Abstract
Tetraspanin protein CD9 supports sperm–egg fusion, and regulates cell adhesion, motility, metastasis, proliferation and signaling. The large extracellular loop and transmembrane domains of CD9 engage in functionally important interactions with partner proteins. However, neither functional nor biochemical roles have been shown for the CD9 C-terminal tail, despite it being highly conserved throughout vertebrate species. To gain new insight into the CD9 tail, three C-terminal amino acids (Glu-Met-Val) were replaced with residues corresponding to C-terminal amino acids from tetraspanin protein CD82 (Pro-Lys-Tyr). Wild-type and mutant CD9 were then stably expressed in MOLT-4, K562, U937, RD and HT1080 cells. Whereas wild-type CD9 inhibited cell adhesion and spreading on fibronectin, mutant CD9 did not. Wild-type CD9 also promoted homotypic cell–cell aggregation and microvilli formation, whereas mutant CD9 did not. Protein interactions of wild-type and mutant CD9 were compared quantitatively using stable isotope labeling with amino acids in cell culture (SILAC) in conjunction with liquid-chromatography–tandem mass spectrometry (LC-MS/MS) technology. SILAC results showed that, despite wild-type and mutant CD9 having identical expression levels, mutant CD9 and its major transmembrane interacting partners were recovered in substantially reduced amounts from 1% Brij 96 lysates. Immunoprecipitation experiments confirmed that mutant CD9 recovery was decreased in Brij 96, but not in more stringent Triton X-100 detergent. Additionally, compared with wild-type CD9 complexes, mutant CD9 complexes were larger and more oligomerized in Brij 96 detergent, consistent with decreased Brij 96 solubility, perhaps due to more membrane domains packing more tightly together. In conclusion, multiple CD9 functions depend on its C-terminal tail, which affects the molecular organization of CD9 complexes, as manifested by their altered solubilization in Brij 96 and organization on the cell surface.
Key words: CD9, Tetraspanin, SILAC, Microvilli, Cell adhesion, Cell spreading
Introduction
The tetraspanin protein family contains 33 distinct members, each with four transmembrane domains, short N- and C-terminal cytoplasmic domains, a small intracellular loop and two extracellular loops (Berditchevski, 2001; Boucheix and Rubinstein, 2001; Hemler, 2003). The larger extracellular loop contains CCG and PXSC motifs, which are hallmarks of the tetraspanin family (Seigneuret et al., 2001). Through the large extracellular loop, tetraspanins interact with themselves and with other proteins, including membrane-bound growth factors, immunoglobulin (Ig) superfamily proteins, signaling enzymes and integrins (Berditchevski, 2001; Levy and Shoham, 2005). These protein–protein interactions underlie the assembly of structural and functional units called tetraspanin-enriched microdomains (TEMs) (Espenel et al., 2008; Hemler, 2005; Nydegger et al., 2006; Yanez-Mo et al., 2009). Within TEMs, tetraspanins can modulate the functions of associated proteins, thereby regulating many physiological and pathological processes, such as fertilization, cell adhesion, motility, tumor invasion and transendothelial migration (Barreiro et al., 2005; Berditchevski and Odintsova, 1999; Miyado et al., 2000; Ono et al., 1999; Zoller, 2009).
CD9, a member of the tetraspanin family, is expressed in multiple cell types, including hematopoietic cells, endothelial cells, epithelial cells, smooth muscle cells, pre-B cells and many tumor cell lines (Boucheix and Rubinstein, 2001; Hemler, 2003). Oocytes lacking CD9 are deficient in sperm–egg fusion (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000), at least partly due to alterations in microvilli on the oocyte surface (Runge et al., 2007). CD9 also regulates myoblast (Tachibana and Hemler, 1999) and monocyte (Takeda et al., 2003) fusion, and HIV-induced syncytia formation (Gordon-Alonso et al., 2006). CD9 has tumor-suppressor-like functions in many tumor cell types, and can inhibit cell invasion and metastasis (Ikeyama et al., 1993; Zoller, 2009). CD9 also contributes to cell signaling (Huang et al., 2004), and can regulate cell adhesion (Masellis-Smith and Shaw, 1994), migration (Anton et al., 1995), apoptosis (Murayama et al., 2004), membrane protein shedding (Shi et al., 2000) and diphtheria toxin binding (Iwamoto et al., 1994). To assist in these diverse functions, CD9 interacts directly with transmembrane proteins EWI-2 (Charrin et al., 2003; Stipp et al., 2001a) and EWI-F (also called CD9P-1 and FPRP) (Charrin et al., 2001; Stipp et al., 2001b). CD9 also interacts with other proteins, including other tetraspanins, a subset of integrins, other adhesion molecules, membrane proteases, choline receptors and G proteins (Le Naour et al., 2006).
Whereas the functional importance of tetraspanin large extracellular loops (EC2) is well recognized, the C-terminal tails have received less attention. The C-terminal tail of tetraspanin CD63 binds to AP-3 adaptor subunit μ3 (Rous et al., 2002) and to a PDZ domain in syntenin-1 (Latysheva et al., 2006), which affects CD63 distribution and trafficking. The CD81 C-terminal tail was suggested to associate directly with ezrin-radixin-moesin (ERM) proteins (Sala-Valdes et al., 2006), whereas a YRSL sequence in the CD151 cytoplasmic domain might determine intracellular trafficking and function (Liu et al., 2007). In addition, the short C-terminal tail of CD151 supports integrin-α6β1-dependent cellular cable formation and adhesion strengthening (Lammerding et al., 2003; Yang et al., 2002). As in other tetraspanins, CD9 contains a C-terminal tail that is short (only eight residues) and highly conserved across several animal species, suggesting functional importance. However, essentially nothing is known about the function and biochemistry of the CD9 C-terminal tail. Here, we have mutated the CD9 C-terminal tail and examined the functional consequences. In addition, we have used a differential mass spectrometry technology called SILAC [stable isotope labeling of amino acids in cell culture (Ong et al., 2002)], together with immunoprecipitation, gel filtration and flow cytometry, to examine the effects of CD9 tail mutation on the molecular organization of CD9 complexes in detergent lysates and on the cell surface. To establish the generality of the conclusions, functional and biochemical studies were carried out using four to five different cell lines, including both adherent and non-adherent types.
Results
Expression and distribution of CD9 and CD9 mutant in different cell lines
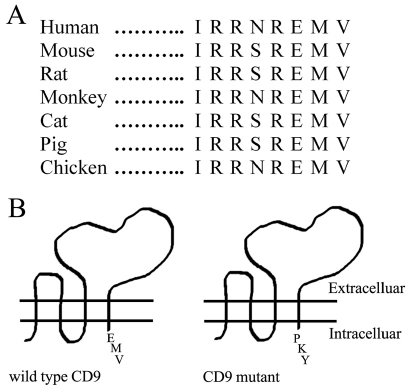
The short C-terminal tail of CD9 (eight residues) is well conserved among seven different vertebrate species (Fig. 1A). To address the functional and biochemical importance of the CD9 tail, the C-terminal EMV motif was replaced with PKY, which corresponds to the C-terminal amino acids of CD82 (Fig. 1B). This particular conservative strategy was chosen because: the C-terminal EMV motif in CD9 partly resembles the functionally important PDZ-domain-interacting C-terminal EVM site in CD63 (Latysheva et al., 2006); more radical CD9 C-tail mutations lead to loss of cell-surface expression (e.g. Ryu et al., 2000); CD9 and CD82 are the two tetraspanins most known for having tumor suppressor functions (Cajot et al., 1997; Huang et al., 1998; Miranti, 2009; Miyake et al., 1995); and various other CD9–CD82 chimeras have been effectively utilized in prior studies (Charrin et al., 2003; Gutierrez-Lopez et al., 2003). Several CD9-deficient cell lines (MOLT-4, K562, RD, HT1080 and U937) were stably transfected with wild-type and mutant CD9, to yield similar total expression levels, as seen by immunoblotting of CD9 (supplementary material Fig. S1A and Fig. S1C, inset). Cell-surface expression was also comparable, as seen by flow cytometry (supplementary material Fig. S1B,C).
Fig. 1.
CD9 C-terminal tail and mutagenesis. (A) CD9 C-terminal tail sequences from seven different animal species. (B) The three C-terminal amino acids (glutamic acid-methionine-valine, EMV) of wild-type CD9 were changed to proline-lysine-tyrosine (PKY) to yield mutant CD9.
CD9 but not CD9 mutant inhibits cell adhesion and cell spreading
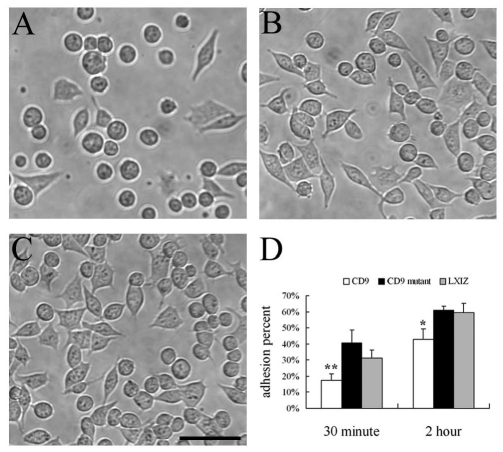
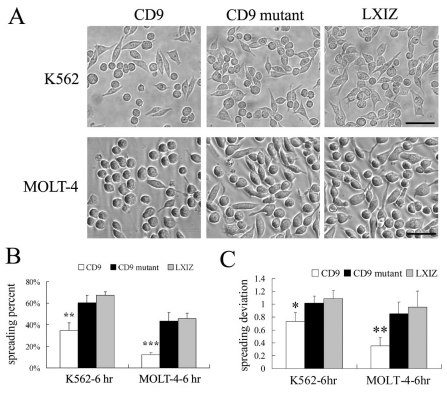
CD9 might regulate integrin-dependent cell adhesion and spreading (Berditchevski, 2001; Deng et al., 2000; Hemler, 2003). Hence, we analyzed cell adhesion and spreading for K562 cells transfected to express wild-type and mutant CD9. Diminished adhesion to fibronectin was seen for wild-type CD9 cells (Fig. 2A) compared to mutant CD9 cells (Fig. 2B) and control K562 cells (Fig. 2C). Quantification (Fig. 2D) confirmed that adhesion was diminished upon expression of wild-type CD9, but not mutant CD9, at both 30 minutes and 2 hours after plating. CD9 also inhibited spreading of both MOLT-4 and K562 cells on fibronectin, as seen in photos (Fig. 3A) and after quantification (Fig. 3B,C). Unlike CD9, mutant CD9 failed to inhibit spreading after 6 hours (Fig. 3), and also after 1 and 3 hours (supplementary material Fig. S2A,B). Likewise, wild-type CD9, but not mutant CD9, inhibited spreading of RD cells at multiple time points after plating (supplementary material Fig. S2C–E).
Fig. 2.
CD9 mutation affects K562 cell adhesion to fibronectin. K562 cells expressing (A) wild-type CD9, (B) mutant CD9 or (C) LXIZ control vector were allowed to adhere to fibronectin for 2 hours, and then photographed. Scale bar: 50 μm. (D) Quantification of K562 cell adhesion results. Error bars indicate means ± s.d. *P<0.05; **P<0.01.
Fig. 3.
CD9 mutation affects cell spreading on fibronectin. (A) K562 and MOLT-4 cells expressing CD9, mutant CD9 and control vector were photographed after spreading on fibronectin for 6 hours. Scale bar: 50 μm. (B) Percent cell spreading was quantified. **P<0.01; ***P<0.001. (C) The deviation of cell spreading was determined by subtracting the value 1.0 (perfect round) from the exact value of cell spreading. At least 50 cells were quantified. Error bars indicate means ± s.d. *P<0.05; **P<0.01.
CD9 but not CD9 mutant promotes cell aggregation
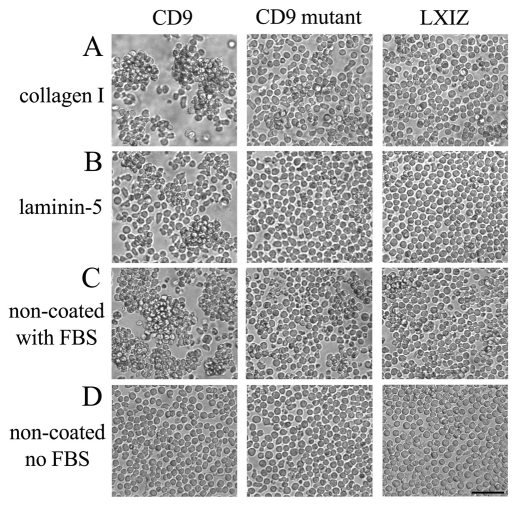
CD9 has been associated with cell–cell aggregation (Letarte et al., 1993; Masellis-Smith et al., 1990). Indeed, expression of wild-type CD9 promoted MOLT-4 cell–cell aggregation when cells were plated for 2 hours on collagen I (Fig. 4A) or on laminin-5 (Fig. 4B), or in the presence of 10% fetal bovine serum (FBS) (Fig. 4C). Expression of mutant CD9 had a minimal pro-aggregation effect (Fig. 4A–C). Non-coated plastic with no serum yielded no aggregation, regardless of CD9 expression (Fig. 4D). CD9 promotion of MOLT-4 cell aggregation in the presence of collagen I, laminin-5 or FBS (which contains fibronectin) is consistent with a role for β1 integrins (Letarte et al., 1993; Ziyyat et al., 2006).
Fig. 4.
CD9 and mutant CD9 effects on MOLT-4 cell aggregation. MOLT-4 cells stably expressing CD9, mutant CD9 or control vector were incubated on (A) collagen I, (B) laminin-5, (C) non-coated tissue culture plastic with FBS or (D) non-coated tissue culture plastic. Photos were obtained after 2 hours. Scale bar: 50 μm.
CD9 but not CD9 mutant promotes microvilli formation
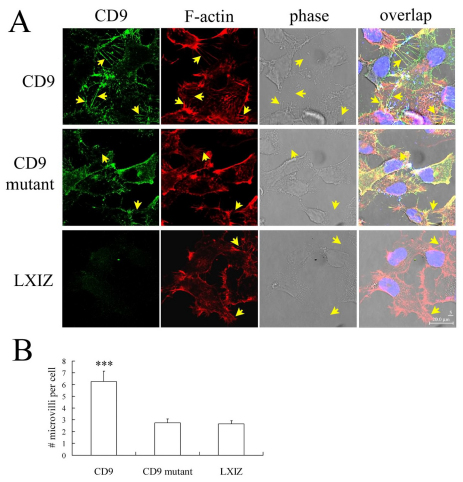
CD9 was previously observed within filopodia and microvilli, in proximity to F-actin (Kaji et al., 2001; Runge et al., 2007; Stipp et al., 2003). To gain clues regarding functional differences between wild-type and mutant CD9, we analyzed staining patterns on adherent cell lines RD (Fig. 5) and HT1080 (supplementary material Fig. S3). In both cell types, wild-type CD9 expression correlated with formation of abundant filopodia and microvilli-like projections, with CD9 within the projections (Fig. 5A, arrows) colocalized with F-actin (Fig. 5A; supplementary material Fig. S3A). By contrast, RD and HT1080 cells transfected with mutant CD9 did not show microvilli-like projections (Fig. 5A, arrows), as seen by staining of either CD9 or F-actin. The density and lengths of microvilli-like structures in CD9 mutant cells closely resembled those in cells transfected with control vector (Fig. 5A; supplementary material Fig. S3A). Quantification of the number of microvilli per cell confirmed that microvilli formation was promoted by CD9, but not by mutant CD9 (Fig. 5B; supplementary material Fig. S3B).
Fig. 5.
CD9 and mutant CD9 effects on microvilli formation in RD cells. (A) RD cells were stained for CD9 using mAb MM2/57 labeled with AlexaFluor488 (green) and for F-actin using AlexaFluor594–phalloidin (red). Cell nuclei are stained with DAPI (blue). Microvilli-like structures are marked with arrows. Scale bar: 20 μm. (B) Counting of microvilli (greater than 5 μm) was completed using Scion Image software, with quantification for each cell type from 10 random photos, each containing multiple cells (counted by staining of nuclei). Error bars indicate means ± s.d. ***P<0.001.
CD9 mutation alters sensitivity to Brij 96 detergent
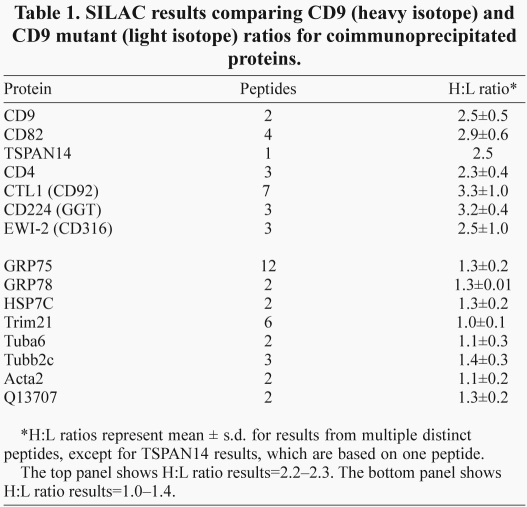
To compare wild-type and mutant CD9 with respect to molecular interactions, they were expressed in a U937 cell variant, which lacks both CD9 and CD81. Thus, we hoped to avoid indirect CD81-dependent interactions between various partner proteins and CD9. CD9 complexes then were analyzed using SILAC, a differential mass spectrometry quantification method (Ong et al., 2002). Wild-type and mutant CD9 were expressed at comparable levels in U937 cells, as demonstrated by cell surface flow cytometry (supplementary material Fig. S1C) and by immunoblotting from Triton X-100 whole-cell lysates (supplementary material Fig. S1C, inset). Wild-type CD9-transfected cells were then grown for 10 doublings in 13C-leucine heavy isotope, whereas mutant CD9-transfected cells were grown in medium containing normal leucine (light isotope). Equal numbers of cells were mixed, lysed together in 1% Brij 96 and then CD9 complexes were recovered, using beads coated with anti-CD9 monoclonal antibody (mAb) ALB6. Mass spectrometry coupled with quantitative SILAC analysis showed that peptides from eight different proteins were coimmunoprecipitated in comparable amounts [heavy (H):light (L) ratios~1.0–1.4] with wild-type and mutant CD9 (Table 1, bottom). However, recovery of CD9 itself and six of its known partner binding proteins was notably amplified for wild-type CD9 compared with for mutant CD9 (H:L ratios=2.3–3.3; Table 1, top). Although recovery of the major CD9 partner protein EWI-2 was increased for wild-type CD9 compared with mutant CD9 (Table 1), cell surface expression of EWI-2 on U937 cells (data not shown), K562 and RD cells (supplementary material Fig. S4) was unaltered by CD9 tail mutation.
Table 1.
SILAC results comparing CD9 (heavy isotope) and CD9 mutant (light isotope) ratios for coimmunoprecipitated proteins.
CD9 mutation changes protein solubilization and organization in Brij 96
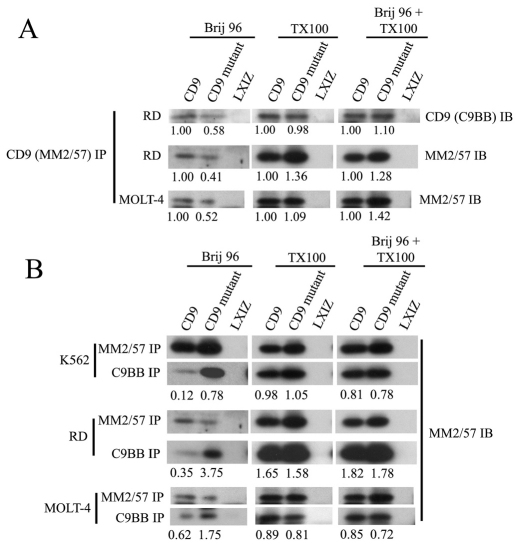
Results obtained in Table 1 indicated that, even though wild-type and mutant CD9 were expressed at comparable levels on the surface of U937 cells and recovered at nearly identical levels in Triton X-100 lysates, recovery from Brij 96 lysates was diminished for complexes containing mutant CD9. Consistent with this interpretation, mutant CD9 was immunoprecipitated from RD and MOLT-4 Brij 96 lysates at levels that were 40–60% less than those of wild-type CD9 (Fig. 6A, left panels). By contrast, wild-type and mutant CD9 were recovered at nearly identical levels from Triton X-100 lysates (Fig. 6A, middle panels). Furthermore, when 1% Brij 96 cell lysates were subsequently supplemented, post lysis, to contain also 1% Triton X-100, the difference in CD9 recoveries disappeared (Fig. 6A, right panels). These results suggest that mutant CD9 was not simply left behind in the cell pellet, but rather was reorganized to become partially unavailable (i.e. less soluble) within the Brij 96 lysate.
Fig. 6.
CD9 mutation affects recovery of CD9 complexes in Brij 96 detergent. (A) RD and MOLT-4 cells were lysed in 1% Brij 96, 1% Triton X-100, or 1% Brij 96 subsequently supplemented to also contain 1% Triton X-100. After immunoprecipitation (IP) using anti-CD9 mAb MM2/57 and SDS-PAGE resolution, CD9 was detected by immunoblotting using mAb C9BB or MM2/57. Numbers indicate recovery of mutant CD9 relative to that of wild-type CD9. Note that the six lower panels are used again in B for a different purpose. (B) After detergent lysis as in A, immunoprecipitations were carried out using anti-CD9 mAb MM2/57 or C9BB as indicated. CD9 was then detected by immunoblotting using mAb MM2/57. Numbers indicate recovery of CD9 (wild type or mutant) using mAb C9BB relative to mAb MM2/57. Note that the third and fifth panel rows are identical to images used in the second and third rows of A.
In contrast to the recognition of total CD9 by high-affinity anti-CD9 mAb (i.e. MM2/57 and ALB6), low-affinity anti-CD9 mAb C9BB preferentially recognizes clustered CD9 (Yang et al., 2006). Whereas immunoprecipitation with anti-CD9 mAb MM2/57 typically yielded more wild-type CD9 (e.g. see Fig. 6A), immunoprecipitation with mAb C9BB consistently yielded more mutant CD9 from Brij 96 lysates (Fig. 6B, left panels). Hence, C9BB:MM2/57 recovery ratios (Fig. 6B, numbers below panels) were consistently higher for mutant CD9 relative to wild-type CD9. These ratios show that mAb C9BB, compared to MM2/57, has a 3–10-fold preference for mutant CD9 over wild-type CD9 in K562, RD and MOLT-4 cells. This result is consistent with mutant CD9 complexes being more clustered (Yang et al., 2006) and less well solubilized in Brij 96 detergent. Preferential recovery of mutant CD9 by mAb C9BB disappeared upon solubilization in Triton X-100 or upon post-lysis addition of Triton X-100 to lysates already containing Brij 96 (Fig. 6B; middle and right panels).
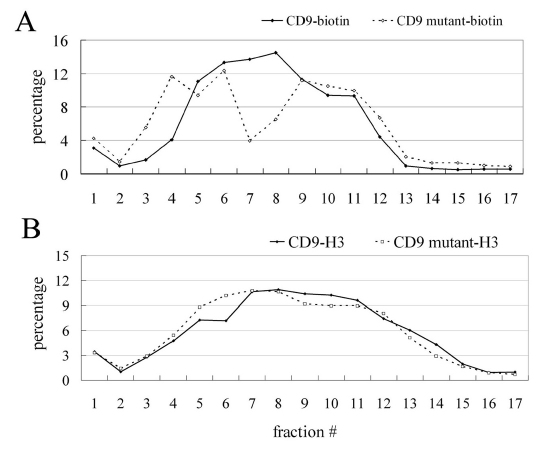
To assess further the differences between mutant and wild-type CD9 complexes in Brij 96 lysates, gel filtration experiments were carried out. Consistent with diminished mutant CD9 solubility in 1% Brij 96 and increased mutant CD9 clustering, a subset of surface biotin-labeled mutant CD9 complexes were distinctly larger than the largest wild-type CD9 complexes (Fig. 7A, fractions 3,4). The gel filtration size difference was not as obvious when cells were metabolically labeled with 3[H]-palmitate, largely because newly synthesized CD9 did not have time to assemble into larger complexes. Nonetheless, mutant CD9 complexes were still also a little larger than wild-type CD9 complexes, as particularly evident in column fractions 4–6 (Fig. 7B).
Fig. 7.
Gel filtration analyses of CD9 and mutant CD9 complexes. (A) U937 cells stably expressing CD9 or mutant CD9 were surface labeled with biotin and then lysed in 1% Brij 96. Lysates were fractionated using a sepharose CL6B gel filtration column and biotin-labeled CD9 was detected as described in the Materials and Methods. (B) U937 cells were metabolically labeled using 3H-palmitate, and then CD9 and mutant CD9 were fractionated and detected as described in the Materials and Methods. The sum of densitometry quantifications for each set of samples is 100%.
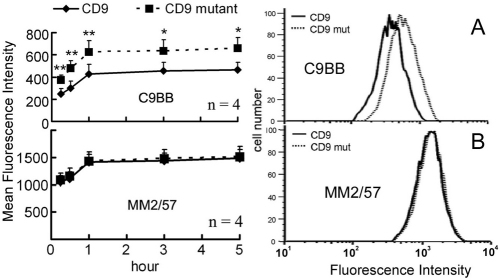
Results in Figs 6 and 7 suggest that our CD9 mutation might also alter the organization of CD9 complexes on the surface of intact cells. As shown in Fig. 8A, mutant CD9 again showed enhanced binding of mAb C9BB, a probe for homooligomerization (Yang et al., 2006). In a control experiment, surface expression of total CD9, assessed using mAb MM2/57, was not altered (Fig. 8B).
Fig. 8.
CD9 mutation effects on cell-surface organization. RD cells expressing CD9 or mutant CD9 were incubated with 4.0 μg low-affinity anti-CD9 antibody (C9BB, A) or 1.0 μg high-affinity antibody (MM2/57, B) for the indicated times, and CD9 mean fluorescence intensities (MFIs) were plotted. Right panels show representative flow cytometry profiles from the 1 hour time point.
Discussion
A functional role for the CD9 C-terminal tail
To assess the functional role of the highly conserved CD9 C terminus, a chimeric protein was designed, replacing the CD9 C-terminal Glu-Met-Val with Pro-Lys-Tyr from CD82. Expression of wild-type CD9 (in cells otherwise lacking CD9) inhibited cell adhesion and spreading on fibronectin, but promoted homotypic aggregation and microvilli formation. By contrast, mutant CD9 did not inhibit adhesion or spreading, and did not promote homotypic aggregation and microvilli formation.
Involvement of CD9 in integrin-dependent adhesion, spreading and homotypic aggregation has previously been observed (Masellis-Smith et al., 1990; Masellis-Smith and Shaw, 1994; Shaw et al., 1995). CD9-dependent formation of microvilli has also been observed (Runge et al., 2007) and anti-CD9-antibody-induced microvilli ‘zippers’ might promote cell–cell associations, leading to homotypic aggregation (Singethan et al., 2008). We suggest that microvilli formation could interfere with cell adhesion and spreading, thus accounting for CD9 inhibition of these processes. Therefore, because mutant CD9 did not form microvilli, it was less able to promote homotypic aggregation and to inhibit adhesion and spreading. Our CD9 C-terminal mutation severely hindered each of the four different CD9 functions tested, and these results were seen in four different cell lines (two adherent, two nonadherent). Hence, the C-terminal tail is also likely to be crucial for other CD9-dependent processes, including platelet functions (Goschnick and Jackson, 2007), tumor suppression (Cajot et al., 1997; Huang et al., 1998; Miyake et al., 1995), oocyte fertilization (Le Naour et al., 2000; Miyado et al., 2000), MHC protein organization (Kropshofer et al., 2002; Unternaehrer et al., 2007), cell–cell fusion (Tachibana and Hemler, 1999; Takeda et al., 2003), formation and/or maintenance of paranodal junctions (Ishibashi et al., 2004), HIV1-induced membrane fusion (Gordon-Alonso et al., 2006) and HIV1 cell–cell transmission (Krementsov et al., 2009).
Specific residues within the large extracellular loop of CD9 contribute to gamete fusion (Zhu et al., 2002) and to upregulation of diphtheria toxin binding (Hasuwa et al., 2001), and specific transmembrane residues contribute to CD9 homodimerization (Kovalenko et al., 2005). However, until now there was little, if any, evidence for the CD9 C-terminal tail being functionally important. Although a role for the CD9 C-terminal tail was not previously established, there is precedent for the C termini of other tetraspanins being functionally important. For example, the C-terminal tail of CD151 supports integrin-dependent adhesion strengthening, cell morphology (Lammerding et al., 2003; Zhang et al., 2002) and intracellular trafficking (Liu et al., 2007), and the C-terminal tail of CD63 regulates its internalization (Latysheva et al., 2006; Rous et al., 2002), together with a cell-surface partner protein (Duffield et al., 2003).
Biochemical consequences of CD9 C-terminal tail mutation
Tetraspanin CD63, with a C-terminal G-Y-E-V-M sequence, interacts with a PDZ domain in syntenin-1 (Latysheva et al., 2006), a transmembrane and cytoplasmic connector protein. However, CD9 does not directly interact with syntenin-1 (Latysheva et al., 2006), and the C-terminal R-N-R-E-M-V sequence of CD9 does not closely match any of 16 consensus PDZ-interaction motifs (Tonikian et al., 2008). The C-terminal tail of CD81, a tetraspanin similar to CD9, might interact directly with ERM proteins (Sala-Valdes et al., 2006). However, the C terminus of CD9 did not directly interact with ERM proteins (Sala-Valdes et al., 2006). To gain an unbiased assessment of CD9 protein associations affected by C-terminal tail mutation, we undertook a SILAC mass spectrometry approach, applying it to the study of multimolecular tetraspanin CD9 protein complexes. Although we mixed equal numbers of cells expressing equal amounts of wild-type CD9 (heavy-isotope labeled) and mutant CD9 (light-isotope labeled), we recovered 2.3–3.3-fold more wild-type CD9 and associated partner proteins from 1% Brij 96 lysates. Notably, CD9 and mutant CD9 were present in equal amounts both on the cell surface (as indicated by flow cytometry) and in the total cell lysate (as indicated by immunoblotting of Triton X-100 lysates). Furthermore, eight different background proteins, not known to be associated with CD9, were recovered at similar levels (H:L ratios=1.0–1.4). Hence, complexes containing mutant CD9 and six of its associated proteins (but not the eight background proteins) were selectively less available to the anti-CD9 mAb-coated beads used to retrieve CD9 complexes from 1% Brij 96 lysates. These results point to a selective decrease in Brij 96 solubility for complexes containing mutant CD9 from U937 cells. Reduced recovery of mutant CD9 from 1% Brij 96 lysates was seen in two additional cell lines (RD and MOLT-4), expressing CD9 and mutant CD9 at comparable levels on the cell surface and in Triton X-100 lysates.
The post-lysis addition of Triton X-100 to Brij 96 lysates restored CD9 and mutant CD9 to a state of equal recovery, thus establishing that mutant CD9 was not left in the Brij 96 insoluble pellet. Instead, mutant CD9 seems to be reorganized into larger complexes in Brij 96 lysates, with skewed mAb recognition profiles. This idea is reinforced by mutant CD9 in lysates showing selectively decreased recognition by anti-CD9 mAb such as MM2/57 and ALB6, and showing selectively enhanced recognition by mAb C9BB, which preferentially binds to oligomerized CD9 (Yang et al., 2006). Also, mutant CD9 seems to be larger than wild-type CD9, as seen by gel filtration of 1% Brij 96 lysates. Consistent with results seen in detergent lysates, mutant CD9 on the surface of intact cells again showed selectively enhanced mAb C9BB binding, indicative of increased homooligomerization. In a prior study, molecular alterations of tetraspanin CD81 complexes resulted in major size changes (as seen by gel filtration) and major differences in recognition by selected anti-CD81 antibodies, both on the cell surface and in detergent lysates, with the latter differences disappearing upon addition of Triton X-100 (Kolesnikova et al., 2004).
Regarding CD9 molecular organization, consistent results were seen in five different cell lines (two adherent, three nonadherent), each expressing CD9 and mutant CD9 at comparably moderate levels. Hence, conclusions seem to be generally applicable and not due to either excessive CD9 overexpression or specialized tetraspanin microdomain conditions only found in one or a few cell types.
We suggest that mutation of the CD9 tail leads to altered and/or unregulated assembly of CD9 complexes, with enhanced CD9 homooligomerization, such that transmembrane domain packing becomes more difficult to disrupt by the oleate sidechain of Brij 96. By contrast, wild-type CD9 probably engages in a specific (but not yet identified) interaction with the cytoskeleton, which leads to assembly of complexes that are less extensively organized in the lateral dimension and that seem more likely to contribute to assembly of microvilli.
CD9-associated proteins with high H:L ratios include CD82, TSPAN14, CD4, CTL1, CD224 and EWI-2. CD9 interactions with other tetraspanin proteins, including CD82 and TSPAN14, are well established (e.g. see Le Naour et al., 2006; Rubinstein et al., 1996). CD9 associations with CD4, CD224 (γ-glutamyl transferase) and CTL1 (a choline transporter) have also been previously noted (Le Naour et al., 2006; Toyo-Oka et al., 1999), but their functional implications are unknown. CD9 associates directly with EWI-2 (Charrin et al., 2003; Stipp et al., 2001a); EWI-2 might contribute to CD9-dependent inhibition of cell migration and glioblastoma growth (Kolesnikova et al., 2008). A problem with classical protein mass spectrometry assessment of protein complexes is that it is sometimes difficult to distinguish ‘real’ from adventitious protein associations. In our study, the six CD9 partner proteins identified with high H:L ratios are all therefore at least partly dependent on the CD9 C-terminal tail for assembly into the complex and thus might be validated as ‘real’ CD9 partners. By contrast, eight other proteins, with H:L ratios close to 1.0, associate independent of the C-terminal tail, consistent with them being background proteins.
The C-terminal tail of CD9 and other tetraspanins has several times been covalently tagged for the purpose of studying subcellular distribution and/or tetraspanin dynamics. Findings shown in this paper indicate that such results need to be evaluated cautiously. We show that even a relatively small change to the C-terminal tail can have a major effect on a wide range of functions, while also altering the cell-surface organization of a tetraspanin and nearly all, if not all, of its transmembrane partners.
Materials and Methods
Antibodies, plasmids and cell culture
mAbs to tetraspanin CD9 (MM2/57, ALB6, C9BB), integrin β1 (TS2/16) and EWI-2 (rabbit polyclonal antibody) were referenced elsewhere (Yang et al., 2006). MOLT-4, K562 and U937 cells [American Type Culture Collection (ATCC), Manassas, VA] were cultured in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). RD, HT1080 and ΦNX-ampho packaging cells (ATCC) were maintained in DMEM with 10% FBS plus antibiotics. In CD9 mutant plasmid, the last three amino acids of CD9 (EMV) were replaced by the last three amino acids of CD82 (PKY). CD9 and CD9 mutant were cloned in a C-terminal FLAG-tagged pLXIZ retroviral vector. CD9, CD9 mutant and pLXIZ were transiently transfected into ΦNX-ampho cells and retroviral infection of MOLT-4, K562, U937, RD and HT1080 cells was performed as previously described (Kolesnikova et al., 2004). Stable infectants were selected in medium containing 200 μg/ml zeocin (Invitrogen).
Cell adhesion, spreading and aggregation assays
For fibronectin coating, 12-well or 96-well plates were treated with 10 μg/ml human fibronectin (BD Biosciences, Bedford, MA) in PBS at 4°C overnight, and blocked with 1% BSA in PBS for 1 hour. For adhesion assays, K562 cells (1.0×105) were labeled with 5 μM Calcein AM (Vybrant Cell Adhesion Assay Kit, Invitrogen) for 30 minutes, then washed, resuspended in FBS-free medium and added to fibronectin-coated 12-well plates for 0.5 and 2 hours. Unattached cells were removed by two washes with culture medium. Fluorescence intensity was then measured using a Wallac 1420 workstation (PerkinElmer, Waltham, MA) to detect emission at 494 nm before and after washes. Adhesion percent is calculated as (adherent cell fluorescence – background)/(total cell fluorescence – background). The background is taken as fluorescence of cells without Calcein AM labeling.
For spreading assays, K562, MOLT-4 and RD cells (1.0×105) were plated on fibronectin for 0.5–6 hours, and then photographed. To quantify cell spreading, ‘deviation from round’ was calculated by dividing the theoretical maximum area for a given perimeter (perimeter2/4π) by the observed pixel area (Zhang et al., 2001) obtained using Scion Image software (Scion Corp., Frederick, MD). Cells with values >1.2 were judged to be spread (1.0=value for a perfectly round cell). At least 50 cells from three independent experiments were evaluated for each data point presented.
For cell aggregation assays, 12-well plates were either non-coated or coated with 10 μg/ml collagen I (Santa Cruz) or laminin-5 (produced from human A431 cells). MOLT-4 cells (1.0×105) were dispersed, washed in FBS-free medium, incubated in 12-well plates and then photographed after 2 hours. Cell images were acquired with a monochrome CCD camera (Spot RT; Diagnostic Instruments, Sterling Heights, MI) on a microscope (Axiovert 135; Carl Zeiss MicroImaging, Thornwood, NY). Cell shape was evaluated by the Scion Image Software.
Flow cytometry
MOLT-4, K562, RD, U937 and HT1080 cells (1.0×105) were stained with 1.0 μg specific mAb for 1 hour on ice, followed by 30 minutes with FITC-conjugated secondary antibody (Biosource, Camarillo, CA) and then analyzed using a FACS Calibur (Becton Dickinson, Bedford, MA). At least 10,000 cells were counted per sample. Graphs were processed using FlowJo software (Tree Star, Ashland, OR).
Immunofluorescence
Cells grown on glass cover slips were fixed with 4% paraformaldehyde at 4°C for 15 minutes and permeabilized by 0.1% Triton X-100 in PBS for 5 minutes, and then rinsed with phosphate-buffered saline (PBS). Cells were then blocked (1 hour) with 3% BSA in PBS (w/v), immunolabeled with CD9 primary antibody (1 hour), followed by AlexaFluor488-conjugated goat anti-mouse IgG for 1 hour (Invitrogen). For double staining of CD9 and F-actin, cells were stained with AlexaFluor594–phalloidin (30 minutes) before incubation with CD9 antibody. Cells were then mounted [ProLong Gold antifade mounting media containing DAPI (Invitrogen)] and imaged using a Leica SP5X laser scanning confocal microscope (Leica Microsystems, Chicago, IL). Microvilli greater than 5 μm were counted using Scion Image software and the cells from 10 random pictures were quantified.
Immunoprecipitation and immunoblotting
K562 and RD cells were washed twice with PBS and lysed in buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2) containing 1% Brij 96 or Triton X-100 (Sigma) and EDTA-free protease inhibitors. After 1 hour at 25°C, insoluble material was removed by centrifugation. For immunoprecipitation, lysates were precleared for 1 hour with protein G-sepharose beads (Pharmacia, Uppsala, Sweden). Specific antibodies (1–2 μg) were added to lysates of total protein (500 μg) and incubated at 4°C overnight. Antibody–protein complexes were captured using protein G-sepharose beads, washed three times with lysis buffer, and immune complexes were eluted in 2× Tris-glycine SDS sample buffer. For immunoblotting, samples were resolved by SDS-PAGE, transferred to nitrocellulose membrane (Whatman, Dassel, Germany), blocked with 5% nonfat milk blocking agent, incubated with specific antibody, followed by addition of peroxidase- or HRP-conjugated secondary antibody and chemiluminescence detection (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific, Rockford, IL). For multiple blottings, membranes were stripped with Restore Western Blot Stripping Buffer (Pierce). Band intensities were quantified using Quantity One software (Bio-Rad, Hercules, CA), with non-CD9 band intensities calculated relative to CD9=1.00.
SILAC
U937.7C2 cells lacking CD81 and CD9 (a gift from S. Levy, Department of Medicine, Stanford University, Stanford, CA) were stably transduced to express wild-type CD9 or mutant CD9. U937-CD9 cells were labeled with isotopically heavy leucine (U-13C6-leucine; Cambridge Isotope Labs, Andover, MA) using RPMI labeling medium lacking L-leucine. Mutant CD9 cells were grown in isotopically light L-leucine (Sigma). All cells were maintained in labeling medium [also containing 10% dialyzed fetal calf serum (Hyclone) and penicillin-streptomycin (Invitrogen)] for 10 days to ensure complete labeling. Equal numbers (2×109 cells each) of U937-CD9 (heavy leucine) and U937-CD9 mutant (light leucine) were then mixed, washed three times with PBS and lysed together in lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2) supplemented with 1% Triton X-100 (Roche Applied Science) or 1% Brij 96 (Sigma), and protease inhibitor mixture (Roche Applied Science) at 4°C for 1 hour. Lysates were centrifuged for 30 minutes at 20,000 gand supernatants were pre-cleared stepwise with protein A-sepharose, protein G-sepharose and two irrelevant antibodies covalently coupled to protein A-agarose. After each pre-clearing, the lysates were centrifuged for 5 minutes at 20,000 g. After final pre-clearing, the supernatants were mixed with CD9 antibody (ALB6) that had been covalently coupled to protein G-sepharose beads (using dimethylpilimidate; Sigma) and incubated for 16 hours at 4°C. Beads were then extensively washed with lysis buffer (100 ml), and proteins were eluted with 100 mM glycine, pH 2.5, and the eluted fractions were neutralized with 1.5 M Tris-HCl, pH 8.8. After polyacrylamide gel electrophoresis and silver staining, gel lanes (~2 cm long) were excised, proteins were in-gel digested with trypsin and then analyzed by LC-MS/MS techniques (Haas et al., 2006). Identified peptides were quantified based on the area-under-the-curve ratios for co-eluting heavy and light forms of each peptide using the Vista algorithm (Bakalarski et al., 2008). These ratios are given in Table 1.
Gel filtration
Intact U937-CD9 cells and U937-CD9 mutant cells were each cell-surface labeled with sulfo-NHS-LC biotin (Pierce, Rockford, IL) as previously described (Kolesnikova et al., 2004) and then cells (each at 1×107) were lysed in 1% Brij 96. The same cells were also labeled metabolically, by growing for 16 hours in [3H]-palmitate (NEN Bioscience, Boston, MA) and 10% dialyzed FBS, and then lysed in 1% Brij 96. Lysates were mixed (CD9 biotin + CD9 mutant [3H]-palmitate; CD9 mutant biotin + CD9 [3H]-palmitate) and then passed over a sepharose CL6B gel filtration column. CD9 was then immunoprecipitated from each fraction, using mAb ALB6. Eluted proteins were resolved by SDS-PAGE, transferred to PVDF membrane (BioRad Laboratories) and exposed to BioMax MS film (Kodak, Rochester, NY) at −80°C with an intensifying screen to detect [3H] labeling. Biotin-labeled CD9 samples were transferred to nitrocellulose membrane and immunoblotted with ExtrAvidin-HRP (Sigma). A phosphoimager machine (Storm 860, Molecular Dynamics), together with ImageQuant version 1.2 program, was used for densitometric analysis. The sum of signals from each column fraction was set to equal 100%.
Supplementary Material
Acknowledgments
We thank C. E. Bakalarski for his help with the Vista algorithm. This work was supported by a grant from the National Institutes of Health (GM38903) to M.E.H. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/16/2702/DC1
References
- Anton E. S., Hadjiargyrou M., Patterson P. H., Matthew W. D. (1995). CD9 plays a role in Schwann cell migration in vitro. J. Neurosci. 15, 584-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalarski C. E., Elias J. E., Villen J., Haas W., Gerber S. A., Everley P. A., Gygi S. P. (2008). The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756-4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro O., Yanez-Mo M., Sala-Valdes M., Gutierrez-Lopez M. D., Ovalle S., Higginbottom A., Monk P. N., Cabanas C., Sanchez-Madrid F. (2005). Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 105, 2852-2861 [DOI] [PubMed] [Google Scholar]
- Berditchevski F. (2001). Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114, 4143-4151 [DOI] [PubMed] [Google Scholar]
- Berditchevski F., Odintsova E. (1999). Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J. Cell Biol. 146, 477-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucheix C., Rubinstein E. (2001). Tetraspanins. Cell. Mol. Life Sci. 58, 1189-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajot J. F., Sordat I., Silvestre T., Sordat B. (1997). Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res. 57, 2593-2597 [PubMed] [Google Scholar]
- Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. (2001). The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276, 14329-14337 [DOI] [PubMed] [Google Scholar]
- Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., Rubinstein E. (2003). EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373, 409-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Yeung V. P., Tsitoura D., DeKruyff R. H., Umetsu D. T., Levy S. (2000). Allergen-induced airway hyperreactivity is diminished in CD81-deficient mice. J. Immunol. 165, 5054-5061 [DOI] [PubMed] [Google Scholar]
- Duffield A., Kamsteeg E. J., Brown A. N., Pagel P., Caplan M. J. (2003). The tetraspanin CD63 enhances the internalization of the H,K-ATPase beta-subunit. Proc. Natl. Acad. Sci. USA 100, 15560-15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenel C., Margeat E., Dosset P., Arduise C., Le Grimellec C., Royer C. A., Boucheix C., Rubinstein E., Milhiet P. E. (2008). Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 182, 765-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Alonso M., Yanez-Mo M., Barreiro O., Alvarez S., Munoz-Fernandez M. A., Valenzuela-Fernandez A., Sanchez-Madrid F. (2006). Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 177, 5129-5137 [DOI] [PubMed] [Google Scholar]
- Goschnick M. W., Jackson D. E. (2007). Tetraspanins-structural and signalling scaffolds that regulate platelet function. Mini Rev. Med. Chem. 7, 1248-1254 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Lopez M. D., Ovalle S., Yanez-Mo M., Sanchez-Sanchez N., Rubinstein E., Olmo N., Lizarbe M. A., Sanchez-Madrid F., Cabanas C. (2003). A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J. Biol. Chem. 278, 208-218 [DOI] [PubMed] [Google Scholar]
- Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villen J., Gygi S. P. (2006). Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326-1337 [DOI] [PubMed] [Google Scholar]
- Hasuwa H., Shishido Y., Yamazaki A., Kobayashi T., Yu X., Mekada E. (2001). CD9 amino acids critical for upregulation of diphtheria toxin binding. Biochem. Biophys. Res. Commun. 289, 782-790 [DOI] [PubMed] [Google Scholar]
- Hemler M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion and fusion events, and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397-422 [DOI] [PubMed] [Google Scholar]
- Hemler M. E. (2005). Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801-811 [DOI] [PubMed] [Google Scholar]
- Huang C. I., Kohno N., Ogawa E., Adachi M., Taki T., Miyake M. (1998). Correlation of reduction in MRP-1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am. J. Pathol. 153, 973-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. L., Liu D., Masuya D., Kameyama K., Nakashima T., Yokomise H., Ueno M., Miyake M. (2004). MRP-1/CD9 gene transduction downregulates Wnt signal pathways. Oncogene 23, 7475-7483 [DOI] [PubMed] [Google Scholar]
- Ikeyama S., Koyama M., Yamaoko M., Sasada R., Miyake M. (1993). Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J. Exp. Med. 177, 1231-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T., Ding L., Ikenaka K., Inoue Y., Miyado K., Mekada E., Baba H. (2004). Tetraspanin protein CD9 is a novel paranodal component regulating paranodal junctional formation. J. Neurosci. 24, 96-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto R., Higashiyama S., Mitamura T., Taniguchi N., Klagsbrun M., Mekada E. (1994). Heparin-binding EGF-like growth factor, which acts as a diphtheria toxin receptor, forms a complex with membrane protein DRAP27/CD9, which upregulates functional receptors and diphtheria toxin sensitivity. EMBO J. 13, 2322-2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Oda S., Shikano T., Ohnuki T., Uematsu Y., Sakagami J., Tada N., Miyazaki S., Kudo A. (2000). The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 24, 279-282 [DOI] [PubMed] [Google Scholar]
- Kaji K., Takeshita S., Miyake K., Takai T., Kudo A. (2001). Functional association of CD9 with the Fc gamma receptors in macrophages. J. Immunol. 166, 3256-3265 [DOI] [PubMed] [Google Scholar]
- Kolesnikova T. V., Stipp C. S., Rao R. M., Lane W. S., Luscinskas F. W., Hemler M. E. (2004). EWI-2 modulates lymphocyte integrin alpha4beta1 functions. Blood 103, 3013-3019 [DOI] [PubMed] [Google Scholar]
- Kolesnikova T. V., Kazarov A. R., Lemieux M., Lafleur M. A., Kesari S., Kung A. L., Hemler M. E. (2008). Glioblastoma suppression by tetraspanin partner protein EWI-2 in vitro and in vivo. Neoplasia 11, 77-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko O. V., Metcalf D. G., DeGrado W. F., Hemler M. E. (2005). Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct. Biol. 5, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsov D. N., Weng J., Lambele M., Roy N. H., Thali M. (2009). Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology 6, 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropshofer H., Spindeldreher S., Rohn T. A., Platania N., Grygar C., Daniel N., Wolpl A., Langen H., Horejsi V., Vogt A. B. (2002). Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 3, 61-68 [DOI] [PubMed] [Google Scholar]
- Lammerding J., Kazarov A. R., Huang H., Lee R. T., Hemler M. E. (2003). Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. USA 100, 7616-7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latysheva N., Muratov G., Rajesh S., Padgett M., Hotchin N. A., Overduin M., Berditchevski F. (2006). Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 26, 7707-7718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F., Rubinstein E., Jasmin C., Prenant M., Boucheix C. (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319-321 [DOI] [PubMed] [Google Scholar]
- Le Naour F., Andre M., Greco C., Billard M., Sordat B., Emile J. F., Lanza F., Boucheix C., Rubinstein E. (2006). Profiling of the tetraspanin web of human colon cancer cells. Mol. Cell. Proteomics 5, 845-857 [DOI] [PubMed] [Google Scholar]
- Letarte M., Seehafer J. G., Greaves A., Masellis-Smith A., Shaw A. R. (1993). Homotypic aggregation of pre-B leukemic cell lines by antibodies to VLA integrins correlates with their expression of CD9. Leukemia 7, 93-103 [PubMed] [Google Scholar]
- Levy S., Shoham T. (2005). The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136-148 [DOI] [PubMed] [Google Scholar]
- Liu L., He B., Liu W. M., Zhou D., Cox J. V., Zhang X. A. (2007). Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 282, 31631-31642 [DOI] [PubMed] [Google Scholar]
- Masellis-Smith A., Shaw A. R. E. (1994). CD9-regulated adhesion: anti-CD9 monoclonal antibody induce pre-B cell adhesion to bone marrow fibroblasts through de novo recognition of fibronectin. J. Immunol. 152, 2768-2777 [PubMed] [Google Scholar]
- Masellis-Smith A., Jensen G. S., Seehafer J. G., Slupsky J. R., Shaw A. R. E. (1990). CD9-regulated adhesion: anti-CD9 monoclonal antibodies induce homotypic adhesion of pre-B cell lines by a novel mechanism. J. Immunol. 144, 1607-1613 [PubMed] [Google Scholar]
- Miranti C. K. (2009). Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell. Signal. 21, 196-211 [DOI] [PubMed] [Google Scholar]
- Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., et al. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321-324 [DOI] [PubMed] [Google Scholar]
- Miyake M., Nakano K., Ieki Y., Adachi M., Huang C.-L., Itoi S., Koh T., Taki T. (1995). Motility related protein 1 (MRP-1/CD9) expression: inverse correlation with metastases in breast cancer. Cancer Res. 55, 4127-4131 [PubMed] [Google Scholar]
- Murayama Y., Miyagawa J., Oritani K., Yoshida H., Yamamoto K., Kishida O., Miyazaki T., Tsutsui S., Kiyohara T., Miyazaki Y., et al. (2004). CD9-mediated activation of the p46 Shc isoform leads to apoptosis in cancer cells. J. Cell Sci. 117, 3379-3388 [DOI] [PubMed] [Google Scholar]
- Nydegger S., Khurana S., Krementsov D. N., Foti M., Thali M. (2006). Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173, 795-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376-386 [DOI] [PubMed] [Google Scholar]
- Ono M., Handa K., Withers D. A., Hakomori S. (1999). Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res. 59, 2335-2339 [PubMed] [Google Scholar]
- Rous B. A., Reaves B. J., Ihrke G., Briggs J. A., Gray S. R., Stephens D. J., Banting G., Luzio J. P. (2002). Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 13, 1071-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E., Le Naour F., Lagaudrière-Gesbert C., Billard M., Conjeaud H., Boucheix C. (1996). CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA antigens. Eur. J. Immunol. 26, 2657-2665 [DOI] [PubMed] [Google Scholar]
- Runge K. E., Evans J. E., He Z. Y., Gupta S., McDonald K. L., Stahlberg H., Primakoff P., Myles D. G. (2007). Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev. Biol. 304, 317-325 [DOI] [PubMed] [Google Scholar]
- Ryu F., Takahashi T., Nakamura K., Takahashi Y., Kobayashi T., Shida S., Kameyama T., Mekada E. (2000). Domain analysis of the tetraspanins: studies of CD9/CD63 chimeric molecules on subcellular localization and upregulation activity for diphtheria toxin binding. Cell Struct. Funct. 25, 317-327 [DOI] [PubMed] [Google Scholar]
- Sala-Valdes M., Ursa A., Charrin S., Rubinstein E., Hemler M. E., Sanchez-Madrid F., Yanez-Mo M. (2006). EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 281, 19665-19675 [DOI] [PubMed] [Google Scholar]
- Seigneuret M., Delaguillaumie A., Lagaudriere-Gesbert C., Conjeaud H. (2001). Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276, 40055-40064 [DOI] [PubMed] [Google Scholar]
- Shaw A. R. E., Domanska A., Mak A., Gilchrist A., Dobler K., Visser L., Poppema S., Fliegel L., Letarte M., Willett B. J. (1995). Ectopic expression of human and feline CD9 in a human B cell line confers β1 integrin-dependent motility on fibronectin and laminin substrates and enhanced tyrosine phosphorylation. J. Biol. Chem. 270, 24092-24099 [DOI] [PubMed] [Google Scholar]
- Shi W., Fan H., Shum L., Derynck R. (2000). The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced EGF receptor activation and cell proliferation. J. Cell Biol. 148, 591-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singethan K., Muller N., Schubert S., Luttge D., Krementsov D. N., Khurana S. R., Krohne G., Schneider-Schaulies S., Thali M., Schneider-Schaulies J. (2008). CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic 9, 924-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. (2001a). EWI-2 is a major CD9 and CD81 partner, and member of a novel Ig protein subfamily. J. Biol. Chem. 276, 40545-40554 [DOI] [PubMed] [Google Scholar]
- Stipp C. S., Orlicky D., Hemler M. E. (2001b). FPRP: a major, highly stoichiometric, highly specific CD81 and CD9-associated protein. J. Biol. Chem. 276, 4853-4862 [DOI] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003). EWI-2 regulates {alpha}3{beta}1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana I., Hemler M. E. (1999). Role of transmembrane-4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J. Cell Biol. 146, 893-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Tachibana I., Miyado K., Kobayashi M., Miyazaki T., Funakoshi T., Kimura H., Yamane H., Saito Y., Goto H., et al. (2003). Tetraspanins CD9 and CD81 function to prevent the fusion of mononuclear phagocytes. J. Cell Biol. 161, 945-956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., et al. (2008). A specificity map for the PDZ domain family. PLoS Biol. 6, e239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyo-Oka K., Yashiro-Ohtani Y., Park C. S., Tai X. G., Miyake K., Hamaoka T., Fujiwara H. (1999). Association of a tetraspanin CD9 with CD5 on the T cell surface: role of particular transmembrane domains in the association. Int. Immunol. 11, 2043-2052 [DOI] [PubMed] [Google Scholar]
- Unternaehrer J. J., Chow A., Pypaert M., Inaba K., Mellman I. (2007). The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc. Natl. Acad. Sci. USA 104, 234-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M., Barreiro O., Gordon-Alonso M., Sala-Valdes M., Sanchez-Madrid F. (2009). Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434-446 [DOI] [PubMed] [Google Scholar]
- Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. (2002). Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell 13, 767-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Kovalenko O. V., Kolesnikova T. V., Andzelm M. M., Rubinstein E., Strominger J. L., Hemler M. E. (2006). Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J. Biol. Chem. 281, 12976-12985 [DOI] [PubMed] [Google Scholar]
- Zhang X. A., Bontrager A. L., Stipp C. S., Kraeft S.-K., Bazzoni G., Chen L. B., Hemler M. E. (2001). Phosphorylation of a conserved integrin α3 chain QPSXXE motifs regulates signaling, motility, and cytoskeletal engagement. Mol. Biol. Cell 12, 351-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. A., Kazarov A. R., Yang X., Bontrager A. L., Stipp C. S., Hemler M. E. (2002). Function of the tetraspanin CD151-a6b1 integrin complex during cellular morphogenesis. Mol. Biol. Cell 13, 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G. Z., Miller B. J., Boucheix C., Rubinstein E., Liu C. C., Hynes R. O., Myles D. G., Primakoff P. (2002). Residues SFQ (173-175) in the large extracellular loop of CD9 are required for gamete fusion. Development 129, 1995-2002 [DOI] [PubMed] [Google Scholar]
- Ziyyat A., Rubinstein E., Monier-Gavelle F., Barraud V., Kulski O., Prenant M., Boucheix C., Bomsel M., Wolf J. P. (2006). CD9 controls the formation of clusters that contain tetraspanins and the integrin {alpha}6{beta}1, which are involved in human and mouse gamete fusion. J. Cell Sci. 119, 416-424 [DOI] [PubMed] [Google Scholar]
- Zoller M. (2009). Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40-55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.