Abstract
We previously showed that gamma interferon (IFNγ) and its receptor subunit, IFNGR1, interacted with the promoter region of IFNγ-activated genes along with transcription factor STAT1α. Recent studies have suggested that activated Janus kinases pJAK2 and pJAK1 also played a role in gene activation by phosphorylation of histone H3 on tyrosine 41. This study addresses the question of the role of activated JAKs in specific gene activation by IFNγ. We carried out chromatin immunoprecipitation (ChIP) followed by PCR in IFNγ treated WISH cells and showed association of pJAK1, pJAK2, IFNGR1, and STAT1 on the same DNA sequence of the IRF-1 gene promoter. The β-actin gene, which is not activated by IFNγ, did not show this association. The movement of activated JAK to the nucleus and the IRF-1 promoter was confirmed by the combination of nuclear fractionation, confocal microscopy and DNA precipitation analysis using the biotinylated GAS promoter. Activated JAKs in the nucleus was associated with phosphorylated tyrosine 41 on histone H3 in the region of the GAS promoter. Unphosphorylated JAK2 was found to be constitutively present in the nucleus and was capable of undergoing activation in IFNγ treated cells, most likely via nuclear IFNGR1. Association of pJAK2 and IFNGR1 with histone H3 in IFNγ treated cells was demonstrated by histone H3 immunoprecipitation. Unphosphorylated STAT1 protein was associated with histone H3 of untreated cells. IFNγ treatment resulted in its disassociation and then re-association as pSTAT1. The results suggest a novel role for activated JAKs in epigenetic events for specific gene activation.
Keywords: cytokine signaling, JAK/STAT pathway, nuclear translocation, interferon
1. INTRODUCTION
Receptor endocytosis and translocation to the nucleus upon ligand stimulation is well documented in signaling events that involve the JAK/STAT pathway [1, 2]. In spite of the evolutionary conservation of polycationic nuclear localization sequences (NLSs) in ligands and/or receptors that utilize the JAK/STAT pathway [3–5], the prevailing view is that the ligand activates the cell solely via interaction with the extracellular domain of the receptor complex with no particular role for the NLSs [1]. This interaction in turn results in the activation of receptor or receptor-associated tyrosine kinases primarily of the JAK kinase family, leading to phosphorylation and dimerization of the STAT transcription factors, which then dissociate from the receptor cytoplasmic domain and translocate to the nucleus via intrinsic nonclassical NLSs [1]. This view ascribes no further role of the ligand, the receptor, or the JAKs in the signaling process [6–8].
Most of the STAT proteins form homodimers in response to ligand stimulation, with few exceptions [3–5]. Given the limited number of STAT family members and their preference for homodimerization for transcription, it is difficult to explain the various specificities of the over 60 proteins that utilize STATs in signal transduction [3–5].
We have previously shown that IFNγ and one of its receptor subunits, IFNGR1, are translocated to the nucleus, together with activated STAT1α as one macromolecular complex via the classical importin-dependent pathway [9]. We have further shown that IFNγ and IFNGR1 are recruited to the IFNγ-activated sequence (GAS) element at the promoter site of IFNγ activated genes [9]. The direct association of IFNGR1 with the promoter region of IFNγ-activated genes suggested a transcriptional/cotranscriptional role for IFNGR1 as well as its possible role in determining the specificity of gene activation by IFNγ.
The presence of activated JAK2 in the nucleus of wild-type cells only after PDGF2 or IL-3 treatment raises the question of a possible nuclear role of the activated JAK2 in gene activation by the ligands [10]. There remains, however, important issues such as the mechanism of nuclear import of the activated JAK2 and its role in specific gene activation that is associated with the particular ligand. We were thus interested in the possible association of activated JAK2 and JAK1 in the IFNγ/IFNGR1/pSTAT1α complex formed in cells treated with IFNγ at the cytoplasmic, nuclear, and specific gene level. Insight into these events could provide a key to the mechanism of specific gene activation by IFNγ specifically and other ligands that use JAK/STAT signaling generally. The studies presented here provide a mechanism for direct interaction of activated JAK2 with histone H3 at a gene that is specifically activated by IFNγ. The findings have implications for specific gene activation by IFNγ.
2. MATERIALS AND METHODS
2.1. Cell culture and Antibodies
WISH cells were purchased from American Type Culture Collection (ATCC) and were grown in MEME with 10% FBS and antibiotics. For all experiments, cells were serum starved for at least 4 hours, washed twice with PBS and then given regular media with or without 250 ng/ml IFNγ (Invitrogen). The following polyclonal antisera were purchased from Santa Cruz Biotech (Santa Cruz, CA): IFNGR1, STAT1, pSTAT1, pJAK1, pJAK2, normal rabbit IgG, β -tubulin, β-lamin, and histone H3. Antibody to acetylhistone was from Active Motif. Antibody to JAK2 was from Millipore. Antibody to tyrosine phosphorylated histone H3 was from Abcam.
2.2. Chromatin immunoprecipitation (ChIP) assay
WISH cells were treated or not with IFNγ for 1 hr. Cells were then washed twice with cold PBS and treated with 1% formaldehyde for 10 min at 37°C. The rest of the procedure was conducted using the ChIP kit from Millipore (Temecula, CA), as previously described, including IRF-1 and β-actin primers [9].
2.3. Nuclear Fractionation and nuclear JAK2 activation
Following treatment, WISH cells were washed twice in cold PBS, removed by scraping in lysis buffer (10 mM HEPES pH 7.9, 100 mM KCl, 1% Triton X-100, 1 mM NaF, 1 mM Na3VO4, 2 mM MgCl2, 1 mM DTT, and 1 mM PMSF), and pelleted via low speed centrifugation. The supernatant was discarded while the pellet, containing intact nuclei, was gently resuspended in lysis buffer. Details of nuclear isolation are as previously described by us [9].
2.4. Indirect immunofluorescence assay and confocal microscopy
Immunofluorescence and confocal microscopy were performed exactly as previously described using a Zeiss Axiovert 200M confocal microscope [9].
2.5. Analysis of proteins bound to biotinylated GAS promoter DNA
To identify the proteins associated with the GAS promoter, a nucleotide sequence from human IRF-1 promoter containing the GAS motif, 5’-TGATTTCCCCGAAATG -3’, were chosen. An oligonucleotide containing five copies of this sequence and another oligonucleotide containing the complementary sequence were synthesized. The two strands were biotinylated and annealed, followed by incubation with whole cell lysates, as described [9].
3. RESULTS
3.1. pJAK2 and pJAK1 are recruited to the GAS element in the IRF1 promoter
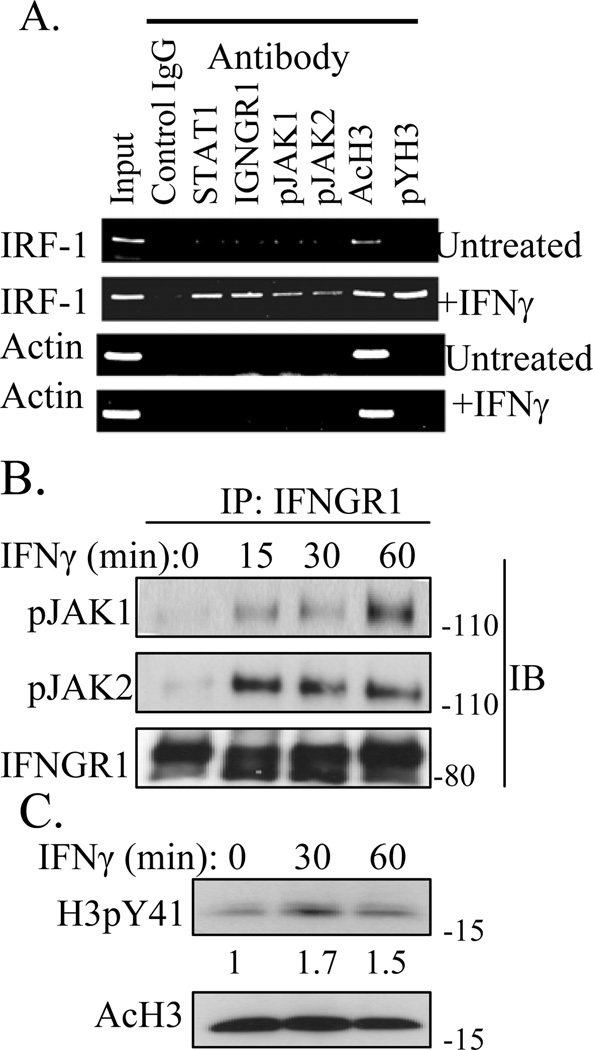
We previously showed by ChIP analysis that treatment of cells with IFNγ resulted in binding of IFNγ, IFNGR1, STAT1α, but not IFNGR2 to the GAS element in the promoter region of the IRF-1 gene [9]. A similar ChIP analysis was performed for activated JAK2 (pJAK2) and JAK1 (pJAK1) using WISH cells treated with IFNγ for 1 hour. Sonicated chromatin containing approximately 500-bp fragments were immunoprecipitated with antibodies to STAT1, IFNGR1, pJAK1, pJAK2, acetylated histone H3 (AcH3), and histone H3 phosphorylated on tyrosine 41 (H3pY41), followed by PCR with primers flanking the GAS element from IRF-1. The PCR product selected for amplification extended from nt −403 to −222 in the promoter of the IRF-1 gene. As a control, PCR product for the promoter of β-actin gene, −967 to −844, was chosen. As shown in Fig. 1A, STAT1, IFNGR1, pJAK1, pJAK2, AcH3, and H3pY41 were associated with the GAS element of the IRF-1 promoter in IFNγ treated cells. As we previously showed the presence of IFNGR1 and STAT1 at the GAS promoter [9], their presence here can also be considered as an internal positive control. Phosphorylation of AcH3 on Y41, H3pY41, results in disassociation of the transcription repressor heterochromatin protein 1α (HP1α) from chromatin, resulting in transcription of genes such as IRF1, which are repressed by HP1α (14). Untreated cells showed IRF-1 promoter only in input and anti-AcH3 precipitated chromatin. β-actin promoter was precipitated only by anti-AcH3 in IFNγ treated cells. Thus, in addition to the presence of STAT1 and IFNGR1, IFNγ treated cells also showed the presence of pJAK1 and pJAK2 in the promoter of the IRF-1 gene. pJAK2 has been shown to phosphorylate histone H3 at Y41 (H3pY41) [10].
Fig. 1.
IFNγ stimulation induces the association of STAT1, IFNGR1, pJAK1, pJAK2, and H3pY41 with the IRF-1 promoter. A) ChIP assay. WISH cells were treated with or without IFNγ (250 ng/ml) for 1 hour, then treated with 1% formaldehyde for 10 minutes. Details of the ChIP assay are as we previously described [9]. B) The IFNγ induced association of pJAK1 and pJAK2 with IFNGR1 correlates with the tyrosine phosphorylation of histone H3. WISH cells were incubated with or without IFNγ (250 ng/ml) for the indicated times, then whole cell lysates were obtained. Equal amounts of protein were immunoprecipitated using IFNGR1 antibody. Immunoprecipitated material was then washed, subject to PAGE, and then Western blotted using the indicated antibodies. C) Phosphorylation of histone H3 on tyrosine 41 (H3pY41) of IFNγ treated cells. Ten μg of whole cell lysates were analyzed by Western blot for H3pY41, then stripped and re-probed for AcH3. Numbers at the bottom of pYH3 blot represent relative intensity of bands as measured by using Image J program (NIH). The results are representative of three experiments.
3.2. Association of pJAK1 and pJAK2 with IFNGR1 in cells treated with IFNγ
IFNγ treatment of cells results in the formation of a complex consisting of IFNγ, pSTAT1α, and IFNGR1 where pSTAT1α is activated STAT1α [9]. This complex is actively transported into the nucleus where the nuclear localization sequence (NLS) is provided by the IFNγ. We were interested in determining if pJAK1 and pJAK2 were associated with IFNGR1 in the complex as this would provide a mechanism for the specific presence of the JAKs at the IRF-1 promoter. Accordingly, WISH cells were treated with IFNγ and whole-cell lysates were immunoprecipitated (IP) with antibodies to IFNGR1 and analyzed by Western blots. As shown in Figure 1B, both pJAK1 and pJAK2 bound to IFNGR1 stably over 15 to 60 minutes with similar levels of pJAK2 over the time period and maximal pJAK1 binding at 60 minutes. pJAK2 has been shown to bind to IFNGR1 in IFNγ treated cells [1]. These observations provide the mechanism for activation of the JAKs and for their specific presence at the promoter site of an IFNγ activated gene.
Cell lysates were also Western blotted for tyrosine phosphorylation of histone H3 (H3pY41) after IFNγ treatment. As shown in Fig. 1C, increased H3pY41 was observed at 30 and 60 minutes, with a peak at 30 minutes. These results are consistent with nuclear pJAK2, and perhaps pJAK1, phosphorylation of histone H3 at Y41, which is associated with specific gene activation.
3.3. pJAK2 is present in the nucleus of cells only after treatment with IFNγ
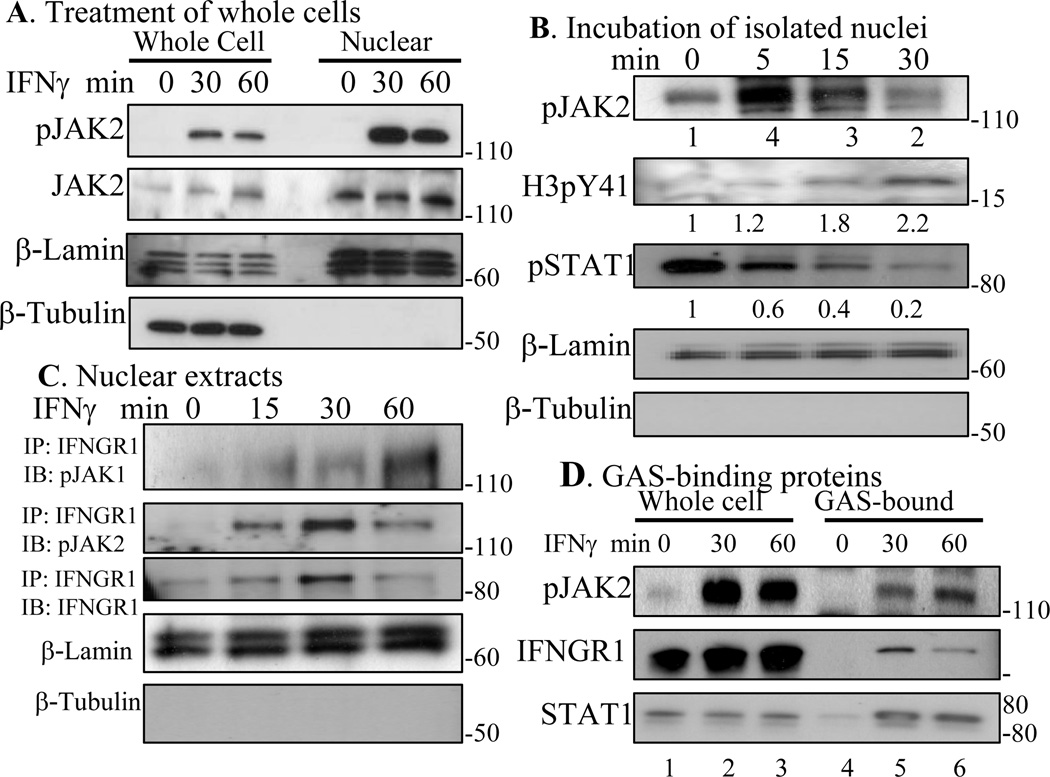
In order to further determine if pJAK2 was present in the nucleus of WISH cells solely as a result of IFNγ treatment, we analyzed whole cell and nuclear lysates for both pJAK2 and JAK2. pJAK2 was present in the nucleus only after treatment of cells with IFNγ, while JAK2 was present constitutively (Fig. 2A). JAK2 contains a classical cationic NLS, which may allow movement in an inactive state between the cytoplasm and nucleus. To ascertain the purity of nuclear fractionation, β-tubulin and β-lamin were used as markers of nuclear and cytoplasmic fractions, respectively. pJAK2 appeared in the nucleus in association with IFNGR1, which we have previously shown to function as a transcription/cotranscription factor in IFNγ activated genes [9]. This does not preclude activation of JAK2 in the nucleus via nuclear IFNGR1.
Fig. 2.
JAK2 is constitutively present in the nucleus and is activated upon IFNγ treatment. A) WISH cells were incubated with or without IFNγ (250 ng/ml) for the indicated times, then whole cell (WL) and nuclear lysates were independently obtained (see Materials and Methods), and Western blotted. B) WISH cells were treated with IFNγ (250 ng/ml) for 10 min and their nuclei were purified (see Materials and Methods). Isolated nuclei were then incubated at 37° C in kinase buffer for the indicated times. Lysates were obtained and subjected to Western blotting for the indicated proteins. Numbers at the bottom represent relative intensity of bands as measured by using Image J program (NIH). C) Cells were treated with 250 ng/ml of IFNγ for the indicated times and isolated nuclear extracts were immunoprecipitated with antibody to IFNGR1. Eluted proteins were Western blotted with antibodies to pJAK1, pJAK2, and IFNGR1. D) pJAK2, IFNGR1, and STAT1 are associated with the GAS promoter. A biotinylated double-stranded oligomer containing five copies of the GAS element taken from the IRF-1 promoter was incubated with equal amounts of whole cell lysates (WL) from WISH cells that were untreated or treated with IFNγ (250 ng/ml) for 30 min or 60 min. This complex was then added to Neutravidin conjugated to agarose (GAS probe). The bound proteins were washed, eluted, electrophoresed, and probed sequentially with antibodies to pJAK2, IFNGR1, and STAT1. The results are representative of at least two experiments.
In order to determine possible activation of JAK2 in the nucleus in addition to the known activation in the cytoplasm, we treated WISH cells with IFNγ for 10 minutes, isolated the nuclei, and determined if there was an increase in nuclear pJAK2 over time. As shown in Fig. 2B, pJAK2 was present in the nucleus at 0 minutes of nuclear isolation of IFNγ treated cells by Western blot. The level of nuclear pJAK2 increased at 5 and 15 minutes and decreased at 30 minutes. Thus, there was further activation of JAK2 in the isolated nucleus, which suggests that the nuclear IFNGR1 of IFNγ treated cells was capable of phosphorylation of nuclear JAK2. These activation events are consistent with our demonstration of an IFNγ/IFNGR1 complex in the nucleus of IFNγ treated cells, but are inexplicable in the models where IFNγ treatment of cells results only in the presence of pSTAT1 in the nucleus. It is noteworthy that pSTAT1 levels were maximal at 0 minutes of isolated nuclei and decreased thereafter over time. By comparison, pY41H3 levels increased in nuclei over time. Immunoprecipitation of IFNGR1 from isolated nuclei of IFNγ treated cells showed IFNGR1 and pJAK2 association, which would be expected in nuclear IFNGR1 involvement with nuclear activation of JAK2 (Fig. 2C). Thus, the increase in pJAK2 appears to be related to nuclear events other than further nuclear activation of pSTAT1α.
3.4. pJAK2, IFNGR1, and STAT1 directly associate with GAS promoter element of cells treated with IFNγ
We have previously shown that IFNGR1 and STAT1 directly associate with the GAS promoter element in IFNγ treated cells [9]. In order to verify the association of pJAK2, a biotinylated GAS promoter was generated and incubated with whole cell lysates from WISH cells treated with IFNγ. Following incubation with lysates, the mixture was then incubated with Neutravidin conjugated to agarose. Following washing, the bound proteins were eluted, electrophoresed, and probed with antibodies to pJAK2, IFNGR1, and STAT1. As shown in Fig. 2D, all these proteins were associated with the GAS promoter of IFNγ treated cells at 30 and 60 minutes. Thus, pJAK2, IFNGR1, and STAT1 are associated with the GAS promoter element only after treatment of cells with IFNγ.
3.5. Immunofluorescence of pJAK2 in the nucleus of cells treated with IFNγ
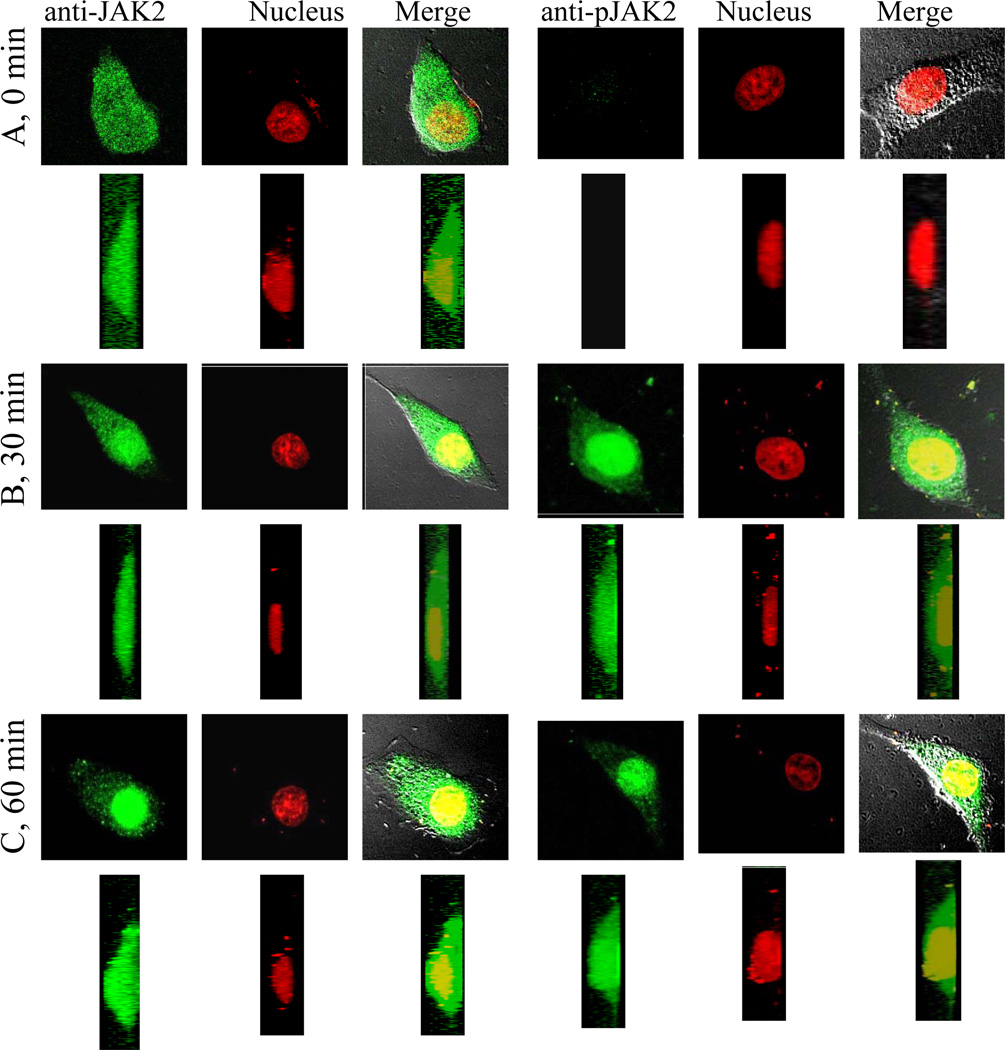
To complement the ChIP analysis and Western blots, we performed immunofluorescence using confocal microscopy on WISH cells treated with IFNγ. Cells were stained for JAK2 and pJAK2. As shown in Figure 3A, JAK2 was present throughout the cell, but pJAK2 was absent from untreated cells. By contrast, pJAK2 was observed in the cells after treatment with IFNγ for 30 minutes, with a strong presence in the nucleus (Fig. 3B). The predominant presence of pJAK2 in the nucleus was also observed at 60 minutes post IFNγ treatment (Fig. 3C). Thus, similar to ChIP and Western blots, JAK2 was present in both the cytoplasm and nucleus of untreated and IFNγ treated cells. pJAK2 was present in the cytoplasm and nucleus only after treatment of cells with IFNγ.
Fig. 3.
JAK2 is constitutively present in nuclei, while pJAK2 appears in nuclei following IFNγ treatment. 2D and 3D volume reconstruction of JAK2 and pJAK2 in WISH cells either left untreated (A) or treated with IFNγ (250 ng/ml) for 30 (B) or 60 (C) minutes. Following immunofluorescence, images of cells were obtained via confocal microscopy. 3D reconstruction of images from 2D images was done using 17 merged image sections from the stack, 4 μm above and below the focal plane through the nucleus of the cells (a 0.5 μm displacement each along the Z-axis). Sections were merged to render a 3D reconstruction of the cells. The resulting images were projected by rotation at 0° along the X-axis, 90° along the Y-axis, and presented below each un-rotated image. All image processing was done using Pascal (Microsoft) software attached to a LSM 5 Pascal workstation.
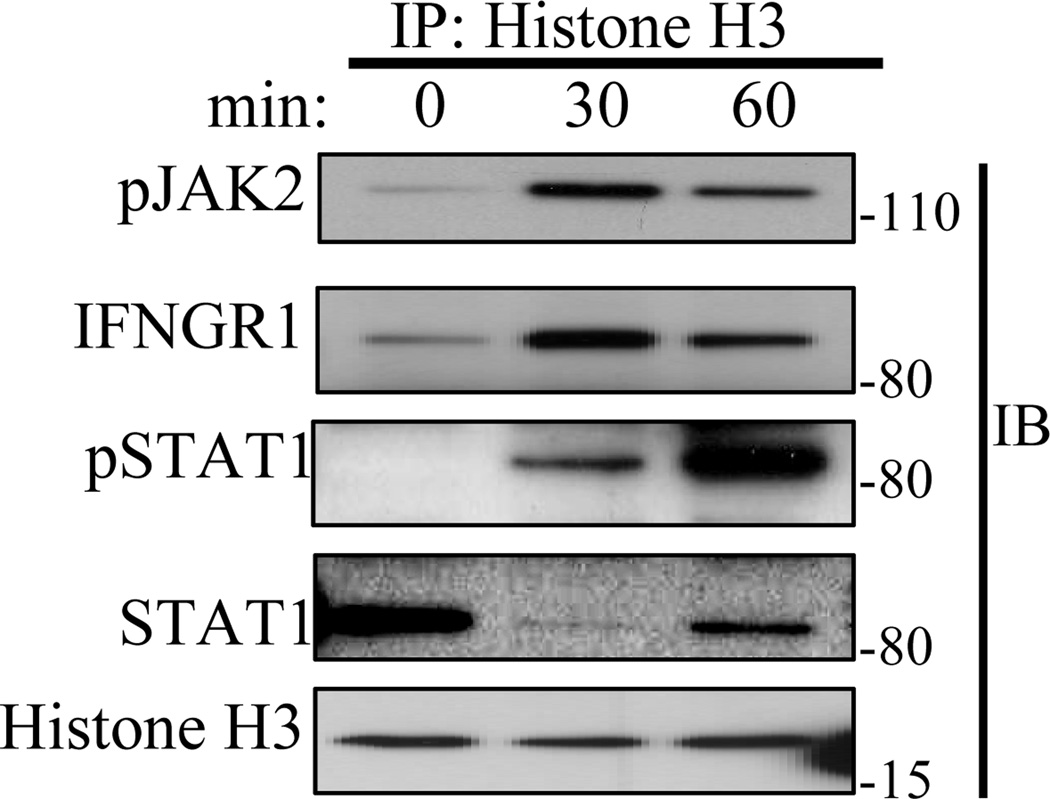
3.6. pJAK2 and IFNGR1 are associated with histone H3 in IFNγ treated cells
We showed in Fig. 1C that treatment of WISH cells with IFNγ resulted in phosphorylation of histone H3 at tyrosine 41. We test here for the association of pJAK2, IFNGR1, and pSTAT1 with histone H3 in IFNγ treated cells. WISH cells were treated with IFNγ for 30 and 60 minutes and whole cell lysates were immunoprecipitated against histone H3 followed by Western blotting for the indicated proteins. As shown in Fig. 4, pJAK2, IFNGR1, and pSTAT1 were associated with histone H3 at 30 and 60 minutes of IFNγ treatment, but not to any significant extent in untreated cells. Interestingly, STAT1 protein was associated with histone H3 in untreated cells, decreased at 30 minutes of IFNγ treatment and then increased proportionally to increased pSTAT1. This would suggest that unphosphorylated STAT1 protein is associated with histone H3 in untreated cells, exits after IFNγ treatment, and returns as pSTAT1. This is consistent with studies in Drosophila that show that unphosphorylated STAT is present in the nucleus of cells and functions as a heterochromatin stabilizer. Exit from the nucleus or disassociation from histone H3/heterochromatin was associated with heterochromatin destabilization and gene activation [11]. The association of pJAK2 and IFNGR1 with histone H3 is consistent with pJAK2 phosphorylation of tyrosine 41 on the protein.
Fig. 4.
pJAK2, IFNGR1, and pSTAT1, are induced to associate with histone H3 in response to IFNγ stimulation while the association of unphosphorylated STAT1 is constitutively associated. Equal amounts of whole cell lysates obtained from WISH cells treated with IFNγ (250 ng/ml) for 0, 30, and 60 minutes were subjected to immunoprecipitation against histone H3 antibody. The bound proteins were washed, eluted, electrophoresed, and probed sequentially with antibodies to pJAK2, IFNGR1, pSTAT1, STAT1, and histone H3. The results are representative of at least two experiments.
4. DISCUSSION
It has recently been acknowledged that the classical model of JAK/STAT signaling was over simplified in its original form [12]. In the case of IFNγ, complexity beyond simple JAK/STAT activation in signal transduction is indicated in the relatively recent demonstration that other pathways, including MAP kinase, PI3 kinase, Cam kinase II, NF-KB, and others cooperate with or act in parallel to JAK/STAT signaling to regulate IFNγ effects at the level of gene activation and cell phenotypes [12]. All of these pathways are generic in the sense that a plethora of cytokines with functions different from those of IFNγ also activate them.
It has been suggested that JAK tyrosine kinases, including the mutant JAK2V617F, play an important role in the epigentics of gene activation in addition to STAT activation in the cytoplasm [10, 13–16]. Leukemic cells with a JAK2V617F gain-of-function mutation have constitutively active JAK2V617F in the nucleus. This leads to tyrosine phosphorylation on Y41 on histone H3, which results in disassociation of heterochromatin protein 1α, HP1α. The heterochromatin remodeling was associated with exposure of euchromatin for gene activation. Although present in the nucleus, wild-type JAK2 was only activated when K562 cells were treated with PDGF or LIF, or when BaF3 cells were treated with IL-3. The question of how a ligand/receptor interaction resulted in the presence of pJAK2 in the nucleus was not addressed, nor its targeting mechanism to discrete genomic sites and promoters. There is evidence, however, that even in the case of JAK2V617F, receptor association may play a role in specific gene activation that is associated with the particular myeloproliferative disorder. Expression of homodimeric type I cytokine receptors has been shown to be required for JAK2V617F-mediated cell transformation [17, 18]. Further, the expression of JAK2V617F with a particular type I cytokine receptor correlated with the myeloproliferative phenotype. Thus, the activation of particular genes by JAK2V617F may be done in the context of associated nuclear receptors, analogous to that of IFNGR1 and JAK2 that we have shown here.
We felt that our discovery of the IFNγ /IFNGR1/pSTAT1α complex and its movement to the nucleus provided a logical mechanism for transport of pJAK1 and pJAK2 not only to the nucleus, but also to histone H3 regions of genes activated by IFNγ. It has previously been shown that JAK2 moves from receptor subunit IFNGR2 to IFNGR1 in IFNγ treated cells [1]. Thus, ChIP followed by PCR in IFNγ treated cells showed the association of pJAK1, pJAK2, IFNGR1, and STAT1α on the same DNA sequence of the IRF-1 gene promoter. Similar to the pJAK2 findings above, pJAK1 has recently been shown to phosphorylate Y41 on histone H3 in in vitro experiments [19]. The β-actin gene, which is not activated by IFNγ, did not show the above associations. These findings were confirmed by biotinylated GAS promoter binding and confocal microscopy. Consistent with leukemic cell studies, the presence of activated JAKs in the nucleus was associated with phosphorylated Y41 on histone H3 in the region of the GAS promoter. We further showed that nuclear JAK2 is activated to pJAK2, probably by the IFNγ/IFNGR1 complex in the nucleus and that this activation is related to epigenetic function and not nuclear STAT activation.
Recent studies in Drosophila have shown that unphosphorylated STAT is associated with HP1α and plays a role in heterochromatin stability [10]. Further, activation/phosphorylation of STAT to pSTAT causes it to disassociate from heterochromatin and binding to cognitive sites in euchromatin. Moreover, these events correlate with unphosphorylated STAT association with stable heterochromatin and gene silencing, while pSTAT was associated with heterochromatin destabilization and gene expression. It has been reported that unphosphorylated STATs 1 and 3 function as transcription factors by mechanisms distinct from those of phosphorylated STATs [20]. The unphosphorylated STATs have been proposed to be involved in prolonged transcription events of several days. Similar to the Drosophila results, we showed that unphosphorylated STAT1α was associated with histone H3 in untreated WISH cells. Treatment of the cells with IFNγ resulted in the disassociation of unphosphorylated STAT1α from histone H3 and its return, possibly at a different site in the activated pSTAT1α form. These observations are consistent with regulated epigenetic events in the region of genes that are activated by IFNγ. Thus, we propose that the complex of IFNγ /IFNGR1/pSTAT1α/pJAK1/pJAK2 contains the transcription/co-transcription signals for specific gene activation as well as the activated JAK activity for the associated epigenetics of histone H3 phosphorylation. We estimate that the aggregate molecular weight of the complex to be approximately 530 kDa, a feasible cargo size for active transport through the nuclear pore complex [21, 22]. The results of our study provide insight into the mechanism of IFNγ signaling including the role of the JAK/STAT pathway in the specificity of such signaling.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI 056152 to HMJ.
REFERENCES
- 1.Levy DE, Darnell JE. STATs: Transcriptional control and biological impact. Nature Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 2.Jans DA DA, Hassan G. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? BioEssays. 1998;20:400–411. doi: 10.1002/(SICI)1521-1878(199805)20:5<400::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 3.Subramaniam PS, Torres BA, Johnson HM. So many ligands, so few transcription factors: A new paradigm for signaling through the STAT transcription factors. Cytokine. 2001;15:175–187. doi: 10.1006/cyto.2001.0905. [DOI] [PubMed] [Google Scholar]
- 4.Johnson HM, Subramaniam PS, Olsnes S, Jans DA. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. BioEssays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- 5.Johnson HM, Ahmed CM. Gamma interferon signaling: Insights to development of interferon mimetics. Cell Mol Biol. 2006;52:71–76. [PubMed] [Google Scholar]
- 6.McBride KM, McDonald C, Reich NC. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 2000;19:6196–6206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear transport. J. Biol. Chem. 2001;276:16447–16455. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 8.Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of STAT1 is regulated by a leucine-rich export signal in the coiled coil domain. Proc Natl Acad Sci USA. 2000;97:10418–10423. doi: 10.1073/pnas.190318397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed CM, Johnson HM. IFNγand its receptor subunit IFNGR1 are recruited to the IFNγ-activated sequence element at the promoter site of IFNγ-activated genes: Evidence of transactivational activity in IFNGR1. J. Immunol. 2006;177:315–321. doi: 10.4049/jimmunol.177.1.315. [DOI] [PubMed] [Google Scholar]
- 10.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan SJ, Lim SJ, Shi S, Dutta P, Li WX. Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J. 2010;25:232–241. doi: 10.1096/fj.10-169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gough DJ, Levy DE, Johnsotone RW, Clarke CJ. IFN gamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19:383–394. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends in Cell Biol. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi S, Calhoun HC, Xia F, Li J, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, Harr MW, Levine RL, Xu H, Tefferi A, Deblasio A, Hatlen M, Menendez S, Nimer DS. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19:283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmer RA, Panchoo M, Dertien JS, Bhakta SM, Hewetson A, Chilton BS. Prolactin-induced JAK2 phosphorylation of RUSH: A key element on JAK/RUSH signaling. Mol Cellular Endocrin. 2010;325:143–149. doi: 10.1016/j.mce.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Levine R, Tong W, Wering G, Pikman T, Zarnegar S, Gilliland DG, Lodish H. Expression of a homodimeric type I cytokine receptor is required for JAK2B617F-mediated transformation. Proc Natl Acad Sci USA. 2005;102:18962–18967. doi: 10.1073/pnas.0509714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Huang LJ, Lodish HF. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. J Biol Chem. 2008;283:5258–5266. doi: 10.1074/jbc.M707125200. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths DS, Li J, Dawson MA, Trotter MW, Cheng YH, Smith AM, Liu P, Kouzarides T, Nichols J, Bannister AJ, Green AR, Gottgens B. LIF-independent JAK signaling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13:13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheon H, Yang J J, Stark GR. The functions of signal transducers and activators of transcription 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jans DA. Nuclear transport in development and disease: The importance of importins. Semin Cell Dev Biol. 2009;20:575–590. doi: 10.1016/j.semcdb.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L. Influence of cargo size on Ran and energy requirements for nuclear protein import. J Cell Biol. 2002;159:55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]