrine and exocrine pancreas were enhanced. It should be noted that no enhancement of pancreatic gene expression was observed following addition of doxycycline to day 2 or 4 EBs for 48 h (data not shown), suggesting that cellular context and duration of induction at this early stage are important to exert a positive effect on subsequent pancreatic differentiation. Consistent with this notion, a recent study demonstrates that overexpression of Sox17 in the common progenitors of bile duct and pancreas (E8.5 Pdx1+ domain) inhibits pancreatic potential.55 Together, these results demonstrate that a short 24-h overexpression of Sox17 between days 2 and 3, but not later EBs, resulted in enhanced pancreatic differentiation.
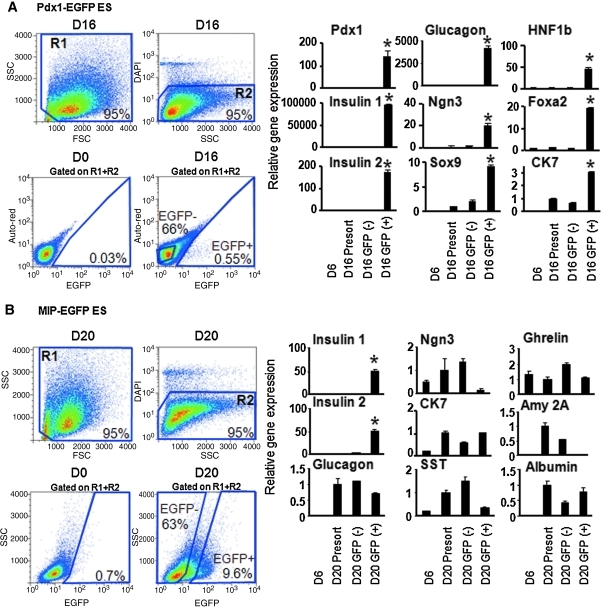
Fig. 3.
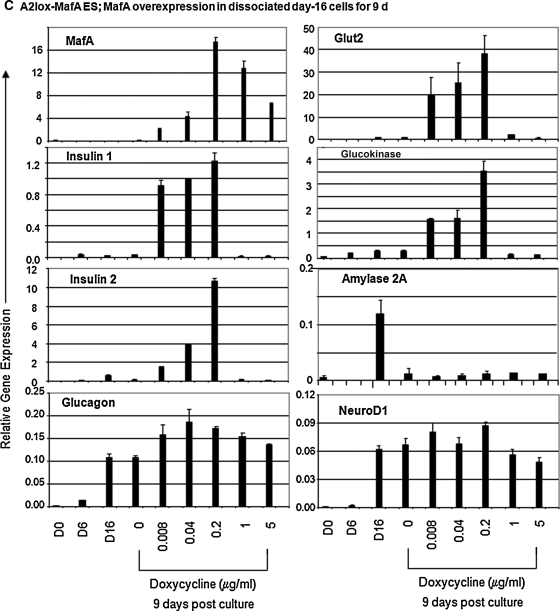
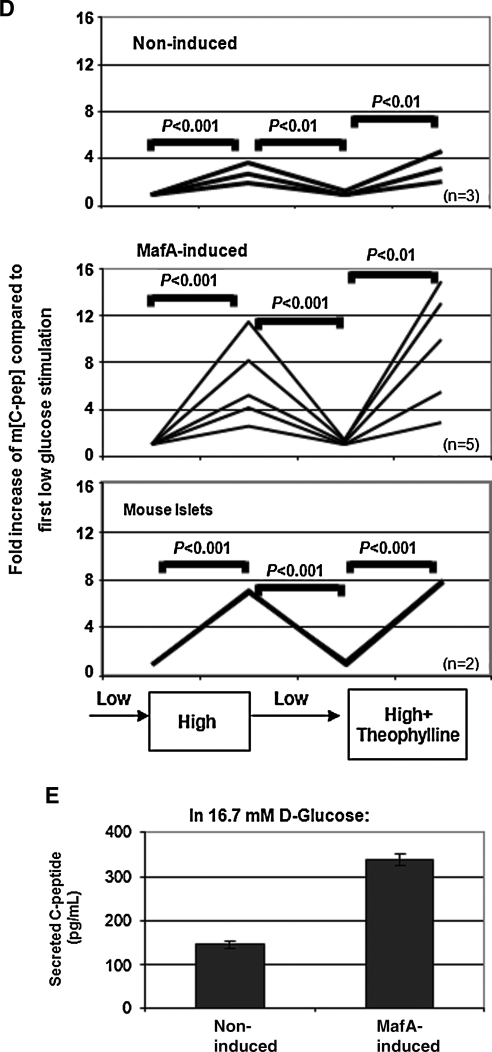
Timed overexpression of Sox17, Ngn3, or MafA and their consequences in pancreatic gene activation in culture. (A) Gene expression analyzed by quantitative RT-PCR of day 20 AinV-Sox17 ES cells after doxycycline-induced expression of Sox17 between days 2 and 3 of culture. Data represent the mean and standard deviation from duplicated samples. (B) Gene expression analyzed by quantitative RT-PCR of day 20 AinV-Ngn3 ES cells after doxycycline-induced expression of Ngn3 at designated time points. (C–E) Effects of forced expression of MafA in A2lox-MafA ES cells. (C) Gene expression analyzed by quantitative RT-PCR of cultures initiated with dissociated day 16 cells that were incubated in semisolid media with designated doses of doxycycline. (D) In vitro glucose challenge assay. Data were expressed as the fold change of C-peptide concentration compared with the basal level (first low glucose incubation) in each well examined. Fold change is different between analyzed sets of samples at P < 0.01 or 0.001, as indicated. (E) As in D, average C-peptide concentrations after the first 16.7 mM d-glucose stimulation was compared between noninduced (n = 3) or MafA-induced (n = 5) cells. The error bars represent standard deviation. Ngn3, neurogenin3.
When driven by the Pdx1 promoter in a transgenic mouse model, Ngn3 has been demonstrated to induce endocrine cell differentiation at E12.5, as shown by the large numbers of glucagon-expressing cells.19 Thus, we tested the hypothesis that overexpression of Ngn3 may enhance glucagon gene expression. Doxycycline (1 μg/mL) was added to either day 10, 13, or 16 cultures differentiated from AinV-Ngn3 ES cells for 24 h; cells were analyzed on day 20 by quantitative RT-PCR. We found that forced expression of Ngn3 consistently led to higher expression levels of glucagon, but not amylase 2A, compared with untreated controls (Fig. 3B). These results are consistent with gene expression analyses (Figs. 1 and 2) and suggest that days 10–16 cultures contain pancreatic-like progenitor cells that could respond to overexpression of Ngn3 in glucagon induction.19
MafA is implicated in beta cell maturation.24 We then tested the hypothesis that forced expression of MafA in our late stage culture could increase the levels of beta cell maturation markers, such as insulin 1, insulin 2, Glut2, and glucokinase. Dissociated day 16 cells derived from A2lox-MafA ES cells were incubated with graded doses of doxycycline and in semisolid media that support further differentiation.27 Gene expression patterns were analyzed at 9 days after induction. Levels of insulin 1, insulin 2, glucokinase, and Glut2, but not amylase 2A, were enhanced when 0.008–0.2 μg/mL, but not 1–5 μg/mL, of doxycycline was added to the culture (Fig. 3C). In contrast to the 30.6- and 78.8-fold enhancement of insulin 1 and 2 genes, respectively, expression of glucagon was increased to a significant albeit lesser degree (1.6-fold) in response to 0.2 μg/mL doxycycline. Insulin 1 expression reached nearly maximal levels when as little as 0.008 μg/mL doxycycline was added, whereas the pattern for insulin 2 gene enhancement closely mimicked MafA induction between doses of 0.008 and 0.02 μg/mL doxycycline. We speculate that these differences may be due to different activation mechanisms by MafA at the promoter regions of these two insulin genes. Consistent with other reports that suggest that MafA lies downstream and interacts with NeuroD1 to induce insulin 2 gene expression,23,56 overexpression of MafA did not enhance NeuroD1 messages (Fig. 3C). Together, these results demonstrate that MafA specifically enhances the expression of genes for endocrine, but not exocrine-lineage cells.
To further confirm whether endocrine cells were the target population for the action of MafA, day 16 cells were sorted into Ngn3-EGFP+ and Ngn3-EGFP− cells, as previously described,27 and then incubated (8 days) with an adenoviral vector expressing MafA. We previously reported that sorted day 16 Ngn3-EGFP+ cells are enriched for gene expression of insulin and glucagon, but not amylase 2A.27 Ngn3-EGFP+, but not EGFP−, cells responded to overexpression of MafA and resulted in enhanced expression of insulin 1 and insulin 2, as expected (Supplementary Fig. 4). Glucagon expression was again enhanced. Although MafA can bind to the glucagon promoter,56 it does not activate its expression in alpha or beta cell lines or in E18.5 pancreata.56,57 It is not clear presently whether the MafA-induced glucagon expression in our system represents an indirect effect secondary to the production of other genes, a nonspecific binding to glucagon promoter due to overexpression, or a direct binding of MafA to glucagon promoter in other ES-derived cells. However, the data suggest that only endocrine-like cells may respond to MafA overexpression.
The enhanced expression of insulin 1, insulin 2, glucokinase, and Glut2 in MafA-induced cells raised the possibility that these cells may also have enhanced glucose-responsive insulin secretion. To test this, nondissociated day 16 cells were incubated with or without doxycycline (0.2 μg/mL) for 9 days. Cells were then subjected to sequential incubation of low (2.7 mM), high (16.7 mM), and low and then high concentrations of d-glucose plus theophylline (10 mM), a cyclic adenosine monophosphate potentiator for maximal stimulation of insulin secretion.58 C-peptide was used as a surrogate marker to indicate de novo synthesized insulin and to rule out exogenous insulin uptake from culture media.59 Secreted C-peptide concentration in buffer from each well after treatment was determined using enzyme-linked immunosorbent assay. We found that both noninduced and MafA-induced cells responded to high-glucose stimulation in C-peptide secretion (Fig. 3D), although 3 of 5 induced wells had similar levels of insulin release compared with the noninduced cells. This could be due to different degrees of MafA overexpression in individual wells or suboptimal time for MafA to exert its maturation programs. Importantly, C-peptide secretion returned to basal levels when the buffer was changed to low concentrations of glucose, suggesting a capability of dynamic response to glucose stimulation in these cells. The lack of C-peptide secretion from cells when exposed to low concentrations of glucose for the second time was not due to cell death, because they secreted C-peptide after incubation with high concentrations of glucose and theophylline. Average concentrations of secreted C-peptide in buffer after stimulation with 16.7 mM glucose differed between noninduced (143.28 ± 7.98 pg/mL) and MafA-induced cells (337.53 ± 13.88 pg/mL) (P < 0.05) (Fig. 3E), suggesting that MafA overexpression in day 16 cells enhanced glucose-stimulated insulin secretion.
Exogenous FGF10 Enhances Pdx1 Expression
FGF10 was shown to expand early pancreatic Pdx1-expressing cells.25 To test if our culture system could respond to exogenous regulators, recombinant FGF10 was added to day 10 cultures derived from Pdx1-EGFP ES cells, at a time when Pdx1 message was relatively low but soon to be upregulated (Fig. 1D and Supplementary Fig. 1A). Cells were analyzed by flow cytometry and quantitative RT-PCR analysis at 3 days postincubation. There was a dose-dependent effect of FGF10 on enhancing the percentage of Pdx1-EGFP–expressing cells (Fig. 4A), consistent with what has been reported.6 Quantitative RT-PCR analysis also confirmed that messages for Pdx1 and Sox9, both of which are early pancreatic progenitor markers, were enhanced by addition of FGF10 (Fig. 4B). These results demonstrate that exogenous growth factor can enhance the development of pancreatic-like progenitors in our culture.
Fig. 4.
Effects of exogenous FGF10 on Pdx1 expression. Pdx1-EGFP ES cells were differentiated. (A) Fluorescent flow cytometric analysis of Pdx1-EGFP expression after addition of designated doses of FGF10 to day 10 culture for a total of 3 days. (B) Gene expression analysis by quantitative RT-PCR. Data represent the mean and standard deviation from duplicated samples. FGF, fibroblast growth factor. Color images available online at www.liebertonline.com/adt
Further Development of Pancreatic-Like Cells In Vitro and In Vivo from Day 20 Culture
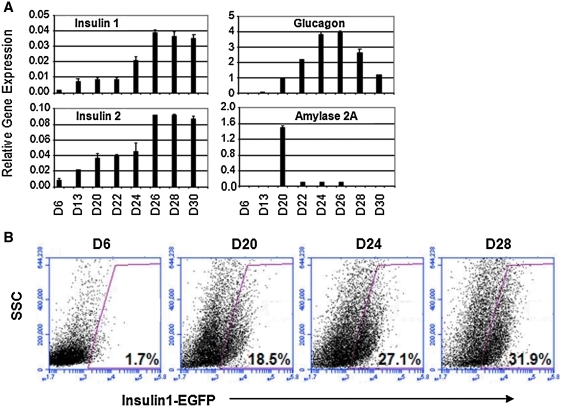
Gene expression analysis from Figure 1 suggests that in vitro generated cells might continue to mature after day 20. To test this, R1 ES cells were allowed to differentiate further to day 30 and gene expression was analyzed (Fig. 5A). We found that expression of insulin 1 and insulin 2 was significantly higher between days 26 and 30 compared with day 20 (Fig. 5A). This result was also supported by the use of MIP-EGFP ES cells (Fig. 5B), in which an increase of the percentage of insulin 1-EGFP+ cells was observed after day 20 in culture. Expression of glucagon was higher between days 24 and 26, but by day 30, its expression was reduced, similar to day 20 levels (Fig. 5A). Amylase expression was reduced after day 20. These results suggest that endocrine cells before day 20 are still at an relatively early phase of differentiation, which may have contributed to the variability of the RT-PCR data before day 20 for insulin 1 and insulin 2 (Fig. 1E and Supplementary Fig. 1A) because of low abundance of these messages.
Fig. 5.
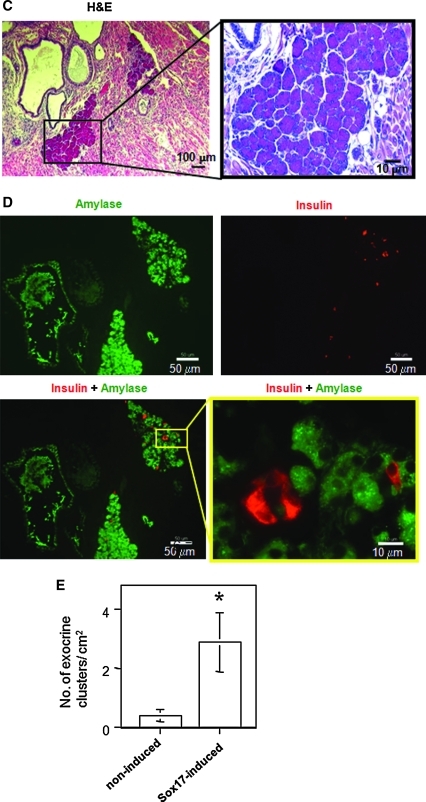
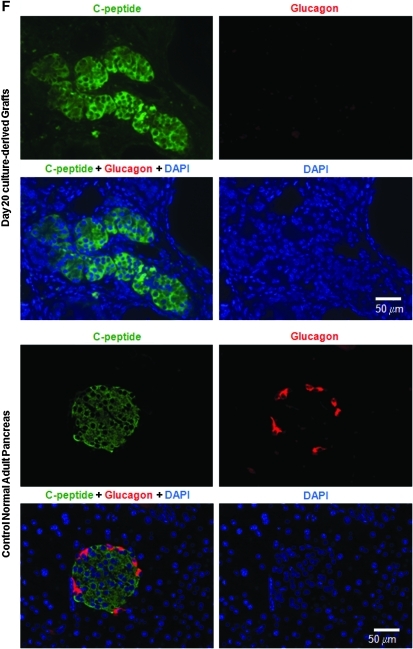
Further development of pancreatic-like cells in vitro and in vivo from day 20 culture. (A) R1 ES cells were differentiated for up to day 30, and cells at the designated age were analyzed by quantitative RT-PCR. Data represent the mean and standard deviation from duplicated samples. (B) MIP-EGFP ES cells were differentiated, and cells at the designated age were analyzed by flow cytometry for EGFP expression. (C–F) In vivo development of grafts at 5 weeks posttransplantation. (C) Representative clusters derived from a day 20 culture of AinV-Sox17 ES cell origin and treated with 1 μg/mL doxycycline between days 2 and 3 are shown. (D) Sections adjacent to those in C were examined by double staining for amylase (green) and insulin (red). Control slides with only secondary antibody showed negative staining (not shown). (E) Average number of acinar-like clusters, as identified by H&E staining, per square centimeter of graft area examined was determined. Data represent the mean and standard deviation from quadruplicated samples. *Number of clusters is different at P < 0.01, compared with control. (F) Double staining for C-peptide (green) and glucagon (red) from representative grafts derived from Sox17-induced cells showed single-hormone–positive cells (upper panels). H&E, hematoxylin and eosin.
We next tested whether day 20 cells could further differentiate in an in vivo microenvironment by transplanting them into the renal capsule of STZ-treated diabetic SCID-beige mice. The STZ injury model was chosen because it was shown to support the in vivo formation of pancreatic-like cells from undifferentiated ES cells.60 We found that clusters of organized pancreatic-like tissues were readily recognizable in H&E-stained sections prepared from grafts at 5 weeks posttransplantation in two different experiments and from either noninduced or Sox17-induced (1 μg/mL doxycycline added on day 2 for 24 h) day 20 cells. Figure 5C shows photomicrographs of those clusters. To confirm that these pancreatic-like clusters expressed amylase and insulin, double immunofluorescent staining was performed. Exocrine-like cells identified in the H&E sections stained positive for amylase (Fig. 5D). Insulin+ cells were interspersed within the acinar-like clusters but did not overlap with the amylase+ cells (Fig. 5D). Quantification (from H&E staining) of acinar-like clusters showed that Sox17-induced cells gave rise to higher numbers of these clusters, compared with the noninduced controls (Fig. 5E), consistent with the results from in vitro experiments (Fig. 3A). Thus, although not supported in vitro (Fig. 5A), acinar-like cells from our day 20 culture could further differentiate in vivo.
Finally, double immunostaining for C-peptide and glucagon was performed to discern whether first- or second-wave–like endocrine cells were generated. We found that none of the C-peptide+ cells coexpressed glucagon in the grafts (Fig. 5F), suggesting that second-wave beta-like cells were generated. It should be noted that all of the recipient mice remained hyperglycemic (data not shown), suggesting that the total mass of the endocrine cells transplanted was not sufficient to revert diabetes, and further enrichment of beta-like cells before transplantation would be required.
Discussion
In the drug discovery field, the “bottom-up” approach, based on structural considerations of known targets, has not been as fruitful as was once promised; in 2008, only 21 new drugs (i.e., small molecule) were approved by the United States Food and Drug Administration and 19 in 2009. Until recently, transformed human cell lines were used as the major cellular platform for pharmaceutical drug screening, sometimes referred to as the “top-down” approach. The major concern with the use of transformed cell lines is that compounds identified as “hits” may not have direct relevance to the normal biological processes being targeted; as a result, false leads consume resources set aside for the subsequent testing on animal models and in clinical trials. Use of normal primary human somatic cells for large-scale screening is not currently feasible because of the limited number of cells obtained from biopsy and subsequent propagation in vitro. Thus, there is a great need to develop large-scale screening platforms using nontransformed cell lines.
ES cells offer a potential solution to this bottleneck. ES cells can be grown in large numbers and maintained in a pluripotent state in vitro. They can also be induced in culture to differentiate into cells from all three germ layers in a relatively normal fashion that is faithful to development in vivo. Three properties make ES cells an ideal platform for drug discovery. First, ES cells can provide virtually inexhaustible quantities of target cells, which is necessary for screening of large numbers of molecules. Second, ES cells can differentiate into mature cells with phenotypes that mimic their counterparts in vivo. Third, compared with immortalized cell lines, ES cells and their derivatives will provide a much more accurate platform for the “top-down” drug screening approach.
Type 1 or 2 diabetes is caused by destruction of pancreatic insulin-secreting beta cells as a result of autoimmunity or functional loss from peripheral insulin resistance, respectively. Currently, relatively little is known about the cell nonautonomous regulations for beta cell proliferation and maturation. Using an ES cell platform to screen and identify growth factors, peptides or small molecule effectors that direct beta cell proliferation or maturation from their immediate progenitors has the potential to significantly impact diabetes therapy. To date, attempts to perform large-scale screening of small molecules for pancreatic-lineage cells from ES cells have only yielded chemicals that enhance the proliferation or commitment of the very early Pdx1-expressing progenitors from definitive endoderm.6 This is partially due to a lack of reliable and cost-effective assays to generate the later glucose-responsive insulin-expressing cells from ES cells.
In this study, we demonstrate that our ES cell to pancreatic differentiation protocol can provide a reproducible and cost-effective cellular assay for screening of beta cell effectors. Pancreatic-like cells derived from our system not only have correct gene expression patterns and glucose-responsive insulin secretion in vitro, but they also respond as expected in gain-of-function studies, suggesting our culture protocol produces committed beta-like cells and their immediate progenitors, which could serve as screening targets.
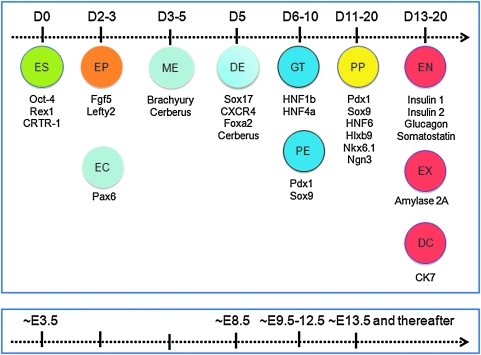
An important goal of the present study was to define target cell stages for future screening. The diagram in Figure 6 correlates the sequential timed gene activation of murine ES cells in our in vitro differentiation protocol (upper box) to development in vivo (lower box). Undifferentiated ES cells (equivalent to ICM at ∼E3.5) became committed to epiblast-like cells around culture days 2–3. This was followed by the formation of mesendodermal progenitors around days 3–5. Definitive endoderm-like cells were formed around in vitro day 5 (equivalent to ∼E8.5). From day 6 to 10 of culture (equivalent to ∼E9.5 to 12.5), gut tube and pancreatic endoderm-like cells were present. Starting from in vitro day 11 (equivalent to ∼E13.5 and thereafter), pancreatic progenitor cells emerged, followed by the appearance of various lineage-committed cells, such as exocrine, endocrine, and ductal cells (culture days 13–20).
Fig. 6.
Cell stage map for our in vitro ES cell differentiation assay. Upper panel depicts stages of in vitro differentiation. The stage designation is based primarily on information obtained from kinetic studies from two murine ES cell lines in this report (Fig. 1 and Supplementary Fig. 1). Lower panel depicts stages of in vivo differentiation according to literatures. EP, epiblast; EC, ectoderm; ME, mesendoderm; DE, definitive endoderm; GT, gut tube; PE, pancreatic endoderm; PP, pancreatic progenitor; EN, endocrine; EX, exocrine; DC, duct. Color images available online at www.liebertonline.com/adt
To screen beta cell effectors, day 16 and later culture would be appropriate targets, because cells at this stage are amenable for further maturation. This is supported by the observations that cells at this stage displayed an upward trend for a number of pancreatic genes, including those for endocrine, exocrine, ductal, and pancreatic transcription factors (Fig. 1 and Supplementary Fig. 1). Additionally, insulin messages and percentage of insulin 1-EGFP+ cells continued to increase after day 20 in vitro (Fig. 5A, B). Finally, day 20 cultures gave rise to well-organized pancreatic-like cells in vivo (Fig. 5C–F). As well, cells from this stage responded to MafA induction in expressing maturation markers (Fig. 3C) and had increased secretion of C-peptide in response to glucose stimulation in vitro (Fig. 3D, E). Together, these data demonstrate that the function of beta-like cells developed in our later stage of culture can be further enhanced by maturation factors and that these cells could be suitable targets for screening of beta cell effectors.
Published protocols9,61–63 for in vitro differentiation of beta-like cells utilize several recombinant growth factors and expensive chemicals to stimulate the formation of definitive endoderm, gut endoderm, and pancreatic cells. Compared with these protocols, our ES cell differentiation system offers several advantages for screening experiments. First, our protocol utilizes EB formation in the early differentiation stage (first 6 days), which can be easily scaled-up for a lower cost, in contrast to the adhesion monolayer culture with microcarrier technologies, which needs to be further investigated. Second, during the specific differentiation stages, relatively inexpensive chemicals or factors were used sequentially to induce endoderm and pancreatic development, including monothioglycerol (from day 0 to 6), exendin-4, and nicotinamide (used at day 13 and thereafter). The only relatively expensive item is the recombinant activin βB (used at day 13 and thereafter), and we are currently testing whether activin βB can be omitted or replaced by small molecules in this assay system. Third, compared with the human ES cell platform, the murine differentiation system provides a shorter project turnover time and ultimately offers advantages in cost-effectiveness, because glucose-responsive insulin-secreting cells from human ES cells require a longer time period to differentiate and mature in vitro (unpublished results). In addition, in some countries, the use of the human ES cell platform carries patent issues and political concerns that may inevitably affect the progress of research. Lastly, our culture protocol generates beta-like cells that do not coexpress glucagon (Fig. 5F) and respond dynamically to glucose stimulation in vitro (Fig. 3D), suggesting that functional beta-like cells were generated. Our data strongly suggest that the use of these cultures as targets for screening will result in a high likelihood of identifying positive hits for bona fide beta-cell regulators. We recognize that the use of murine cells is a limitation of our system, given that the positive hits may not necessarily translate to a similar function in human cells. However, this may be overcome by a secondary screening assay using human cells in vitro. In conclusion, our ES cell differentiation assay is a valuable tool that can be used as a primary screening platform, which has the potential to significantly expedite the productivity of drug discovery in the treatment of diabetes.
Supplementary Material
Abbreviations
- CK
cytokeratin
- CRTR-1
Cp2-related transcriptional repressor-1
- DAPI
4',6-diamidino-2-phenylindole
- EB
embryoid body
- EGFP
enhanced green fluorescent protein
- ES
embryonic stem
- FGF
fibroblast growth factor
- FCS
fetal calf serum
- Glut2
glucose transporter 2
- H&E
hematoxylin and eosin
- HNF
hepatocyte nuclear factor
- HTS
high-throughput screening
- ICM
inner cell mass
- LoxP
locus of X-over P1
- MOI
multiplicity of infection
- Ngn3
neurogenin3
- Pdx1
pancreatic and duodenal homeobox 1
- STZ
streptozotocin
Acknowledgments
The authors thank Dr. Ed Stanley at the Monash University, Australia, for providing Pdx1-EGFP ES cells, Drs. Kohei Ishiyama, Keiko Omori, and Yoko Mullen for providing murine islets, Ms. Lucy Brown and members at the Analytical Cytometry Core at City of Hope for providing cell sorting services, and Dr. Keely Walker, Dr. Silvia DaCosta, Mr. Michael Winkler, and Ms. Nancy Trieu for editing the manuscript. This work was supported in part by NIH grants (R21DK069997 and R01DK081587 to H.T.K.). L.S. is supported by a postdoctoral fellowship from the California Institute of Regenerative Medicine (CIRM).
Disclosure Statement
No competing financial interests exist.
References
- 1.Barbaric I. Gokhale PJ. Andrews PW. High-content screening of small compounds on human embryonic stem cells. Biochem Soc Trans. 2010;38:1046–1050. doi: 10.1042/BST0381046. [DOI] [PubMed] [Google Scholar]
- 2.Desbordes SC. Placantonakis DG. Ciro A. Socci ND. Lee G. Djaballah H. Studer L. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding S. Wu TY. Brinker A. Peters EC. Hur W. Gray NS. Schultz PG. Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci USA. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu S. Wurdak H. Wang J. Lyssiotis CA. Peters EC. Cho CY. Wu X. Schultz PG. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4:416–426. doi: 10.1016/j.stem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Barbaric I. Gokhale PJ. Jones M. Glen A. Baker D. Andrews PW. Novel regulators of stem cell fates identified by a multivariate phenotype screen of small compounds on human embryonic stem cell colonies. Stem Cell Res. 2010;5:104–119. doi: 10.1016/j.scr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Chen S. Borowiak M. Fox JL. Maehr R. Osafune K. Davidow L. Lam K. Peng LF. Schreiber SL. Rubin LL. Melton D. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 7.McNeish J. Roach M. Hambor J. Mather RJ. Weibley L. Lazzaro J. Gazard J. Schwarz J. Volkmann R. Machacek D. Stice S. Zawadzke L. O'Donnell C. Hurst R. High-throughput screening in embryonic stem cell-derived neurons identifies potentiators of AMPA-type glutamate receptors. J Biol Chem. 2010;285:17209–17217. doi: 10.1074/jbc.M109.098814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Amour KA. Agulnick AD. Eliazer S. Kelly OG. Kroon E. Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 9.D'Amour KA. Bang AG. Eliazer S. Kelly OG. Agulnick AD. Smart NG. Moorman MA. Kroon E. Carpenter MK. Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 10.Kroon E. Martinson LA. Kadoya K. Bang AG. Kelly OG. Eliazer S. Young H. Richardson M. Smart NG. Cunningham J. Agulnick AD. D'Amour KA. Carpenter MK. Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 11.Oliver-Krasinski JM. Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai-Azuma M. Kanai Y. Gad JM. Tajima Y. Taya C. Kurohmaru M. Sanai Y. Yonekawa H. Yazaki K. Tam PP. Hayashi Y. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 13.Clements D. Woodland HR. Changes in embryonic cell fate produced by expression of an endodermal transcription factor, Xsox17. Mech Dev. 2000;99:65–70. doi: 10.1016/s0925-4773(00)00476-7. [DOI] [PubMed] [Google Scholar]
- 14.Gu G. Dubauskaite J. Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 15.Guz Y. Montminy MR. Stein R. Leonard J. Gamer LW. Wright CV. Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 16.Offield MF. Jetton TL. Labosky PA. Ray M. Stein RW. Magnuson MA. Hogan BL. Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 17.Ahlgren U. Jonsson J. Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 18.Sommer L. Ma Q. Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 19.Apelqvist A. Li H. Sommer L. Beatus P. Anderson DJ. Honjo T. Hrabe de Angelis M. Lendahl U. Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 20.Gradwohl G. Dierich A. LeMeur M. Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97:1607–1111. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka K. Han SI. Shioda S. Hirai M. Nishizawa M. Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka TA. Artner I. Henderson E. Means A. Sander M. Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci USA. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L. Guo M. Matsuoka TA. Hagman DK. Parazzoli SD. Poitout V. Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C. Moriguchi T. Kajihara M. Esaki R. Harada A. Shimohata H. Oishi H. Hamada M. Morito N. Hasegawa K. Kudo T. Engel JD. Yamamoto M. Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart AW. Baeza N. Apelqvist A. Edlund H. Attenuation of FGF signalling in mouse beta-cells leads to diabetes. Nature. 2000;408:864–888. doi: 10.1038/35048589. [DOI] [PubMed] [Google Scholar]
- 26.Ku HT. Zhang N. Kubo A. O'Connor R. Mao M. Keller G. Bromberg JS. Committing embryonic stem cells to early endocrine pancreas in vitro. Stem Cells. 2004;22:1205–1217. doi: 10.1634/stemcells.2004-0027. [DOI] [PubMed] [Google Scholar]
- 27.Ku HT. Chai J. Kim YJ. White P. Purohit-Ghelani S. Kaestner KH. Bromberg JS. Insulin-expressing colonies developed from murine embryonic stem cell-derived progenitors. Diabetes. 2007;56:921–929. doi: 10.2337/db06-0468. [DOI] [PubMed] [Google Scholar]
- 28.Holland AM. Micallef SJ. Li X. Elefanty AG. Stanley EG. A mouse carrying the green fluorescent protein gene targeted to the Pdx1 locus facilitates the study of pancreas development and function. Genesis. 2006;44:304–307. doi: 10.1002/dvg.20214. [DOI] [PubMed] [Google Scholar]
- 29.Milewski WM. Temple KA. Wesselschmidt RL. Hara M. Generation of embryonic stem cells from mouse insulin I promoter-green fluorescent protein transgenic mice and characterization in a teratoma model. In Vitro Cell Dev Biol Anim. 2009;45:1–5. doi: 10.1007/s11626-008-9142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuoka TA. Zhao L. Artner I. Jarrett HW. Friedman D. Means A. Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyba M. Perlingeiro RC. Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 32.Iacovino M. Hernandez C. Xu Z. Bajwa G. Prather M. Kyba M. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 2009;18:783–792. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alpert S. Hanahan D. Teitelman G. Hybrid insulin genes reveal a developmental lineage for pancreatic endocrine cells and imply a relationship with neurons. Cell. 1988;53:295–308. doi: 10.1016/0092-8674(88)90391-1. [DOI] [PubMed] [Google Scholar]
- 34.Teitelman G. Alpert S. Polak JM. Martinez A. Hanahan D. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development. 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 35.Rogers MB. Hosler BA. Gudas LJ. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 36.Pelton TA. Sharma S. Schulz TC. Rathjen J. Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- 37.Haub O. Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development. 1991;112:397–406. doi: 10.1242/dev.112.2.397. [DOI] [PubMed] [Google Scholar]
- 38.Hebert JM. Boyle M. Martin GR. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- 39.Tesar PJ. Chenoweth JG. Brook FA. Davies TJ. Evans EP. Mack DL. Gardner RL. McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 40.Tereshina MB. Zaraisky AG. Novoselov VV. Ras-dva, a member of novel family of small GTPases, is required for the anterior ectoderm patterning in the Xenopus laevis embryo. Development. 2006;133:485–494. doi: 10.1242/dev.02207. [DOI] [PubMed] [Google Scholar]
- 41.Kubo A. Shinozaki K. Shannon JM. Kouskoff V. Kennedy M. Woo S. Fehling HJ. Keller G. Development of definitive endoderm from embryonic stem cell-derived mesendoderm. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 42.Biben C. Stanley E. Fabri L. Kotecha S. Rhinn M. Drinkwater C. Lah M. Wang CC. Nash A. Hilton D. Ang SL. Mohun T. Harvey RP. Murine cerberus homologue mCer-1: a candidate anterior patterning molecule. Dev Biol. 1998;194:135–151. doi: 10.1006/dbio.1997.8812. [DOI] [PubMed] [Google Scholar]
- 43.Yasunaga M. Tada S. Torikai-Nishikawa S. Nakano Y. Okada M. Jakt LM. Nishikawa S. Chiba T. Era T. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 44.Osada SI. Wright CV. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development. 1999;126:3229–3240. doi: 10.1242/dev.126.14.3229. [DOI] [PubMed] [Google Scholar]
- 45.Hallonet M. Kaestner KH. Martin-Parras L. Sasaki H. Betz UA. Ang SL. Maintenance of the specification of the anterior definitive endoderm and forebrain depends on the axial mesendoderm: a study using HNF3beta/Foxa2 conditional mutants. Dev Biol. 2002;243:20–33. doi: 10.1006/dbio.2001.0536. [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi TP. Dumont DJ. Conlon RA. Breitman ML. Rossant J. Flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- 47.Arai A. Yamamoto K. Toyama J. Murine cardiac progenitor cells require visceral embryonic endoderm and primitive streak for terminal differentiation. Dev Dyn. 1997;210:344–353. doi: 10.1002/(SICI)1097-0177(199711)210:3<344::AID-AJA13>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 48.Coffinier C. Barra J. Babinet C. Yaniv M. Expression of the vHNF1/HNF1beta homeoprotein gene during mouse organogenesis. Mech Dev. 1999;89:211–213. doi: 10.1016/s0925-4773(99)00221-x. [DOI] [PubMed] [Google Scholar]
- 49.Duncan SA. Manova K. Chen WS. Hoodless P. Weinstein DC. Bachvarova RF. Darnell JE., Jr. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seymour PA. Freude KK. Tran MN. Mayes EE. Jensen J. Kist R. Scherer G. Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H. Ables ET. Pope CF. Washington MK. Hipkens S. Means AL. Path G. Seufert J. Costa RH. Leiter AB. Magnuson MA. Gannon M. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev. 2009;126:958–973. doi: 10.1016/j.mod.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison KA. Thaler J. Pfaff SL. Gu H. Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 53.Sander M. Sussel L. Conners J. Scheel D. Kalamaras J. Dela Cruz F. Schwitzgebel V. Hayes-Jordan A. German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 54.Seguin CA. Draper JS. Nagy A. Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3:182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 55.Spence JR. Lange AW. Lin SC. Kaestner KH. Lowy AM. Kim I. Whitsett JA. Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Artner I. Hang Y. Guo M. Gu G. Stein R. MafA is a dedicated activator of the insulin gene in vivo. J Endocrinol. 2008;198:271–279. doi: 10.1677/JOE-08-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kataoka K. Shioda S. Ando K. Sakagami K. Handa H. Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 58.Otonkoski T. Insulin and glucagon secretory responses to arginine, glucagon, and theophylline during perifusion of human fetal islet-like cell clusters. J Clin Endocrinol Metab. 1988;67:734–740. doi: 10.1210/jcem-67-4-734. [DOI] [PubMed] [Google Scholar]
- 59.Rajagopal J. Anderson WJ. Kume S. Martinez OI. Melton DA. Insulin staining of ES cell progeny from insulin uptake. Science. 2003;299:363. doi: 10.1126/science.1077838. [DOI] [PubMed] [Google Scholar]
- 60.Kodama M. Takeshita F. Kanegasaki S. Ochiya T. Quinn G. Pancreatic endocrine and exocrine cell ontogeny from renal capsule transplanted embryonic stem cells in streptozocin-injured mice. J Histochem Cytochem. 2008;56:33–44. doi: 10.1369/jhc.7A7278.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goicoa S. Alvarez S. Ricordi C. Inverardi L. Dominguez-Bendala J. Sodium butyrate activates genes of early pancreatic development in embryonic stem cells. Cloning Stem Cells. 2006;8:140–149. doi: 10.1089/clo.2006.8.140. [DOI] [PubMed] [Google Scholar]
- 62.Jiang J. Au M. Lu K. Eshpeter A. Korbutt G. Fisk G. Majumdar AS. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells. 2007;25:1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 63.Schroeder IS. Rolletschek A. Blyszczuk P. Kania G. Wobus AM. Differentiation of mouse embryonic stem cells to insulin-producing cells. Nat Protoc. 2006;1:495–507. doi: 10.1038/nprot.2006.71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.