Abstract
How HIV controllers (HICs) maintain undetectable viremia without therapy is unknown. The strong CD8+ T-cell HIV suppressive capacity found in many, but not all, HICs may contribute to long-lasting viral control. However, other earlier defense mechanisms may be involved. Here, we examined intrinsic HIC cell resistance to HIV-1 infection. After in vitro challenge, monocyte-derived macrophages and anti–CD3-activated CD4+ T cells from HICs showed low HIV-1 susceptibility. CD4 T-cell resistance was independent of HIV-1 coreceptors and affected also SIVmac infection. CD4+ T cells from HICs expressed ex vivo higher levels of p21Waf1/Cip1, which has been involved in the control of HIV-1 replication, than cells from control subjects. However, HIV restriction in anti–CD3-activated CD4+ T cells and macrophages was not associated with p21 expression. Restriction inhibited accumulation of reverse transcripts, leading to reduction of HIV-1 integrated proviruses. The block could be overcome by high viral inocula, suggesting the action of a saturable mechanism. Importantly, cell-associated HIV-1 DNA load was extremely low in HICs and correlated with CD4+ T-cell permissiveness to infection. These results point to a contribution of intrinsic cell resistance to the control of infection and the containment of viral reservoir in HICs.
Introduction
So-called HIV controllers (HICs), who represent ∼ 0.25% of HIV-1–infected patients in France, have persistently undetectable viremia (< 50 HIV RNA copies/mL) without antiretroviral therapy.1,2 Plasma viral load in HICs is similar to that observed during effective highly active antiretrovial therapy (HAART), whereas their total HIV DNA load in mononuclear blood cells is lower and stable over time.1,3,4 In many HICs, HIV-1–specific CD8+ T cells are able to suppress HIV-1 replication ex vivo through efficient granzyme B– and perforin-mediated killing of infected CD4+ T cells.5–7 In contrast, other HICs do not have high frequencies of HIV-specific CD8+ T cells, and their CD8 T cells do not strongly suppress HIV,8–10 pointing to the existence of other mechanisms of viral control in these subjects.
Recent studies of HICs indicate that spontaneous viral control occurs during the first year after infection,11 and low plasma viral load has been found during the acute phase of HIV infection in patients who are subsequently identified as HICs.12 This suggests that the mechanisms of viral control come into play rapidly after infection. These mechanisms may include intrinsic restrictions that hamper HIV-1 replication in target cells, as shown in persons who are repeatedly exposed to HIV but remain uninfected.13,14 In particular, mechanisms that block HIV-1 integration in target cells may contribute to limiting the size of the viral reservoirs and to controlling the pathogenic effects of HIV.
Host genes influence the susceptibility of cells to HIV-1 infection. Whereas some factors are necessary to promote infection,13 in the past 10 years it has been shown that mammalian cells have devised an intrinsic immunity to block retroviral replication. Restriction factors reported so far act at different steps of viral replication, including uncoating (Trim5α), reverse transcription (Apobec3G), or viral release (Tetherin). However, HIV-1 has developed effective strategies to counteract these factors in human cells.15 Other factors that are not specific antiviral molecules may also inhibit viral replication. This is the case of p21Cip1/Waf1, a cyclin-dependent kinase inhibitor involved in regulating cell cycle and apoptosis,16,17 which we and others have shown to be able to block HIV replication in human macrophages and hematopoietic stem cells after viral entry and before genomic integration.18,19
We therefore investigated the susceptibility of CD4+ T cells and macrophages from HICs to HIV-1 infection in vitro. To our knowledge there are no data on the susceptibility of HIC macrophages to HIV infection, whereas several teams, including our own, have found that activated CD4+ T cells from HICs are as susceptible to HIV-1 infection as cells from healthy donors.4,7,20 However, the conditions used in these CD4 T-cell experiments (mitogen activation or large viral inocula or both) might have overwhelmed mechanisms capable of inhibiting viral replication in vivo.21–23
Here, we examined HIC CD4+ T-cell susceptibility to HIV-1 infection in various experimental conditions, as well as that of monocyte-derived macrophages (MDMs). Viral replication was lower in both cell types from HICs compared with controls. Although p21Cip1/Waf1 (p21) expression was higher in HIC CD4+ T cells ex vivo than in cells from control groups, p21 did not appear to be directly responsible of HIV restriction in cells from HICs. CD4 T-cell permissiveness to HIV replication could be recovered by parameters enhancing viral infection and spreading, suggesting the implication of a saturable mechanism. Importantly, the level of cell-associated HIV DNA in HICs was inversely associated with the resistance of their CD4+ T cells to HIV infection in vitro. These findings point to a contribution of intrinsic cell resistance to the persistent viral control observed in HICs.
Methods
Subjects
HICs enrolled in the ANRS CO18 cohort are defined as patients infected by HIV-1 for ≥ 5 years who have never received antiretroviral treatment and whose last 5 consecutive plasma HIV RNA values are < 400 copies/mL. Frozen samples from 81 HICs enrolled in the cohort were used to determine total cell-associated HIV DNA levels with the use of an ultrasensitive method. Fifty-five HICs were enrolled in this study, and all agreed to provide a fresh blood sample of 30-50 mL. The patients are described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Blood samples from 50 healthy HIV-seronegative donors (HDs) were obtained from the French blood bank (Etablissement Français du Sang) as part of the Etablissement Français du Sang–Institut Pasteur Convention. Nine HIV-infected patients not on antiretroviral therapy and with HIV plasma viral load > 7500 RNA copies/mL (VIR) and 12 HAART-treated patients with viral load < 50 RNA copies/mL for ≥ 6 months (ART) were recruited among patients monitored at Hôpital Saint Louis and Hôpital Européen Georges Pompidou, Paris, France. The main characteristics of the 3 groups of HIV-1–infected patients are summarized in Table 1.
Table 1.
Characteristics of the groups of HIV-1–infected patients included in the study
| Group | No. | Sex | Last CD4, median (range)* | Last viral load, median (range)† | Months on HAART, median (range)‡ |
|---|---|---|---|---|---|
| HIV controllers | 55 | M:31 F:24 | 680 (266-1665) | < 40 (< 40-366) | — |
| HIV-1 viremics | 9 | M:6 F:3 | 378 (278-654) | 42984 (8048-116 266) | — |
| HAART treated | 12 | M:10 F:2 | 633 (292-886) | < 40 (< 40-51) | 45 (7-98) |
Last CD4+ T-cell count/mm3 of blood.
HIV-1 RNA copies/mL of plasma with standard technique.
Months with successful HAART-related control of viremia before inclusion.
All the subjects gave their written informed consent in accordance with the Declaration of Helsinki, and the study was approved by the Ethics Review Committee (Comité de protection des personnes) Ile de France VII.
HIV DNA quantification
Total DNA was extracted from whole blood with QIAamp DNA minikits (QIAGEN), according to the manufacturer's instructions. HIV-1 DNA was then quantified by ultrasensitive real-time PCR (long terminal repeat [LTR] amplification, ANRS, detection limit 10 copies/million leukocytes). Each extract was tested in 4 PCRs, each using 1 μg of total DNA.24
Primary cell culture
CD4+ and CD14+ cells were purified (> 90%) from freshly isolated PBMCs by positive selection with antibody-coated magnetic beads in a Robosep instrument (Stem Cell Technology). Monocytes were differentiated into macrophages as described elsewhere.25 Briefly, purified CD14+ monocytes were cultured for 7-10 days in hydrophobic Teflon dishes (Lumox; D Dutscher) in macrophage medium (RPMI 1640 supplemented with 200mM l-glutamine, 10 IU/mL penicillin, 10 μg/mL streptomycin, 10mM HEPES, 10mM sodium pyruvate, 50μM β-mercaptoethanol, 1% minimum essential medium, vitamins, 1% nonessential amino acids) supplemented with 15% human AB serum. For experiments, macrophages were harvested and resuspended in macrophage medium containing 10% heat-inactivated FCS. Because of limiting amounts of MDM obtained from available blood samples, some experiments, such as p21 RNAi silencing, could be performed only in CD4 T cells.
Productive HIV-1 infection in vitro in optimal conditions
Purified CD4+ cells were stimulated for 3 days with PHA at 1 μg/mL in the presence of IL-2 (Chiron) at 100 IU/mL. The culture medium was RPMI 1640 containing 10% FCS, penicillin (10 IU/mL) and streptomycin (10 μg/mL). CD4+ T cells (106 cells/mL) were superinfected with HIV-1 BaL (R5) in triplicate at a MOI of 10−3.25 in 96-U-well plates with a spinoculation protocol.26 After challenge, the cells were washed and cultured in 96-U-well plates (106 cells/mL in triplicate) for 14 days. Every 3-4 days the supernatant was recovered and replaced with fresh culture medium containing IL-2 (100 U/mL). Viral replication was monitored by measuring p24 in the supernatant with an ELISA method (Zeptometrix).
Productive infection in vitro in suboptimal conditions
Purified CD4+ cells were stimulated for 5 days with an anti-CD3 monoclonal antibody (0.5 μg/mL; X35 clone; Beckman Coulter) and IL-2 (Chiron) at 50 IU/mL in RPMI 1640 medium supplemented with 10% FCS, penicillin (10 IU/mL), and streptomycin (10 μg/mL). The activated CD4+ T cells were then incubated with HIV-1 BaL (R5), NL 4.3 (X4), or SIVmac251 (10−3.8, 10−3, and 10−3.8 MOI, respectively) for 4 hours at 37°C. The cells were then washed twice and cultured in flat-bottom 48-well plates (2.5 × 105 cells/mL in triplicate) for 14 days. Every 3-4 days culture supernatants were recovered and replenished with fresh culture medium containing IL-2 (50 IU/mL). Viral replication was monitored in supernatants by p24 or p27 (SIVmac251) ELISA as described earlier. For experiments of saturation at high viral inoculum, serial dilutions of HIV-1 BaL concentrated by ultracentrifugation (1017.5 TCID50/mL) were used. All viral titers were calculated on PHA-activated CD4+ T cells with the use of optimal conditions of infection.
Single-round infection
Vesicular stomatitis virus glycoprotein (VSV-G)–pseudotyped HIV-1 particles were produced by cotransfecting (SuperFect; QIAGEN) 293T cells with the proviral pNL-Luc-E-R+27 and the pMD2 VSV-G expression vector. CD4+ T cells and macrophages were challenged in triplicate with HIV-1–pseudotyped particles (8.3 ng of p24/mL) in the suboptimal conditions described earlier. Luciferase activity was determined in cell lysates (Luciferase Reporter 1000 Assay System; Promega) with a Glomax microplate luminometer (Turner BioSystems; Promega) 48 hours after infection, as described.28
PCR quantification of HIV-1 cDNA forms
Twenty-four and 48 hours after challenge with VSV-G–pseudotyped HIV-1 particles the cells were washed in PBS, and total DNA was extracted with the DNeasy Tissue kit (QIAGEN). Early and late reverse transcripts and integrated HIV-1 DNA were quantified by real-time R-U5 PCR, U5-Gag PCR, and Alu-Gag–nested PCR, respectively, on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) as described elsewhere.25 DNA loading was controlled by concurrently amplifying the albumin gene by real-time PCR and quantifying with reference to a control human genomic DNA (Roche). HIV-1 DNA copies from experiments conducted in parallel in the presence of Nevirapine 6.25μM (Boehringer Ingelheim) were subtracted to correct for DNA carryover from viral inoculum. HIC cells were studied in parallel, without in vitro challenge, to take into account the possible contribution of already-integrated HIV-1.
Activation phenotyping
The following antibodies were used: CD3-FITC (clone UCHT1), CD4-ECD (SFCI12T4D11), from Beckman Coulter, and CD25-V450 (M-A251) and CD69-PE(FN50) from BD Biosciences.
CD4+ T cells were incubated with the antibodies for 15 minutes then washed in PBS plus 1% FCS and fixed in 1% paraformaldehyde for flow cytometry on an LSRII device (BD Bioscience). The data were analyzed with FlowJo software (TreeStar Inc).
Quantitative RT-PCR analysis
Total RNA was extracted from macrophages and CD4+ T lymphocytes with the RNeasy kit (QIAGEN) and treated with DNase, following the manufacturer's instructions. RNA was quantified with the GeneQuant method (Amersham) and reverse transcribed with SuperScript II reverse transcriptase (Invitrogen). The amplification program consisted of 10 minutes at 25°C, 50 minutes at 42°C, and 15 minutes at 70°C. PCR amplification of cDNA was performed in duplicate in MicroAmp Optical 96-well reaction plates (30 μL/well), using 15 μL of TaqMan Universal Master Mix, 0.2mM TaqMan, and 1.5 μL of Assays-on-Demand Gene Expression Assay premade mix (GAPDH, Hs99999905_m1; p21, Hs00355782_m1; APOBEC3G, Hs00222415_m1; TRIM5α, Hs01552559_m1; Applied Biosystems). The amplification conditions were as follows: 50°C for 2 minutes and 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The amount of target mRNA in each sample was normalized to GAPDH mRNA as an endogenous reference, and the data were analyzed with the cycle threshold (Ct) method.29 All results were expressed relative to a control cDNA, obtained from CD4+ T cells of a healthy donor, as 2−ΔΔCt, where ΔΔCt = ΔCtSAMPLE − ΔCtCONTROL, and ΔCt = CtTARGET GENE − CtGAPDH.
Western blot analysis
Activated CD4+ T lymphocytes cultured in 24-well plates were lysed in 80 μL of M-PER lysis buffer (Pierce) containing the Complete Protease and Phosphatase Inhibitor Cocktail (Roche). Protein was quantified with the BCA kit (Pierce), and samples were then diluted with Laemmli buffer, boiled for 5 minutes, and loaded (30 μg) in NuPAGE gel 4%-12% (Invitrogen) for electrophoretic separation. Proteins were then blotted onto Hybond-P membranes (Amersham). After blocking with 5% skimmed milk, the membranes were incubated with the primary antibodies as indicated, followed by a secondary HPR-conjugated anti–mouse antibody (R&D Systems). Proteins were shown on Hyperfilms (Amersham) with the use of the ECL chemiluminescent substrate (GE Healthcare) and X-Omat film (Kodak). The anti-p21 mouse monoclonal antibody (1:1000) was from Upstate, and anti-GAPDH (1:5000) was from Abcam.
Small interfering RNA transfection
Small interfering RNA (siRNA) duplexes no. 9 and no. 12 from the SMARTpool for p21 were purchased from Dharmacon, and irrelevant negative control siRNA (5′-CTGCATCGTGCACAGGAGTATCA-3′) was synthesized by QIAGEN. siRNA transfection was performed with the AMAXA human T-cell Nucleofector kit (LONZA), following the manufacturer's instructions. Briefly, 5 days after activation with anti-CD3, 2.5-4 × 106 CD4+ T cells were centrifuged and resuspended in 100 μL of Nucleofector solution. SiRNA was added at 200 pmoles per million cells. The Nucleofector program T-023 was used. The cells were then transferred into 2 mL of culture medium without antibiotics in 12-well plates and incubated at 37°C for 24 hours before infectious challenge. Cell lysates were assayed for mRNA by qualitative RT-PCR to determine the efficiency of gene knockdown at the moment of infection.
Statistical analyses
P values were calculated with the rank sum test. Correlations were identified with simple linear regression analysis and Spearman's rank correlation test. SigmaStat 3.5 software was used (Systat Software Inc).
Results
Macrophages and CD4+ T cells from HICs have low susceptibility to HIV-1 infection
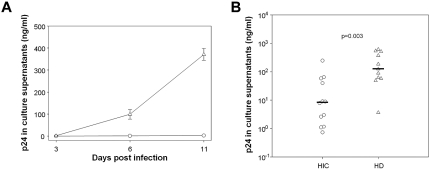
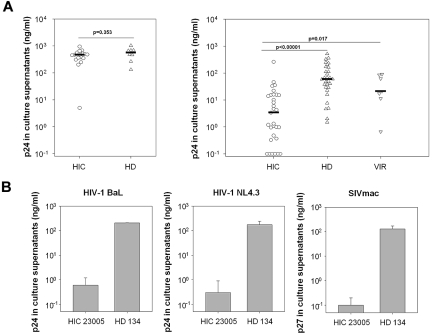
Intrinsic resistance of target cells to HIV-1 infection may contribute to control of infection and, in particular, to the small size of the viral reservoir in HICs. Macrophages are one of the main HIV-1 cell targets, and they play a crucial role in the spread and pathogenesis of HIV-1 infection. However, HIC macrophage susceptibility to HIV-1 infection has not previously been studied. We challenged MDMs from 12 HICs and 11 HDs with HIV-1 BaL. Viral replication was lower in HIC MDMs than in HD MDMs on day 3 after challenge and remained lower thereafter (Figure 1A). Overall, viral replication was far lower in HIC MDMs (median, 8.97 ng p24/mL) than in HD MDMs (median, 125 ng p24/mL; P = .003; Figure 1B). The reduced susceptibility of macrophages to HIV-1 infection prompted us to wonder whether restriction to infection extended to other cell targets in HICs. Previous work, including our own, has suggested that CD4+ T cells from HICs and HDs are similarly susceptible to HIV-1 infection in vitro.4,7,20 However, it is conceivable that the optimized viral challenge conditions used in these studies (potent stimuli or large viral inocula or both) masked restrictive mechanisms operating in vivo.21–23 We therefore reassessed the susceptibility of CD4+ T cells from 31 HICs to HIV-1 superinfection, using CD3 cross-linking (a more physiologic stimulus than PHA) and suboptimal conditions of cell activation and challenge.30 CD4+ T cells from HICs and HDs showed similar susceptibility to HIV-1 in the conditions of activation/challenge used in our previous reports (median, 469.1 and 568.2 ng p24/mL of culture supernatant, respectively; P = .35; Figure 2A left). In suboptimal conditions, however, we noted clear differences in susceptibility between CD4+ T cells from HICs (median, 3.43 ng p24/mL) and HDs (median, 59.3 ng p24/mL; Figure 2A right). HIV-1 also replicated more efficiently in superinfected CD4+ T cells from HIV viremic patients (median, 21.3 ng p24/mL), although replication of endogenous virus may have been a confounding factor. Interestingly, this reduced susceptibility of CD4+ T cells from HICs was observed with both R5-tropic and X4-tropic viruses (see Figure 2B for a representative example and supplemental Figure 1A for the summary of the experiments done with X4 HIV-1 and cells from 11 HICs and 7 HDs). Moreover, CD4+ T cells from HICs showed a remarkable broad resistance to lentiviral infection because we found that they were also less susceptible to infection with SIVmac251 than CD4+ T cells from HDs (Figure 2B shows a representative example and supplemental Figure 1B the summary of the experiments with cells from 8 HICs and 5 HDs). Overall, our results show that both macrophages and CD4+ T cells from HICs have a marked reduced permissiveness to HIV replication in vitro.
Figure 1.
Macrophages from HICs have low susceptibility to HIV-1 infection. (A) Kinetics of HIV-1 BaL replication after infection of MDMs from one representative HD (triangles) and one HIC (circles). Means ± SDs of 3 independent infections are shown. (B) HIV-1 replication on day 10-11 after infection in MDMs from HICs and HDs, after challenge with HIV-1 BaL. Symbols represent the average (n = 3 independent infections) p24 values detected in culture supernatants for each donor. Horizontal lines indicate median values.
Figure 2.
CD4+ T cells from HICs have low susceptibility to HIV-1 infection. (A) PHA-activated CD4+ T cells from HICs and HDs were challenged with replicative HIV-1 BaL, using a protocol optimized to detect viral replication in vitro (left). HIV-1 replication in anti–CD3-activated CD4+ T cells from HICs and HDs (including all those depicted in the left panel) and HIV-1 viremic patients (VIR), after challenge with HIV-1 BaL, using a suboptimal in vitro infection protocol (right). Symbols represent the average (n = 3 independent infections) p24 values detected on day 7 after infection in culture supernatants for each donor. Horizontal lines indicate median values. (B) HIV-1 (BaL [R5], left), NL4.3 ([X4], center), and SIVmac251 (right) replication after infection, in suboptimal conditions, of CD4+ T cells from one representative HD and one HIC whose cells were resistant to infection. Means ± SDs of 3 independent infections are shown.
Reduced HIV-1 susceptibility of cells from HICs is because of restriction occurring during reverse transcription
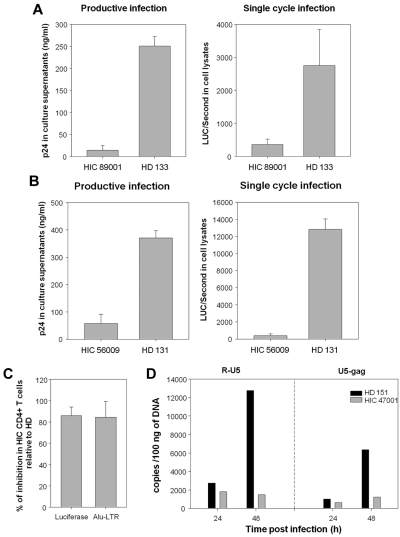
We performed one-round infection experiments with HIV-1 NL4.3Δenv particles carrying the luciferase reporter gene and pseudotyped with the pantropic VSV-G protein. CD4+ T cells from HICs were also less susceptible than cells form HDs to single-cycle HIV-1 replication. A representative example of experiments with cells from 13 HICs and 11 HDs is shown in Figure 3A right. Productive and single-cycle HIV-1 infections were assessed in parallel in CD4+ T cells from 8 HICs and as many HDs. With one exception, the relative levels of viral replication in HIC CD4+ T cells infected with HIV-1 replicative virus and with pseudotyped particles were correlated (Spearman, 0.893; P < .000 01; n = 7 HICs and 7 HDs; Figure 3A for an example). As observed with CD4+ T cells, the HIV restriction in MDMs was already operating at the first cycle of viral replication (Figure 3B for a representative example). These results ruled out the involvement of HIV coreceptors in viral restriction and pointed to a role of factors affecting early steps of the replicative cycle.
Figure 3.
The lower susceptibility of cells from HICs is apparent during the first cycle of infection and related to blockade of HIV-1 during reverse transcription. (A) Comparison of HIV-1 BaL replication on day 7 after infection (left) and single-cycle infection with HIV-1VSV-G, measured 48 hours after infection in terms of luciferase activity (right) in CD4+ T cells from a representative HIC and a HD. Suboptimal conditions were used for infection. Means ± SDs of 3 independent measurements are shown. (B) HIV replication in MDMs from a HIC and a HD on day 10 after infection with productive HIV-1 BaL (left) and 48 hours after infection with single-cycle VSV-G pseudotyped HIV-1 (right). Two additional experiments were performed with MDMs from 2 other HICs and HDs with similar results. Means ± SDs of 3 independent measurements are shown. (C) Levels of luciferase activity and integrated HIV-1 copies (Alu-LTR) in CD4+ T cells from HICs and HDs 48 hours after infection. Means ± SDs of experiments with cells from 7 HICs and 7 HDs are shown. (D) Number of early (R-U5) and late (U5-gag) reverse transcripts determined by qPCR 24 and 48 hours after infection of CD4+ T cells from one HIC ( ) and one HD (■) with VSV-G pseudotyped HIV-1 particles. One representative example of experiments with cells from 5 HICs and 5 HDs is shown.
) and one HD (■) with VSV-G pseudotyped HIV-1 particles. One representative example of experiments with cells from 5 HICs and 5 HDs is shown.
We analyzed whether the block-affected replication before or after viral integration by quantifying at 48 hours after infection the number of integrated provirus (Alu-LTR qPCR) in CD4+ T cells from 7 HICs and 7 HDs. The number of integrated proviruses in cells from HICs was very low compared with HD cells, and this reduction corresponded to the reduction in the level of viral replication measured by luciferase activity (Figure 3C), pointing to a restriction in preintegrative steps of viral replication. To find out if the block occurred during reverse transcription, we quantified early (R-U5) and late (U5-gag) reverse transcripts at 24 and 48 hours after infection in the cells from 5 HICs and 5HDs. One representative example of these experiments is shown in Figure 3D. We found that the kinetics of viral replication were slower in the CD4+ T cells from HDs when infected in suboptimal conditions than what we have previously reported with PHA stimulation and optimal conditions of infection.14 Whereas a sharp accumulation of early transcripts, and proportionate increase in late reverse transcripts, occurs between 24 and 48 hours in the cells from HDs, the reverse transcripts did not accumulate in the cells from HICs. Thus, our results indicate that the block of HIV-1 replication in the cells from HICs occurs during reverse transcription.
CD4+ T cells from HICs express high levels of p21Cip1/Waf1 ex vivo, but this does not appear to explain their reduced susceptibility
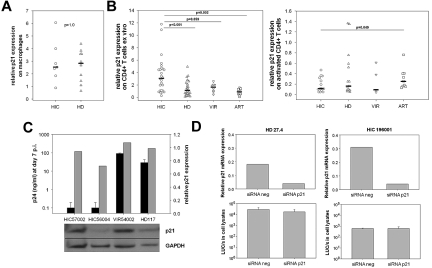
We wondered whether p21 might be responsible for the reduced susceptibility of HIC cells to HIV-1 because we have found that p21 affects reverse transcription.18 We quantified p21 mRNA levels in MDMs at the time of infection and found no difference between MDMs from HICs (median, 2.53; n = 7) and HDs (median, 2.91; n = 11; P = 1.0; Figure 4A), and no correlation was found either between p21 expression and MDM susceptibility to HIV-1 infection (supplemental Figure 2A). In contrast, we found higher levels of p21 mRNA in freshly isolated CD4+ T cells from HICs (median, 3.06 p21 mRNA relative expression; n = 26) than in those from HDs (median, 1.13 p21 mRNA relative expression; P < .0001; n = 29; Figure 4B left). The p21 levels were still higher in fresh CD4+ T cells from HICs than in cells from HIV-1 viremic patients and HAART-treated patients with HIV-RNA < 50 copies/mL (1.64, P = .059, n = 8; and 0.86, P = .002, n = 10, respectively; Figure 4B). However, we found no correlation between p21 levels in their freshly isolated CD4+ T cells and the susceptibility of activated CD4+ T cells to HIV-1 infection in HICs and HDs for whom both parameters were available (supplemental Figure 2B). Moreover, p21 mRNA levels dropped sharply on anti-CD3 stimulation of CD4+ T cells from HICs and controls, and p21 mRNA levels at the time of infection were not higher in cells from HICs than in cells from persons in control groups (Figure 4B right). In addition, The reduced susceptibility of HIC CD4+ T cells to HIV-1 superinfection could not be explained by higher p21 protein levels at the time of infection (Figure 4C). Finally, p21 knockdown with specific siRNA in anti–CD3-activated CD4+ T cells from HICs did not increase their susceptibility to HIV (n = 4). One of these experiments is depicted in Figure 4D. Therefore, although freshly isolated HIC CD4+ T cells carried high levels of p21 mRNA, we found no evidence of a direct role of p21 in the reduced susceptibility of HIC CD4+ T cells and MDMs to HIV-1 infection that we observed in vitro.
Figure 4.
High p21 expression in HIC CD4+ T cells is not directly involved in HIV restriction. (A) Levels of p21 mRNA in MDMs from HICs and HDs at the time of infection (after 7-10 days of differentiation from monocytes). Each symbol represents one person. Median values for each group are shown. (B) Levels of p21 mRNA in nonactivated CD4+ T cells from HICs, HDs, viremic HIV-1–infected patients (VIR) and patients on effective HAART (ART; left). Levels of p21 mRNA in CD4+ T cells from HICs, HDs, VIRs, and ARTs after 5 days of activation with anti-CD3 (right). (A-B) The results are expressed as relative expression levels compared with cDNA in CD4+ T cells from one HD who was used as a common reference throughout the study. (C) Levels of HIV-1 replication 7 days after infection with replicative HIV-1 BaL (■) in CD4+ T cells from 2 HIC, 1 VIR, and 1 HD (top), and levels of p21 protein at the time of infectious challenge (bottom). The ratio of the optical densities of the p21 and GAPDH signals were calculated for each sample with Photoshop CS3 software (Adobe), and reported in the graph ( ) relative to the HD control. (D) Levels of p21 mRNA in anti–CD3-activated CD4+ T cells from one HD and one HIC 24 hours after nucleofection of irrelevant and p21-specific siRNAs (top). Luciferase activity 48 hours after challenge of nucleofected cells with single-cycle HIV-1VSV-G (bottom). Cells were challenged 24 hours after nucleofection. Means ± SDs of 3 independent measures are shown.
) relative to the HD control. (D) Levels of p21 mRNA in anti–CD3-activated CD4+ T cells from one HD and one HIC 24 hours after nucleofection of irrelevant and p21-specific siRNAs (top). Luciferase activity 48 hours after challenge of nucleofected cells with single-cycle HIV-1VSV-G (bottom). Cells were challenged 24 hours after nucleofection. Means ± SDs of 3 independent measures are shown.
We then examined CD4+ T-cell expression of Apobec3G and Trim5α, two restriction factors that might interfere with early steps of viral replication. No difference was found between HICs, HDs (supplemental Figure 3A), and HAART-treated patients (supplemental Figure 3B), either before or after CD3 stimulation.
Reduced HIV-1 susceptibility of cells from HICs is overcome by high viral inocula
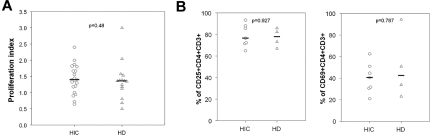
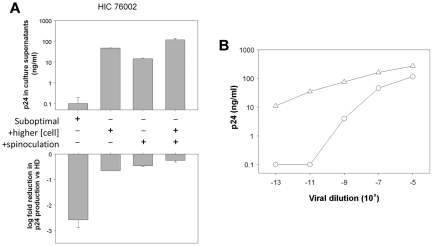
We wondered whether the reduced susceptibility of CD4+ T cells from HICs might be associated with a different response (activation or proliferation) to anti-CD3 stimulation. CD4+ T cells from HICs and HDs showed similar proliferation before HIV challenge (Figure 5A) and expressed similar levels of CD25 and CD69 (Figure 5B). In our previous work,7 in addition to PHA, we used spinoculation to enhance the binding of viral particles to CD4+ T cells,26 together with high cell densities to favor cell-to-cell HIV-1 transmission.31 Here, we explored the effect of each of these factors on HIC CD4+ T-cell susceptibility to infection. Used separately, a higher cell density and spinoculation both enhanced viral replication in CD4+ T cells, especially in cells from HICs, in which viral replication was otherwise barely detectable (Figure 6A). These conditions diminished the difference between CD4+ T cells from HICs and HDs (Figure 6A). This effect was even more pronounced when spinoculation and a high cell density were used simultaneously. These results confirmed that the reduced susceptibility of HIC CD4+ T cells to HIV-1 may be overcome by favoring binding of virus to the target cells or cell-to-cell transmission, thus pointing to a saturable mechanism. To confirm this point, we performed infections of CD4+ T cells from 5 HICs and 6 HDs with the use of serial dilutions of HIV-1 BaL and suboptimal conditions of infection (a representative experiment is shown in Figure 6B). In all cases more virus was necessary to detect viral replication in the cells from HICs (median dilution for first positive p24 value, 10−5 [10−3 to 10−9] and 10−11 [10−7 to 10−13] for HICs and HDs, respectively). However, at higher viral inocula the level of viral replication in CD4+ T cells from HICs approached the one in cells from HDs. This result further corroborates the presence of a saturable mechanism of HIV-1 restriction in the cells from HICs.
Figure 5.
Similar levels of activation and proliferation of CD4 T cells from HICs and HDs in response to anti-CD3 stimulation. (A) Proliferation index of CD4+ T cells from HICs and HDs, calculated as the ratio between the cell density before (106 cells/mL) and after 5 days of anti-CD3 stimulation. Each symbol represents one person. Medians for each group are shown. (B) Expression of the CD25 and CD69 activation markers was measured by flow cytometry on the surface of CD4+ T cells from HICs and HDs 5 days after stimulation with anti-CD3.
Figure 6.
HIC CD4 T-cell resistance is overcome by increasing viral inoculum or cell-to-cell transmission. (A) HIV-1 BaL replication on day 7 after infection in anti–CD3-activated CD4+ T cells from one representative HIC (top) and the logarithmic fold reduction in viral replication compared with cells infected in parallel from a HD (bottom) in various experimental conditions. Cells were challenged as follows: in suboptimal conditions (first column), with a higher cell density (106 cells/mL; second column), using a spinoculation protocol to facilitate viral binding to the cell surface (third column), and using both a higher cell density and spinoculation (fourth column). Means ± SDs are shown (in the bottom panel, standard deviations are relative to the mean infection level measured in cells from HDs). (B) Levels of p24 in culture supernatants of CD4+ T cells from one HIC (circles) and one HD (triangles) at day 9 after infection in suboptimal conditions with serial dilutions of HIV-1 BaL.
HIC CD4+ T-cell permissiveness to HIV-1 replication correlates with the size of the viral reservoir in vivo
Finally, we examined whether the reduced susceptibility of HIC cells to HIV-1 infection in vitro might contribute to limiting the infection in vivo and, particularly, the size of their HIV-1 reservoir. The HIV-1 DNA level in blood cells is a stable parameter reflecting the size of the total HIV-1 reservoir, being proportional to and correlating with HIV-DNA levels in gut-associated lymphoid tissue.32 We quantified total HIV-1 DNA in blood cells from 81 HICs enrolled in the ANRS CO18 cohort. PBMC-associated HIV-1 DNA levels ranged from undetectable (< 11 HIV-DNA copies/106 PBMCs or < 1.04 log) to 728 copies/106 PBMCs (2.86 log). Overall, HIV-1 DNA levels were far lower (1.45 log copies/106 PBMCs; range, < 1.34-1.65) in HICs than in patients with primary infection (ANRS PRIMO cohort: 3.30 log copies/106 PBMCs; range, 1.84-4.93 log copies/106 PBMCs; n = 674),33 in patients with chronic infection (ANRS SEROCO cohort: 2.86 log copies/106 PBMCs; interquartile range [IQR], 2.45-3.21 log copies/106 PBMCs; n = 271),34 in patients on effective HAART (SALTO study: 2.3 log copies/106 PBMCs; IQR, 1.9-2.8 log copies/106 PBMCs; n = 116),35 and om long-term nonprogressors (ANRS ALT cohort: 2.29 log copies/106 PBMCs; IQR, 1.53-2.89 log copies/106 PBMCs; n = 50).36 Thus, our results on a large group of HICs extend previous observations and confirm that HICs have a remarkably small HIV reservoir.
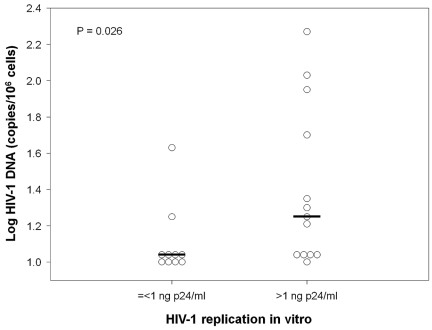
We then compared blood cell viral DNA load in 23 HICs who have been studied for susceptibility to infection in vitro and the level of HIV-1 replication in their CD4+ T cells and found a positive correlation (Spearman, 0.42, P = .044; n = 23; not shown). In other words, those HICs whose CD4+ T cells were least susceptible to HIV-1 infection ex vivo were also those with the lowest levels of total cell-associated HIV-1 DNA in vivo (Figure 7). These results support a role of target cell resistance in the control of HIV-1 infection in HICs.
Figure 7.
Resistance of HIC CD4+ T cells to HIV-1 is associated with low total cell-associated HIV-1 DNA levels. Total cell-associated HIV-1 DNA levels were compared between HICs whose CD4+ T cells yielded ≤ 1 ng of p24/mL of culture supernatant on day 7 after infection with HIV-1 BaL and HICs whose CD4+ T cells yielded > 1 ng of p24/mL of culture supernatant. The samples used to calculate cell-associated HIV-1 DNA load and susceptibility to infection were obtained at the same time. Each symbol represents one person. Medians for each group are shown.
Discussion
Extraordinarily low levels of cell-associated HIV DNA are found in HICs, suggesting that these persons possess mechanisms that hamper the formation of viral reservoirs. We and others have previously described a strong and effective cytotoxic T-cell response that probably plays a role in maintaining viral load at low or undetectable levels.5–7,37 However, other mechanisms may participate in the observed viral control. Simian studies show that the viral reservoir is constituted very early during primary SIV or SHIV infection, when adaptive T-cell responses may not yet be fully developed.38 HIV-1 DNA levels may already be low during primary infection in patients who are later identified as HICs.12 In addition, a strong CTL response is found only in part of HICs8–10 (A.S.-C., G.P., unpublished data on > 150 HIC followed up in the ANRS CO18 cohort since 2009). Here, we show that the main cellular targets of HIV-1, namely CD4+ T lymphocytes and macrophages, exhibit far lower susceptibility to HIV infection in HICs than in the general population or in HIV-1–infected patients noncontrolling infection. Importantly, low CD4+ T-cell permissiveness in vitro was associated with low levels of viral DNA in vivo, suggesting that this intrinsic cell resistance contributes to restricting the size of the viral reservoir in HICs. We found that HIV-1 replication in target cells from HICs was restricted independently of viral tropism for cell receptors and that the mechanism is saturable and affects reverse transcription. Moreover, CD4+ T cells from HICs were also resistant to SIVmac251 infection.
Viral replication in MDMs from most HICs was severely impaired. Macrophages contribute to viral dissemination in body tissues, including “sanctuaries” such as the brain, and to the establishment of viral reservoirs.39–41 A major role of macrophages in HIV-1 spread and pathogenesis is suggested by numerous studies, including one showing that infection of macaques with an SIV(SM) PBj virus unable to replicate in macrophages is associated with milder infection and low viral load.42 Therefore, inhibition of HIV-1 replication in macrophages might have important consequences for the human infection.
CD4+ T cells from HICs were also less susceptible to HIV-1 infection in vitro. However, restriction of viral replication in HIC CD4+ T cells was overcome by spinoculation, by increasing the target cell density or by high virus inoculum. This “saturability” may explain why previous studies, including our own, failed to show reduced susceptibility of CD4+ T cells from HICs in optimized conditions of HIV-1 challenge.4,7 This may also account for the apparent discrepancy between our results that suggest a restriction during reverse transcription in HIC and the results reported very recently by Rabi et al,20 who found no differences in HIV-1 entry and early steps of viral replication between CD4+ T cells from HICs and uninfected donors. In addition to other experimental differences (we used CD3-activated CD4+ T cells, whereas Rabi et al20 used unstimulated CD4+ T cells), they used spinoculation to infect cells with pseudotyped HIV-1 viruses, a technique that increases viral concentration on the cell surface and can overcome viral restriction, as we show here. While we were reviewing the manuscript, a report by Graf et al43 showed that CD4+ T cells from HICs carry in vivo low levels of integrated HIV DNA, which is in agreement with our results showing a restriction before viral integration. However, the investigators could not found a restriction of HIV-1 replication in vitro, but the use of spinoculation may have influenced these experiments. Graf et al43 also show an accumulation of 2-LTR nonintegrated forms in the cells from HICs. In our experiments restriction of viral replication occurred during reverse transcription; however, because of the limited number of persons in which this aspect could be assessed, we cannot discard that viral replication may be blocked at different stages of viral replication in other HIC.
Our results suggest that HIV-1 restriction mechanisms in HICs can be saturated, as can TRIM5α-mediated restriction.22,23 However, TRIM5α is species specific and cannot account for the inhibition of the HIV-1 replicative cycle in cells from HICs. This is also supported by the lack of modulation of TRIM5α expression in our experiments (supplemental Figure 3). The phenotype of HIV-1 restriction found in cells from HICs is reminiscent of that induced by APOBEC3G, which also affects preintegration steps.44,45 A negative correlation between plasma viral load in HIV-1–infected patients and APOBEC3G mRNA levels has been reported,46 but more recent studies failed to confirm this result47 or showed a positive correlation.48 APOBEC3G-induced hypermutation of HIV-1 proviral DNA was not found to be higher in HICs than in HAART-treated patients.47 In addition, APOBEC3G-mediated inhibition is effective in the absence of Vif, and we used Vif+ replicative HIV-1 strains and pseudotypes. Finally, APOBEC3G expression was not significantly different in CD4+ T cells from HICs than in those from HDs (supplemental Figure 3). Overall, our results do not support the involvement of APOBEC3G in the restriction of viral replication in cells from HICs.
Another candidate factor possibly explaining the relative resistance of HIC cells to HIV-1 is p21. Indeed, p21-mediated inhibition of HIV-1 replication in macrophages and hematopoietic stem cells affects preintegrative steps, leading to a strong reduction in the level of integrated proviruses.18,19 In addition, while reviewing the manuscript, Chen et al49 reported a resistance of CD4+ T cells from HICs to HIV-1 infection. Those investigators showed that resistance in their setting was because of elevated levels of p21 in the cells from HICs.49 In our study p21 expression in MDMs did not differ between HICs and uninfected controls, even though viral replication was also far lower in HIC MDMs. Although we found that p21 expression was higher in CD4+ T cells from HICs than from healthy controls, it did not correlate with CD4+ T-cell susceptibility to HIV-1 infection. Furthermore, on CD3 activation, at infectious challenge, p21 mRNA levels in CD4+ T cells were down-regulated and did not differ between HICs and controls. Neither p21 protein expression correlated with the level of HIV-1 replication. Finally, siRNA-mediated p21 knock-down did not restore HIV replication in HIC CD4+ T cells. Overall, the results of the present study argue against a direct role of p21 in the HIV-1 restriction observed here and indicate that additional cell restriction factors are responsible for the resistance of HIC MDMs and CD4 T cells that we found. Our results do not exclude, however, that p21 may have played an indirect role by modulating cellular factors involved in HIV-1 replication or that p21 may play a role in regulating HIV-1 replication in CD4+ T cells in vivo.50 Alternatively, the elevated levels of p21 observed in CD4+ T cells from HICs might be related to other activities of this protein such as its capacity to regulate cell survival or T-cell homeostasis.51,52 Along these lines, van Grevenynghe et al53 have shown that memory CD4+ T cells from HICs are resistant to apoptosis. Further work is needed to understand the possible influence of elevated p21 expression on HIC CD4+ T-cell susceptibility to HIV.
In conclusion, our data suggest that HIV target cells in HICs are intrinsically resistant to infection, possibly contributing to the observed control of viral replication in vivo. In particular, mechanisms blocking viral DNA integration in CD4+ T cells and macrophages appear to hinder the formation of the viral reservoir, as suggested by the negative correlation between the degree of CD4+ T-cell resistance and the level of cell-associated HIV-1 DNA. Although the restriction observed in vitro can be overcome by optimizing the conditions of viral challenge in vitro, it is probably an effective barrier to viral spread in vivo. This intrinsic resistance could cooperate with specific T-cell responses in controlling HIV infection and might have a prominent role in HICs.
Supplementary Material
Acknowledgments
The authors thank Jeannine Delgado, Hanane Mehawej, and Pr Laurence Weiss for precious help with the recruitment of patients in control groups. We also thank all the other physicians and nurses who cared for the patients included in the ANRS CO18 cohort. They especially thank the subjects who participated in this study for their cooperation.
This study was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS). G.P. received funds from Inserm. A.B. was recipient of a postdoctoral fellowship from Sidaction. C.H. received a predoctoral fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche (MESR). David Young, a medical English editor supported by funding from ANRS, provided English editorial assistance to the authors during preparation of this manuscript. Nevirapine was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.S.-C., C.H., A.B., A.D., P.V., and A.M. performed research; A.S.-C., C.H., A.B., C.R., and G.P. analyzed and interpreted data; F.B. and O.L. coordinated inclusion of patients; F.B. collected clinical data from patients; F.B.-S. and O.L. discussed results; A.S.-C. and G.P. designed the research; and A.S.-C. and G.P. wrote the manuscript. All the authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A list of the members of the ANRS CO18 Cohort is available in the supplemental Appendix.
Correspondence: Asier Sáez-Cirión, Unité de Régulation des Infections Rétrovirales, Institut Pasteur, 25 rue du Dr Roux, 75725 Paris Cedex 15, France; e-mail: asier.saez-cirion@pasteur.fr.
References
- 1.Lambotte O, Boufassa F, Madec Y, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41(7):1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 2.Saez-Cirion A, Pancino G, Sinet M, Venet A, Lambotte O. HIV controllers: how do they tame the virus? Trends Immunol. 2007;28(12):532–540. doi: 10.1016/j.it.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Dinoso JB, Kim SY, Siliciano RF, Blankson JN. A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy. Clin Infect Dis. 2008;47(1):102–104. doi: 10.1086/588791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julg B, Pereyra F, Buzon MJ, et al. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin Infect Dis. 2010;51(2):233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersperger AR, Pereyra F, Nason M, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6(5):e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migueles SA, Osborne CM, Royce C, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29(6):1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saez-Cirion A, Lacabaratz C, Lambotte O, et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A. 2007;104(16):6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emu B, Sinclair E, Hatano H, et al. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82(11):5398–5407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197(4):563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 10.Saez-Cirion A, Sinet M, Shin SY, et al. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol. 2009;182(12):7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 11.Okulicz JF, Marconi VC, Landrum ML, et al. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J Infect Dis. 2009;200(11):1714–1723. doi: 10.1086/646609. [DOI] [PubMed] [Google Scholar]
- 12.Goujard C, Chaix ML, Lambotte O, et al. Spontaneous control of viral replication during primary HIV infection: when is “HIV controller” status established? Clin Infect Dis. 2009;49(6):982–986. doi: 10.1086/605504. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 14.Saez-Cirion A, Versmisse P, Truong LX, et al. Persistent resistance to HIV-1 infection in CD4 T cells from exposed uninfected Vietnamese individuals is mediated by entry and post-entry blocks. Retrovirology. 2006;3:81. doi: 10.1186/1742-4690-3-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8(1):55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Asada M, Yamada T, Ichijo H, et al. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 1999;18(5):1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper JW AG, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 18.Bergamaschi A, David A, Le Rouzic E, Nisole S, Barre-Sinoussi F, Pancino G. The CDK inhibitor p21Cip1/WAF1 is induced by FcgammaR activation and restricts the replication of human immunodeficiency virus type 1 and related primate lentiviruses in human macrophages. J Virol. 2009;83(23):12253–12265. doi: 10.1128/JVI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J SD, Crumpaker CS. Primitive hematopoietic cells resist HIV-1 infection via p21Waf1/Cip1/Sdi1. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabi SA, O'Connell KA, Nikolaeva D, et al. Unstimulated primary CD4+ T cells from HIV type 1 positive elite suppressors are fully susceptible to HIV-1 entry and productive infection. J Virol. 2011;85(2):979–986. doi: 10.1128/JVI.01721-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begaud E, Chartier L, Marechal V, et al. Reduced CD4 T cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology. 2006;3:35. doi: 10.1186/1742-4690-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A. 2002;99(18):11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99(21):13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avettand-Fenoel V, Chaix ML, Blanche S, et al. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01). J Med Virol. 2009;81(2):217–223. doi: 10.1002/jmv.21390. [DOI] [PubMed] [Google Scholar]
- 25.David A, Saez-Cirion A, Versmisse P, et al. The engagement of activating FcgammaRs inhibits primate lentivirus replication in human macrophages. J Immunol. 2006;177(9):6291–6300. doi: 10.4049/jimmunol.177.9.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 28.Sáez-Cirión A, Nicola MA, Pancino G, Shorte SL. Quantitative real-time analysis of HIV-1 gene expression dynamics in single living primary cells. Biotechnol J. 2006;1(6):682–689. doi: 10.1002/biot.200600045. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Salkowitz JR, Purvis SF, Meyerson H, et al. Characterization of high-risk HIV-1 seronegative hemophiliacs. Clin Immunol. 2001;98(2):200–211. doi: 10.1006/clim.2000.4969. [DOI] [PubMed] [Google Scholar]
- 31.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol. 2007;81(2):1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avettand-Fenoel V, Prazuck T, Hocqueloux L, et al. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. AIDS. 2008;22(14):1880–1882. doi: 10.1097/QAD.0b013e32830fbdbc. [DOI] [PubMed] [Google Scholar]
- 33.Ghosn J, Deveau C, Chaix ML, et al. Despite being highly diverse, immunovirological status strongly correlates with clinical symptoms during primary HIV-1 infection: a cross-sectional study based on 674 patients enrolled in the ANRS CO 06 PRIMO cohort. J Antimicrob Chemother. 2010;65(4):741–748. doi: 10.1093/jac/dkq035. [DOI] [PubMed] [Google Scholar]
- 34.Rouzioux C, Hubert J-B, Burgard M, et al. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192(1):46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- 35.Piketty C, Weiss L, Assoumou L, et al. A high HIV DNA level in PBMCs at antiretroviral treatment interruption predicts a shorter time to treatment resumption, independently of the CD4 nadir. J Med Virol. 2010;82(11):1819–1828. doi: 10.1002/jmv.21907. [DOI] [PubMed] [Google Scholar]
- 36.Xie J, Lu W, Samri A, et al. Distinct differentiation profiles of HIV-Gag and Nef-specific central memory CD8+ T cells associated with HLA-B57/5801 and virus control. AIDS. 2010;24(15):2323–2329. doi: 10.1097/QAD.0b013e32833e5009. [DOI] [PubMed] [Google Scholar]
- 37.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura Y, Sadjadpour R, Mattapallil JJ, et al. High frequencies of resting CD4+ T cells containing integrated viral DNA are found in rhesus macaques during acute lentivirus infections. Proc Natl Acad Sci U S A. 2009;106(19):8015–8020. doi: 10.1073/pnas.0903022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aquaro S, Calio R, Balzarini J, Bellocchi MC, Garaci E, Perno CF. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 2002;55(2):209–225. doi: 10.1016/s0166-3542(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 40.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276(5320):1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 41.Sharova N, Swingler C, Sharkey M, Stevenson M. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 2005;24(13):2481–2489. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirsch V, Sharkey M, Brown C, et al. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4(12):1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf EH, Mexas AM, Yu JJ, et al. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7(2):e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 45.Mbisa JL, Barr R, Thomas JA, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81(13):7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79(17):11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82(6):3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loke P, Favre D, Hunt PW, et al. Correlating cellular and molecular signatures of mucosal immunity that distinguish HIV controllers from noncontrollers. Blood. 2010;115(15):e20–32. doi: 10.1182/blood-2009-12-257451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Li C, Huang J, et al. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121(4):1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu W, Kehn-Hall K, Pedati C, et al. Drug 9AA reactivates p21/Waf1 and Inhibits HIV-1 progeny formation. Virol J. 2008;5:41. doi: 10.1186/1743-422X-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arias CF, Ballesteros-Tato A, Garcia MI, et al. p21CIP1/WAF1 controls proliferation of activated/memory T cells and affects homeostasis and memory T cell responses. J Immunol. 2007;178(4):2296–2306. doi: 10.4049/jimmunol.178.4.2296. [DOI] [PubMed] [Google Scholar]
- 52.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1(8):639–649. [PubMed] [Google Scholar]
- 53.van Grevenynghe J, Procopio FA, He Z, et al. Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection. Nat Med. 2008;14(3):266–274. doi: 10.1038/nm1728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.