Abstract
Kisspeptin, the product of the KiSS1 gene, has emerged as a key component of the mechanism by which the hypothalamus controls puberty and reproductive development. It does so by stimulating the secretion of gonadotropin releasing hormone (GnRH). Little is known about the transcriptional control of the KiSS1 gene. Here we show that a set of proteins postulated to be upstream components of a hypothalamic network involved in controlling female puberty regulates KiSS1 transcriptional activity. Using RACE-PCR we determined that transcription of KiSS1 mRNA is initiated at a single transcription start site (TSS) located 153–156 bp upstream of the ATG translation initiation codon. Promoter assays performed using 293 MSR cells showed that the KiSS1 promoter is activated by TTF1 and CUX1-p200, and repressed by EAP1, YY1, and CUX1-p110. EAP1 and CUX-110 were also repressive in GT1-7 cells. All four TFs are recruited in vivo to the KiSS1 promoter and are expressed in kisspeptin neurons. These results suggest that expression of the KiSS1 gene is regulated by trans-activators and repressors involved in the system-wide control of mammalian puberty.
Keywords: gene transcription, EAP1, CUX1, TTF1, YY1, KiSS1, transcription factors
1. Introduction
It is well established that an increase in pulsatile gonadotropin-releasing hormone (GnRH) release from GnRH neurons is required for the initiation of puberty. The pubertal increase in GnRH secretion is determined by changes in transsynaptic (Kordon et al. 1994; Ojeda and Terasawa 2002) and glial (Ojeda et al. 2003; Ojeda and Terasawa 2002) inputs to the GnRH neuronal network. The transsynaptic control of GnRH neurons has two components: an excitatory arm that is activated at puberty and an inhibitory arm that loses strength at the time when the excitatory inputs to GnRH neurons are increasing (Ojeda and Terasawa 2002; Plant and Witchel 2006; Terasawa and Fernandez 2001).
Although glutamatergic neurons have been known for many years to be major contributors to the excitatory transsynaptic system controlling GnRH secretion (Brann 1995; Ojeda and Skinner 2006; Plant and Witchel 2006), compelling evidence now exists demonstrating the existence of another, more critical set of neurons providing excitatory inputs to the GnRH neuronal network. These neurons use a peptide known as kisspeptin for neurotransmission, and the G protein-coupled receptor GPR54 for signaling. While inactivating mutations of the GPR54 gene result in hypogonadotrophic hypogonadism (HH) in humans (De Roux et al. 2003; Seminara et al. 2003; Tenenbaum-Rakover et al. 2007; Semple et al. 2005), activating mutations and certain types of polymorphisms of this gene have been reported to be associated with precocious puberty (Luan et al. 2007; Teles et al. 2008). Both Kiss1 and Gpr54 null mice fail to undergo sexual development; they are infertile and have reduced gonadotropin levels (Seminara et al. 2003; Lapatto et al. 2007). These and other observations demonstrate that an intact KiSS1/GPR54 signaling system is mandatory for normal reproductive maturation and fertility in mammals [reviewed in (D’Anglemont de Tassigny et al. 2007; Oakley et al. 2009)].
Kisspeptin is a product of the KiSS1 gene which encodes a 145 amino acid precursor. Post-translational modifications of this peptide result in a C-terminally amidated 54-amino acid peptide and several shorter fragments (e.g. kisspeptin-10, kisspeptin-13, kisspeptin-14) (Ohtaki et al. 2001). The first biological actions assigned to the KiSS1 system were its capacity to inhibit cell migration and tumor metastasis (Ohtaki et al. 2001; Lee et al. 1996). For this reason the 54-amino acid peptide is also known as metastin.
Although kisspeptin plays a pivotal role of in the hypothalamic control of reproductive function, the transcription factors controlling KiSS1 expression have not been identified. Using a high-throughput approach (DNA arrays) we used rats and rhesus monkeys to provide evidence for the existence of a gene network that, according to our results, contributes to the hypothalamic control of puberty (Roth et al. 2007). Genes composing this network have diverse cellular functions, but they share the common feature of having been earlier identified as involved in tumor suppression/tumor formation (Roth et al. 2007). Computational analysis revealed that the KiSS1 gene is a subordinate member of this tumor suppressor gene (TSG) network. As such, KiSS1 was predicted to be under the transcriptional control imposed by central TSG hubs and to be regulated by non-TSG genes. These non-TSG genes, which are also connected to the TSG network, are involved in the transcriptional regulation of the pubertal process.
In the present study, we verified this computational prediction by determining if the human KiSS1 gene is transcriptionally regulated by two postulated central hubs of the TSG network, CCAAT displacement protein (CDP, also known as CUTL1 and CUX1) and Ying Yang 1 (YY1) (Roth et al. 2007), in addition to two non-TSGs previously shown to be transcriptional regulators of puberty, Enhanced at Puberty 1 (EAP1) (Heger et al. 2007) and Thyroid Transcription Factor 1 (TTF1) (Mastronardi et al. 2006). CUTL1 (henceforth called CUX1) is an evolutionary conserved homeobox gene encoding a protein that functions as a transcriptional repressor in several systems (Nepveu 2001), but that can be proteolytically processed into isoforms with trans-activational capabilities (Sansregret and Nepveu 2008). Due to this, CUX1 can either repress (Dufort and Nepveu 1994; Superti-Furga et al. 1989; Valarche et al. 1993) or activate gene transcription (Harada et al. 2008; Kim et al. 1997). Cux1-null mutant male mice are subfertile (Luong et al. 2002). In our studies we utilized two isoforms of CUX1, p200 and p110, which have been previously shown to have divergent transcriptional activity in breast cancer cells (Goulet et al. 2002). YY1 is a member of the Polycomb (PcG) group silencing complex and as such, it plays an important role in gene silencing, because it recruits PcG repressive proteins in addition to histone 3 trimethylated at lysine 27 (H3K27me3) to gene promoters to silence transcription (Wilkinson et al. 2006; Woo et al. 2010). EAP1 is a newly identified transcription factor that displays mostly repressive activity, and that is required for both the timing of puberty and normal female reproductive cyclicity (Heger et al. 2007; Roth et al. 2007). Finally, TTF1 (NKX2.1) is a member of the NKX family of homeobox genes (Pera and Kessel 1998; Price et al. 1992) required for embryonic development of the diencephalon and pituitary gland (Kimura et al. 1996). Studies involving peripubertal female rats demonstrated that Ttf1 expression persists in hypothalamic regions involved in the control of reproductive development (Lee et al. 2001). Within these regions, kisspeptin neurons were later identified as cells that express Ttf1 (Mastronardi et al. 2004). Conditional deletion of the Ttf1 gene from neurons lead to a delayed onset of puberty and reduced fertility in female mice (Mastronardi et al. 2004).
Here we report that TTF1, YY1, EAP1 and CUX1 interact with the KiSS1 promoter in vivo, and that Eap1, Yy1, and Cux1 are expressed in kisspeptin neurons of the rodent hypothalamus. We also show that the KiSS1 gene is trans-activated by TTF1 and repressed by YY1, EAP1 and CUX1-p110. CUX-p200 exhibits a dual transcriptional activity depending on the cell context. These results suggest that KiSS1 expression depends on a balance between trans-activators and repressors involved in the system-wide hypothalamic control of puberty.
2. Materials and Methods
2.1. Identification of the human KiSS1 Transcription Start Site
To map the Transcription Start Site (TSS) of KiSS1, total RNA was isolated from human placental tissue (kindly provided by R. Faber, Leipzig, Germany) using the RNeasy Mini Kit (Qiagen, Hilden, Germany). The integrity of the RNA was verified by agarose gel electrophoresis. Five-prime rapid amplification of cDNA ends (RACE) was performed using the GeneRacer kit (Invitrogen, Karlsruhe, Germany). Briefly, 1μg total human placental RNA were treated with calf intestinal phosphatase (provided with the kit) to dephosphorylate truncated mRNAs and non-mRNA species. Subsequent treatment with tobacco acid phosphatase removed the 5′ cap structure from intact, full-length mRNAs. The GenRacer RNA oligodeoxynucleotide was ligated to pre-treated human placental RNA using the T4 RNA ligase provided with the kit. Linked RNA was reversed transcribed (1h at 37°C followed by 5 min at 93°C) using an oligo-DT primer (Omniscript Kit, Qiagen). PCR amplification utilizing HotStarTaq DNA Polymerase (Qiagen) and the primers 1F/R (Table 1) were used for hot start touchdown PCR employing the following conditions: 15 min at 95°C, 30 sec at 94°C, and 4 min at 72°C for 5 cycles, 30 sec at 94°C followed by 4 min at 70°C for 5 cycles, and 30 sec at 94°C followed by 4 min at 68°C for 20 cycles. Final extension was 10 min at 72°C. The amplicon was used as a template for nested PCR (primers 2F/R, Table 1). The PCR product was visualized on a 1% agarose gel and cloned into pCRII® TOPO®-Vector (TOPO TA Cloning Kit, Invitrogen). DH5-α competent cells (Invitrogen) and XL1-Blue super competent cells (Stratagene, Heidelberg, Germany) were used for transformation. Twenty clones were analyzed by sequencing using the primer provided with the kit. To confirm the results, the experiment was repeated using random hexamer primers for reverse transcription, pGEM®-T Easy Vector (Promega, Mannheim, Germany) for PCR product cloning, and XL1-Blue cells for transformation. The use of human tissue has been approved by the Ethical Committee of the University of Leipzig, Germany.
Table 1.
Primer sequences
| Name | Sequence | Position according to accession-no: NT_004487 |
|---|---|---|
|
| ||
| Primer1F | 5′ - CGA CTG GAG CAC GAG GAC ACT GA - 3′ | provided by GenRacer Kit |
| Primer1R | 5′ - CTC TCG GTG CAC GGC AGG CTC T - 3′ | bp 54650216 – 54650237 |
|
| ||
| Primer2F | 5′ - GGA CAC TGA CAT GGA CTG AAG GAG TA - 3′ | provided by GenRacer Kit |
| Primer2R | 5′ - GAG GCC CAG TTC TAG CTG CT - 3′ | bp 54650254 – 54650276 |
|
| ||
| Primer3F | 5′ - AGA ATC TCT GCC ACC ACC AC - 3′ | bp 54656479 – 54656503 |
| Primer3R | 5 ′-GCT GGG CTC CCG GTC TCA AG - 3′ | bp 54655947 – 54655966 |
|
| ||
| Primer4F | 5′ - GGA CAG GCC AAC GTA CAC ATC ATC - 3′ | bp 54657263 – 54657286 |
| Primer4R | 5′ - GCT GGG CTC CCG GTC TCA AG - 3′ | bp 54655947 – 54655966 |
|
| ||
| Primer5F | 5′ - CTT GAA CTT GGA TCA TTG GTT GAG - 3′ | bp 54657899 – 54657922 |
| Primer5R | 5′ - AGG GCA GAG ACT GTT TCT TCT ATC - 3′ | bp 54656972 – 54656995 |
|
| ||
| Position according to TSS=+1 accession-no: NM_002256 | ||
|
| ||
| Primer6F | 5′ - CCC CCG CAC CTT CTC CAT TTG A - 3′ | bp −2456 – −2434 |
| Primer6R | 5′ - CCG CAC TTA GCC AGA TCC CCA GAA - 3′ | bp −2288 – −2264 |
|
| ||
| Primer7F | 5′ - TGT CCC TGT CCT CAA AGT GCT GTA - 3′ | bp −839 – −816 |
| Primer7R | 5′ - CTT CCT TCC TGC TTC CCT TCT TTC - 3′ | bp −562 – −538 |
|
| ||
| Primer8F | 5′ - CGG CCC CGG GTG TCG TT - 3′ | bp 11149 – 11166 |
| Primer8R | 5′ - CTG GGC AGT GTG GGG TTA TTT TCT - 3′ | bp 11225 – 11249 |
Primer 1F/1R and 2F/2R were used for RACE-PCR, Primer 3F/3R, 4F/4R and 5 F/5R for promoter cloning, and Primer 6F/6R to 8F/8R for PCR detection of Chromatin immunoprecipitated DNA.
2.2. Gene reporter constructs
To generate reporter constructs containing different lengths of the KiSS1 5′-flanking region, DNA fragments were amplified from human genomic DNA (Promega) using the primers 3F/R and 4F/R (Table 1). PCR products of a length of 556 bp and 1339 bp were cloned into the pCRII® TOPO®-Vector (Invitrogen). To generate a 1975 bp long DNA fragment, a 951 bp PCR-product, amplified by primers 5F/R (Table 1), was subcloned into the pCRII® TOPO®-vector (Invitrogen). From there a 636 bp fragment was excised with HpaI and KpnI and cloned into the HpaI and KpnI sites of pCRII® TOPO® -KiSS1p1339 utilizing the Rapid Ligation Kit (Fermentas, St. Leon-Rot, Germany). The sequence and orientation of each construct (pCRII® TOPO®-KiSS1p1975, pCRII® TOPO®-KiSS1p1339, and pCRII® TOPO®-KiSS1p556) were verified by automatic sequencing, and the fragments were subcloned into the KpnI and XhoI sites of the luciferase pGL4.10 basic vector (Promega).
2.3. Expression vectors
Human EAP1
To determine if EAP1 regulates KiSS1 gene expression, we used an expression vector containing the coding region of EAP1 mRNA (Heger et al. 2007). A mutated EAP1, lacking the RING finger domain (Heger et al. 2007; Waterman et al. 1999) was used to determine the structural requirements for EAP1 action on KiSS1 transcription.
Human YY1
To express YY1, a construct containing the YY1 coding region tagged with a hemagglutinin epitope (HA) was excised with XbaI and ApaI from pCMV-HAYY1 (kindly provided by Y. Shi, Harvard Medical School, Boston, USA), and cloned into pcDNA3.1+/zeo. The sequence of the construct was verified by DNA sequencing.
Human CUX1
We used two different CUX1 constructs, both cloned into the vector pXJ42 (both kind gifts from A. Nepveu, Goodman Cancer Center and Departments of Oncology, Biochemistry and Medicine, McGill University, Montreal, Canada). The pXJ42/MCH construct contains the full length CUX1 (termed p200); pXJ42/MCH/878-1505 contains CUX1 isoform p110. Both isoforms differ in length and transcriptional activity. P200 has been reported to be inhibitory, while p110 exhibit either repressive or trans-activating activity, depending on the promoter context (Sansregret and Nepveu 2008).
Rat TTF1
rTTF1 was expressed from a pcDNA3.1+/zeo-rTTF1 construct containing the rTTF1 coding region (Lee et al. 2001).
Human SP1
hSP1 (2891 bp) was purchased from Deutsches Ressourcenzentrum für Genomforschung GmbH (RZPD, Berlin, Germany), cleaved out from the pOTB7-hSP1 vector by EcoRI and XhoI, and cloned into pcDNA3.1/zeo.
2.4. Cell culture
GripTite™ 293 MSR cells (Invitrogen) were cultured in a humidified atmosphere containing 5% CO2 and 37°C. They were maintained in Dulbecco’s modified Eagles medium containing high glucose (4.5 g/L; PAA Laboratories, Pasching, Germany), supplemented with 10% fetal bovine serum (FBS; Biochrom; Berlin; Germany), 0.1 mM non-essential amino acids (PAA Laboratories), and 600 μg/ml G-418 Sulphate (PAA Laboratories).
GT1-7 cells (kind gift of Pam Mellon, Department of Reproductive Medicine and Center for Reproductive Science and Medicine, University of California, San Diego, La Jolla, Ca, USA), were cultured in Dulbecco’s modified Eagles medium containing high glucose (4.5 g/L; PAA Laboratories), supplemented with 10% FBS (Biochrom), 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
Hela cells (LGC Standards GmbH, Wesel, Germany were grown in a humidified atmosphere containing 5% CO2 and 37°C. The culture medium was Dulbecco’s modified Eagles medium containing high glucose (4.5 g/L; PAA Laboratories), supplemented with 10% fetal bovine serum, 2mM Glutamine (PAA Laboratories), 100 U/ml penicillin, and 100 μg/ml streptomycin.
2.5. Transient transfection and Dual-Luciferase promoter assays
The luciferase reporter constructs used in these assays contained fragments of the KiSS1 gene 5′-flanking region of different lengths: −1948 to +27 (1975 bp); −1312 to +27 (1339 bp) or −529 to +27 (556 bp). The transregulatory effects of TTF1, EAP1, YY1 and CUX1 on the KiSS1 promoter activity were examined in the human embryonic kidney cell line GripTite™ 293 MSR, and GT1-7 cells (mouse immortalized gonadotropin-releasing hormone neurons). The cells were seeded in antibiotic-free medium at a density of 2 × 105 cells per well in 24-well dishes (Greiner bio-one, Frieckenhausen, Germany) 18 h before transfection. Each of the reporter plasmids was transiently transfected (at 500 ng/well) for 4 h using Nanofectin (PAA Laboratories) for GripTite™ 293 MSR, or TurboFect™ in vitro Transfection Reagent (Fermentas) for GT1-7 cells, in conjunction with either the empty expression vector (pcDNA3.1zeo, Invitrogen at 500 ng/well) or vectors expressing EAP1, TTF1, YY1 or CUX1. Each well received a total of 0.5 ml of culture medium. Transfection efficiency was normalized by co-transfecting the renilla plasmid pGL4.70 [hRluc] (Promega) at 20 ng/well. Cells were harvested 48 h after transfection and lyzed in lysis buffer (Promega). Luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega) in a luminometer according to the manufacturer’s protocol. Relative firefly luciferase activities were calculated following correction for transfection efficiency using the Renilla luciferase activities as the normalizing unit. To ensure that the transcriptional effects observed were exerted on the KiSS1 promoter and not on the pGL4.1 basic vector, basal pGL4.1 luciferase activity was measured after transfecting the cells with each of the four TFs examined. Experiments were performed three times using three or four wells per group each time.
2.6. Chromatin Immunoprecipitation (ChIP) Assay
To determine the association of EAP1 and TTF1 to the KiSS1 promoter, we transfected Hela cells with either an expression vector carrying the EAP1 coding region tagged with an influenza hemagglutinin (HA) epitope (kindly provided by H. Samuels, New York University School of Medicine, New York) or the rat TTF1 construct described in section 2.3. Subsequent immunoprecipitation was performed using antibodies that recognize either the HA tag of the EAP1 construct or rat TTF1 itself (see below). In the case of YY1 and CUX1, we used antibodies against the endogenous YY1 and CUX1 proteins (see below). For transfection, Hela cells (25×106) were seeded in 15 cm diameter plates, and 24 h later 45 μg of each expression vectors were transiently transfected for 5 h using Lipofectamine LTX (Invitrogen). Forty-eight hours after transfection, the cells were harvested for chromatin immunoprecipitation. They were washed once in ice-cold PBS containing a protease inhibitor cocktail (1 mM phenylmethylsulfonylfluoride, 7 μg/ml aprotinin, 0.7 μg/ml pepstatin A, 0.5 μg/ml leupeptin), a phosphatase inhibitor cocktail (1 mM β-glycerophosphate, 1 mM sodium pyrophosphate and 1 mM sodium fluoride), an HDAC inhibitor (20 mM sodium butyrate), and a tyrosine phosphatase inhibitor (1 mM). Cross-linking was performed by incubating the cell suspension in 1% formaldehyde for 10 minutes at room temperature. After two additional washing steps in PBS the cells were lysed with 200 μl SDS buffer (1% SDS, 50 mM Tris-HCl, 10 mM EDTA) containing protease, phosphatase, and HDAC inhibitors and sonicated to yield chromatin fragments of 500–1000 bp using a Kontes Micro Ultrasonic Cell Disruptor (Vineland, NJ, USA) (power 4.5 and tune 1.5). Size fragmentation was confirmed by agarose gel electrophoresis. The sonicated chromatin was clarified by centrifugation at 14,000 rpm for 10 min at 4°C, brought up to 1 ml in Chip Dilution Buffer (CDB) (16.7 mM Tris-HCl, pH 8.1, 150 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, and 0.01% SDS) containing protease, phosphatase, and HDAC inhibitors and stored at −80°C for subsequent immunoprecipitation. For this step, chromatin was pre-cleared with Protein A/G beads (Dynabeads, Invitrogen, Carlsbad, CA) for 1 h at 4°C. One aliquot (200 μl of 1000 μl) of the cleared chromatin was stored at -80°C as input DNA. Aliquots (200 μl) of the remaining chromatin were then incubated with 5 μg of a mouse monoclonal antibody against HA (Covance, Berkeley, CA) to detect EAP1, rabbit polyclonal antibodies against TTF1 (kindly provided by C. Mendelson, Southwestern Medical Center, Dallas, TX, USA), a mouse monoclonal antibody (sc-7341 X, Santa Cruz Biotechnology, Santa Cruz, CA, USA) to detect YY1, or rabbit polyclonal antibodies (sc-13024, rabbit polyclonal, Santa Cruz Biotechnology) to detect CUX1, each reaction in final volume of 1 ml CDB. Two additional aliquots of chromatin were incubated with either normal mouse IgG (sc-2025, Santa Cruz Biotechnology) or rabbit IgG (sc-2027, Santa Cruz Biotechnology) and used as negative controls. Antibody-chromatin complex mixtures were incubated at 4°C overnight with gentle agitation. Immunocomplexes were then collected by adding 25 μl of protein A or G beads solution (Dynabeads) and incubated for 2 h at 4°C with gentle agitation. Immunocomplexes were washed sequentially with 0.5 ml low salt wash buffer (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS), high salt wash buffer (20 mM Tris-HCl, pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS), LiCl buffer (10 mM Tris-HCl, pH 8.1, 250 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate and 1 mM EDTA), and with TE buffer (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA). Immunocomplexes were eluted with 300 μl of 0.1 M NaHCO3 and 1% SDS at room temperature for 30 min. Cross-linking was reversed by adding 20 μl of 5 M NaCl and incubating at 65°C overnight. Proteins were then digested by adding 6 μl of proteinase K (20 mg/ml), 6 μl of 0.5 M EDTA, and 6 μl of Tris-HCl, pH 6.5, and incubating the mixture for 2 h at 55°C. DNA was recovered by using the QIAquick PCR purification Kit (Qiagen, Germantown, MD) and stored at −80°C until subsequent PCR analysis. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA)
2.7. PCR detection of Chromatin Immunoprecipitated DNA
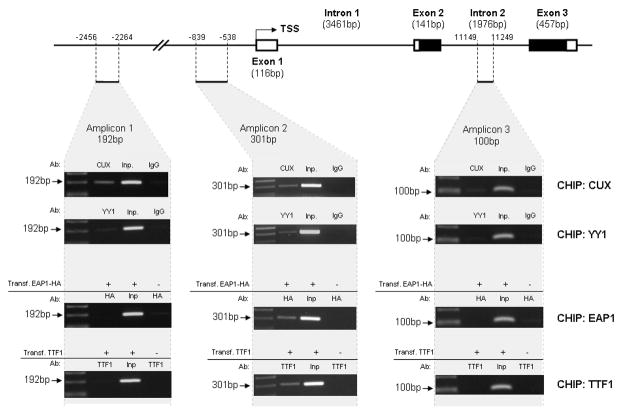
To detect CUX1, EAP1, YY1, and TTF1 binding to the promoter region of the KiSS1 gene we PCR-amplified, using Hot Star Taq polymerase (Qiagen), a DNA fragment derived from the KiSS1 proximal promoter (NM_002256). As controls for the ChIP assays, we amplified an upstream fragment located more than 2,000 bp from the gene’s TSS, and a second segment located in intron 2. The regions amplified were: 1) a 192 bp segment (amplicon 1) located at position −2456 to −2264 from the KiSS1 TSS, 2) a 301bp segment (amplicon 2) located at −839 to −538, and 3), and 3) a 100bp fragment (amplicon 3) located in intron 2 (nucleotides 11149 to 11249 in the KiSS1 genomic sequence). Amplicon 1 was generated using primers 6F/R, amplicon 2 with primers 7F/R, and amplicon 3 with primers 8F/R (Table 1). All primers were purchased from Eurofins MWG Operon, Huntsville, Al. The thermocycling conditions used were 95°C for 5 min, followed by 31 cycles of 95°C for 30 sec, 59°C for 30 sec and 72°C for 1 min. PCR products were resolved in a 1.5% agarose gel, stained with ethidium bromide.
2.8. Immunohistofluorescent detection of EAP1, YY1, and CUX1 in kisspeptin neurons
To determine if kisspeptin neurons in the rat hypothalamus contain Yy1, Eap1, and/or Cux1, we detected kisspeptin neurons with a sheep antibody against kisspeptin (diluted 1:50,000) (Ramaswamy et al. 2008). Eap1 was detected using rabbit polyclonal antibodies (diluted 1:3,000) (Heger et al. 2007); Cux1 and Yy1 were detected using mouse monoclonal antibodies (Abcam, Cambridge, MA, USA; 1:1000, and Santa Cruz; 1:3000, respectively). Frozen sections (30 μm) obtained from brains perfusion-fixed with 4% paraformaldehyde- PBS, pH 7.4, were mounted on Superfrost glass slides, dried for 2 h under an air stream and subjected to an antigen retrieval protocol (Shi et al. 1991a). Thereafter, the sections were incubated for 48 h at 4°C with the primary antibodies indicated above. At the end of this period, kisspeptin immunostaining was visualized using Alexa 488-donkey antisheep IgG (1:500; Invitrogen), Eap1 staining with Alexa 594-donkey antirabbit IgG (1:500), and Cux1 staining with Alexa 568 donkey antimouse IgG (1:500). Yy1 staining was developed using biotinylated donkey antimouse IgGs (1:250; Invitrogen) followed by incubation with Streptavidin-Alexa 568 (1:500; Invitrogen). Before this incubation, the sections were subjected to the biotyramine enhancement method, as previously described (Jung et al. 1997). Fluorescent images were acquired as reported (Heger et al. 2007). The specificity of the TTF1, EAP1 and kisspeptin antibodies used has been reported in earlier publications from either our laboratory (Mastronardi et al. 2006; Heger et al. 2007) or others investigators (Ramaswamy et al. 2008). The specificity of the YY1 and CUX1 antibodies is described by the respective manufacturers (Santa Cruz and AbCAM, respectively). We also verified that the pattern of expression of these two proteins in the hypothalamus corresponds to that of their respective mRNA detected by in situ hybridization (Allan Mouse Brain Atlas; http://mouse.brain-map.org). In the case of CUX1, we stained brain sections using rabbit polyclonal antibodies (Ellis et al. 2001), kindly provided by M. Busslinger (Research Institute of Molecular Pathology, Vienna, Austria) and found a pattern of expression identical to that seen with the commercial monoclonal antibody (not shown). In addition, for every reaction were included sections incubated without the first antibodies. Animal usage was duly approved by the Institutional Animal Care and Use Committee of the Oregon National Primate Research Center, USA.
2.9. PCR detection of transcription factors in different cell lines
We used reverse transcription (RT)-PCR to assess the presence of KiSS1, EAP1, CUX1, YY1, and TTF1 mRNAs in the three cell lines used for study (GripTite™293 MSR, GT1-7, and Hela cells). Total RNA was isolated using the RNeasy Mini Kit (Qiagen), and RNA integrity was assessed by gel electrophoresis. The RT-PCR was performed using 1 μg RNA and the Omniscript RT Kit (Qiagen) according to the manufacturer’s protocol. The PCR reactions were performed using 500 ng RT product, 1 μM primer (Table 2; Primer9F/9R to Primer14F/14R), 2.5 mM dNTPs, 2.5 Dream Taq Buffer which includes 20 mM MgCl2 and 2.5 U Dream Taq Polymerase (Fermentas). The PCR program employed an initial activation step of 3 min at 95°C, and 35 cycles of denaturing at 94°C for 45 sec, annealing at 58°C (KiSS1), 56°C (EAP1), 48°C (CUX1), 62°C (YY1), and 60°C (TTF1), for 45 sec and 1 min extension at 72°C, followed by a final extension of 7 min at 72°C.
Table 2.
Primer sequences for KiSS1 and other transcription factors in cell lines
| Name | Sequence | Primer for PCR amplification in different cell lines: |
|---|---|---|
|
| ||
| KiSS1 | human bp 477– 697 (NM_002256.3) | |
| Primer9F | 5′ - ACC TGC CGA ACT ACA ACT GG - 3′ | |
| Primer9R | 5′ - CTT TTA TTG CCT CGG GTT GG - 3′ | |
|
| ||
| KiSS1 | mouse bp 124– 187 (NM_178260.3) | |
| Primer 10F | 5′ – GAG CCG CTG GCA AAA GTG - 3′ | |
| Primer 10R | 5′ – GCA TAC CGC GAT TCC TTT TC - 3′ | |
|
| ||
| EAP1 | human bp 937 – 1063 (NM_024496) | |
| Primer11F | 5′ - CTT GCT ACC TGT GCG AC - 3′ | mouse bp 952 – 1077 (AF525300) |
| Primer11R | 5′ - GTG CTG TCT CGA TCA CG - 3′ | rat bp 366 – 491 (AY879229) |
|
| ||
| CUX1 | human bp 1574– 2705 (NM_181552) | |
| Primer12F | 5′ - CAG CAC AAA CTC CAT ATC - 3′ | mouse bp 1454– 2576 (NM_009986) |
| Primer12R | 5′ - GTA TGG GGA CTC AGC G - 3′ | rat bp 1825– 2947 (XM_347163) |
|
| ||
| YY1 | human bp 826– 940 (NM_003403.3) | |
| Primer13F | 5′ - GAC GAC TCG GAC GGG - 3′ | mouse bp 454– 568 (NM_009537.3) |
| Primer13R | 5′ - CCA CGG TGA CCA GCG - 3′ | rat bp 343– 457 (NM_173290.1) |
|
| ||
| TTF1 | human bp 203– 278 (NM_001079668) | |
| Primer14F | 5′ - AGC ACA CGA CTC CGT TCT CA - 3′ | mouse bp 630– 705 (NM_009385.3) |
| Primer14R | 5′ - CCC TCC ATG CCC ACT TTC TT - 3′ | rat bp 17– 92 (NM_013093.1) |
2.10. Statistics
Differences between two groups were analyzed using one-way ANOVA and Student’s t-test for individual means. Data are expressed as mean ± SEM. The level of significance was p<0.05.
3. Results
3.1. Transcription of the human KiSS1 gene is initiated at a single TSS
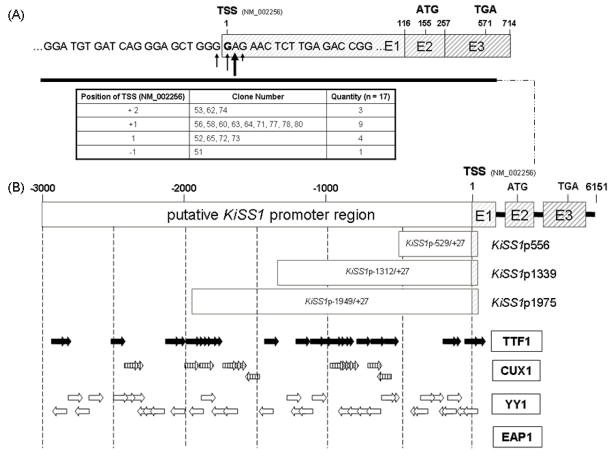
To identify the TSS of KiSS1 we analyzed 17 clones derived from RACE-PCR of human placental cDNA. Nine out of 17 clones exhibited a transcriptional start at 155 bp upstream of the ATG translational start site. All 17 clones mapped to a narrow region of 4 bp, indicating that the KiSS1 gene utilizes a single TSS mapping to a region located between 153 bp and 156 bp upstream of the ATG (Fig. 1A). This finding is consistent with the computer-predicted location of KiSS1 TSS (NM_002256).
Figure 1. Identification of the KiSS1 transcription start site (TSS), and in silico prediction of transcription factor recognition sites in the KiSS1 5′-flanking region.
A, The KiSS1 gene is transcribed from a single TSS. The translational start site (“ATG”) is located within exon 2 (at nt +155 from the TSS) and the translational stop codon is located in exon 3 (at nt +571 from the TSS). Vertical arrows and the table below the sequence illustrate the TSSs found by RACE-PCR. The bold arrow denotes the nucleotide most frequently used as a TSS. B, Three constructs containing KiSS1 5′-flanking region segments of different lengths (KiSS1p556, KiSS1p1339, and KiSS1p1975) were generated to assess KiSS1 transcriptional activity. Putative binding sites for three transcription factors (TTF1, CUX1 and YY1) implicated in the control of puberty were identified in silico (TESS, Vector NTI) and are shown by horizontal wide arrows. The direction of each arrow indicates the presence of each binding site on either the sense or antisense DNA strand. EAP1 binding sites, if present, are currently unknown.
3.2. The 5′-flanking region of KiSS1 contains putative binding sites for transcription factors implicated in the hypothalamic control of female puberty
In silico analysis of the region upstream the KiSS1 TSS, using TESS [http://www.cbil.upenn.edu/cgi-bin/tess/tess; (Schug 2008)], Vector NTI 9.0.0 (Invitrogen; Sep 02, 2003; InforMax, Frederick, USA) and information from literature search (PubMed; http://preview.ncbi.nlm.nih.gov/pubmed/), revealed the presence of several putative TTF1 recognition sites located within 1200 bp upstream of the TSS (Bruno et al. 1995; Guazzi et al. 1990; Son et al. 2003) (Fig. 1B). Several CUX1 binding sites (Harada et al. 1995) were also detected, in addition to recognition sites for YY1 (Fig. 1B), a member of the PcG silencing complex (Park and Atchison 1991; Shi et al. 1991b) that has both repressive and activating functions (Shrivastava and Calame 1994; Thomas and Seto 1999; Wilkinson et al. 2006), and recruits other PcG proteins and histone 3 (H3) trimethylated a lysine 27 to gene promoters to silence transcription (Wilkinson et al. 2006; Woo et al. 2010). TTF1, CUX1 and YY1 have been implicated as upstream transcriptional regulators of the pubertal process (Mastronardi et al. 2006; Roth et al. 2007). We did not search for EAP1 binding sites, because it is not yet known if EAP1 regulates gene transcription by binding directly to DNA. Table 3 provides an overview of the expression of KiSS1, EAP1, TTF1, CUX1 and YY1 mRNAs in the three cell lines used for experiments.
Table 3.
Expression of KiSS1 and transcription factor mRNAs in different cell lines
| KiSS1 | EAP1 | TTF1 | CUX1 | YY1 | |
|---|---|---|---|---|---|
| GripTite™293 MSR | − | + | − | + | + |
| GT1-7 | + | + | − | − | + |
| Hela | + | + | + | + | + |
+= expressed; −= not expressed
The non-neuronal cell line GripTite™293 MSR expresses EAP1, CUX1, and YY1, whereas neuronal GT1-7 cells express KiSS1, EAP1, and YY1. Only Hela cells express all four transcription factors examined in this study, in addition to KiSS1.
3.3. The human KiSS1 promoter is active in both neuronal and non-neuronal cell lines
We used luciferase assays to determine the transcriptional activity of the KiSS1 5′-flanking region using three segments of different lengths (1975 bp, 1339 bp, and 556 bp) (Fig. 1B). These assays showed that the shorter 556 bp fragment was similarly active in non-neuronal GripTite™ 293 MSR cells and neuronal GT1-7 cells, but the longer fragments (1339 bp and 1975 bp) tended to be more active in the non-neuronal cells (Fig. 2). Because of this, all subsequent promoter assays were conducted in GripTite™ 293 MSR cells. We used GT1-7 cells to determine if transcriptional effects on the KiSS1 promoter observed in the non-neuronal cells can be also observed in neuronal cells.
Figure 2. Assessment of transcriptional activity of three KiSS1 promoter constructs using Griptite™ 293 MSR, and GT1-7 cells, and a luciferase reporter system.
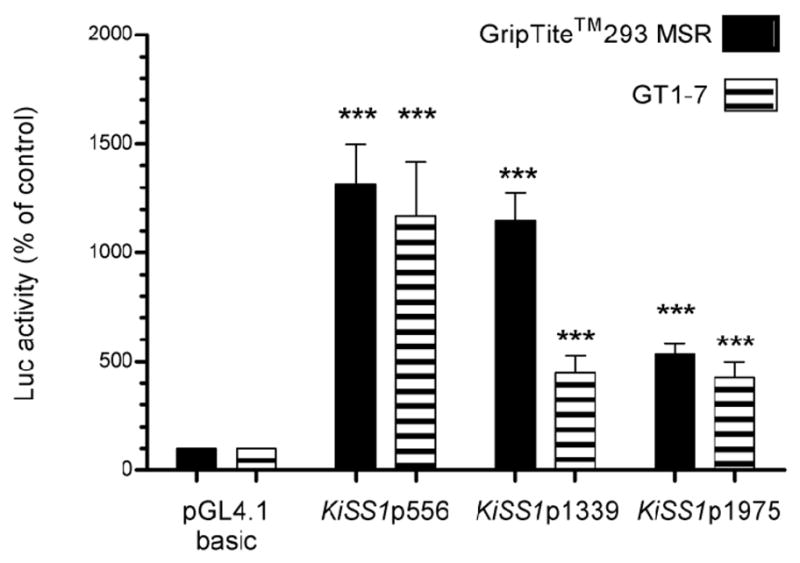
Luciferase activity is shown as percent of the activity displayed by pGL4.10 basic, the luciferase reporter plasmid in which the three KiSS1 constructs were cloned. Bars are means and vertical lines represent SEM. Griptite™ 293 MSR cells n = 3, GT1-7 cells n = 3; *** = P<0.001 compared to pGL4.1 basic vector.
3.4. CUX1, YY1, EAP1 and TTF1 are recruited to the KISS promoter in vivo
CUX1, an evolutionary conserved homeobox transcription factor, has been postulated to be an upstream regulator of female puberty and to target the KiSS1 gene for transcriptional control (Roth et al. 2007). To determine if CUX1 can be recruited to the KiSS1 promoter in vivo, we performed ChIP assays using Hela cells, specific antibodies against CUX1, and the primers listed in Table 1 for PCR amplification. The results show that endogenous CUX1 binds to a region located located ~2,000 bp upstream of the TSS (Fig. 3; Amplicon 1) and to the KISS1 proximal promoter (Amplicon 2), but not to a region located in intron 2 (Amplicon 3). While the PCR product showing CUX1 binding in the KiSS1 proximal promoter contains several putative CUX1 binding sites (Fig. 1B), the more distal (−2456 to −2264 bp) product likely derives from DNA fragments that include putative CUX1 binding sites predicted to be present directly adjacent to the amplified region (Fig. 1B).
Figure 3.
CUX1, YY1, EAP1 and TTF1 are recruited to the KiSS1 promoter in Hela cells, as assessed by ChIP assays. Endogenous CUX1 and YY1, as well as transfected HA-tagged EAP1 and untagged TTF1, are recruited to the proximal promoter region of the KiSS1 gene (Amplicon 2). An upstream region (Amplicon 1) showed Cux1 association, some YY1 binding, and neither EAP1 nor TTF1 association. A downstream region localized in intron 2 (Amplicon 3) showed no binding of any of the four proteins examined. When endogenous proteins (CUX1 and YY1) were immunoprecipitated, we used monoclonal antibodies against each of these proteins and mouse IgGs as a negative control for the immunoprecipitation. When the immunoprecipitating proteins derived from transfected expression vectors (EAP1-HA and TTF1), immunoprecipitation was performed using a mouse monoclonal antibody against HA (for EAP) or rabbit polyclonal antibodies against TTF1. Chromatin prepared from cells transfected with the empty expression vector was used as the negative control. Inp: input DNA, chromatin precleared with protein A beads before immunoprecipitation. Transf = transfected.
YY1 is a transcriptional regulator postulated to be a central node in a gene network involved in the hypothalamic control of female puberty (Roth et al. 2007). As such, YY1 was also predicted by in silico analysis to control a host of subordinate genes, including KiSS1. CHIP assays, using Hela cells, specific antibodies against YY1, and the same primers used to demonstrate CUX1 association to the KiSS1 promoter (Table 1) showed that endogenous YY1 is most efficiently recruited to the KiSS1 proximal promoter (Fig. 3; Amplicon 2), and to a much lesser extent to the upstream −2456 to −2264 bp region (Amplicon 1). In contrast, no YY1 binding to intron 2 (Amplicon 3) was detected.
EAP1 is a transcription factor with trans-activating and repressive activities, previously implicated in the transcriptional control of female puberty (Heger et al. 2007). In the rat, Eap1 is highly expressed in the arcuate nucleus and preoptic area, the two regions where kisspeptin neurons are located (Heger et al. 2007). ChIP assays using Hela cells transfected with an HA-tagged construct expressing the EAP1 coding region demonstrated that EAP1 is exclusively recruited to the KiSS1 proximal promoter (Fig. 3; Amplicon 2)
TTF1 (Nkx2.1, T/ebp) is a homeodomain gene required for basal forebrain morphogenesis (Kimura et al. 1996), but that remains expressed in the hypothalamus after birth (Lee et al. 2001). In the postnatal hypothalamus, TTF1 transactivates genes involved in the stimulatory control of puberty, and represses genes inhibitory to the pubertal process (Mastronardi et al. 2006). ChIP assays using Hela cells transfected with a construct expressing the rat Ttf1 coding region, and the primers described above for PCR amplification, demonstrated that, as is the case of EAP1, TTF1 is only recruited to the KiSS1 proximal promoter (Fig. 3; Amplicon 2).
3.5. EAP1, TTF1, CUX1 and TTF1 have different effects on KiSS1 transcription
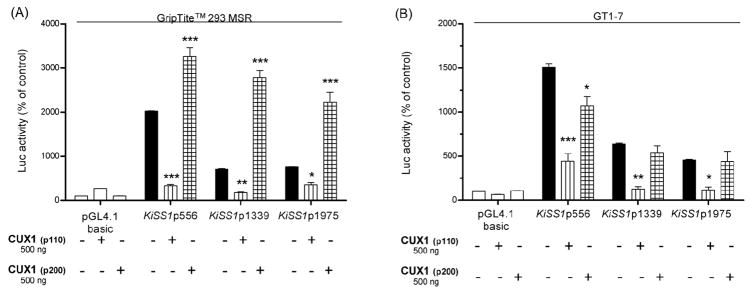
Luciferase assays, using GripTite™ 293 MSR and neuronal GT1-7 cells, demonstrated that the shorter CUX1 isoform (p110) potently inhibited transcription of all three KiSS1 promoter fragments in both non-neuronal and neuronal cells. In contrast, the p200 isoform was strongly trans-activational in GripTite™ 293 MSR cells (Fig. 4A). Surprisingly, p200 was repressive in GT1-7 cells. This activity, however, was limited to the shorter (556 bp) KiSS1 promoter segment (Fig. 4B). These results suggest that CUX1 can either activate or inhibit KiSS1 expression, depending on the predominant isoform present and the cell context in which the protein-DNA interaction takes place.
Figure 4. CUX1 regulates KiSS1 gene expression dually.
A, The p110 isoform of CUX1 inhibits, whereas the full-length CUX1 isoform (p200) trans-activates KiSS1 transcription in GripTite™ 293 MSR cells, n = 3. B, Both isoforms repress KiSS1 transcription in GT1-7 cells, n = 3. Bars are mean and vertical lines are SEM; * = P< 0.05, ** = P< 0.01 and *** = P<0.001 compared to KiSS1 promoter alone.
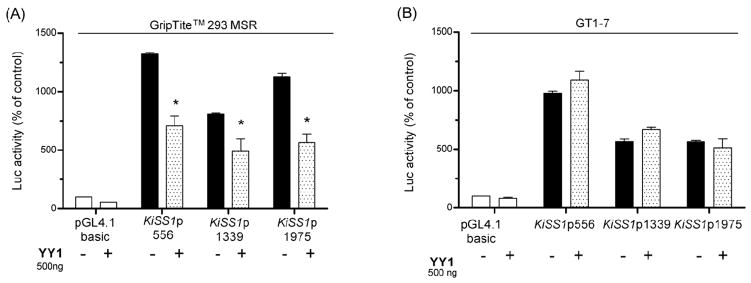
Similar assays performed in GripTite™ 293 MSR, indicate that YY1 significantly inhibited the transcriptional activity of the KiSS1 promoter constructs (Fig. 5A). Because YY1 interacts with Sp1 to regulate gene transcription (Lee et al. 1993), and the KiSS1 proximal promoter has a number of SP1 binding sites (Mitchell et al. 2006; Mitchell et al. 2007), we tested this potential interaction on KiSS1 promoter activity, using GripTite™ 293 MSR cells. The results showed that Sp1 does not affect the repressive effect of YY1 on KiSS1 promoter activity (data not shown). Thus YY1 represses KiSS1 transcription without recruiting Sp1 as a co-repressor. We did not observed an effect of YY1 on KiSS1 gene expression in GT1-7 cells (Fig. 5B).
Figure 5. YY1 inhibits KiSS1 transcriptional activity.
A, YY1 inhibits KiSS1 transcription in GripTite™ 293 MSR cells, n = 3, B, YY1 does not modify KiSS1 transcription in GT1-7 cells. Bars are mean and vertical lines are SEM, n = 3; * = P< 0.05 compared to KiSS1 promoter alone.
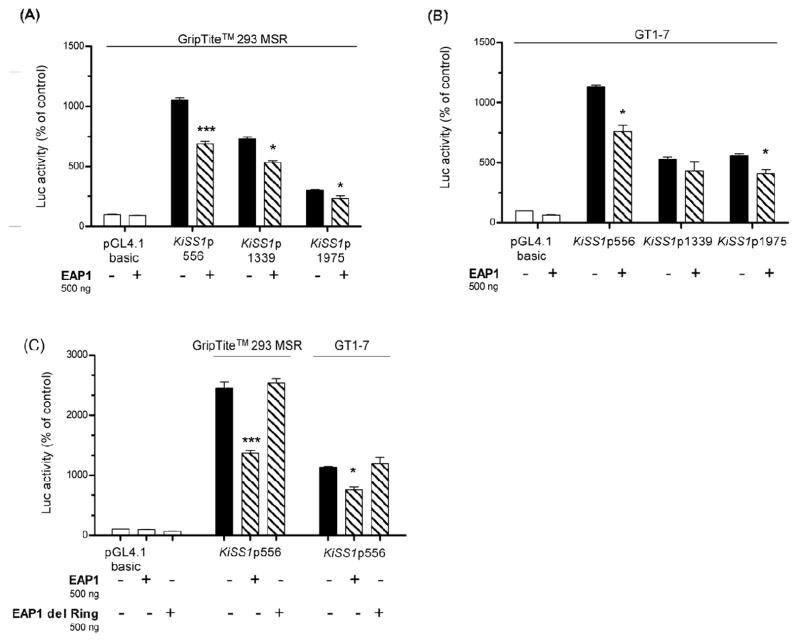
Co-transfection of GripTite™ 293 MSR with pcDNA3.1+/zeo-EAP1 and each of the KiSS1 promoter constructs showed that EAP1 exerts a modest, but significant, inhibitory effect on KiSS1 transcriptional activity of all KiSS1 promoter fragments (Fig. 6A). A similar, though less consistent effect was observed in GT1-7 cells (Fig. 6B). EAP1 lacking the RING finger domain, tested with the 556 bp KiSS1 promoter construct, failed to affect transcriptional activity of this DNA fragment in both non-neuronal cells (GripTite™ 293 MSR) and GT1-7 neurons (Fig. 6C). These results indicate that EAP1 modestly represses KiSS1 transcriptional activity in two different cell lines and that the RING finger domain of EAP1 is required for this effect.
Figure 6. EAP1 represses KiSS1 transcriptional activity in non-neuronal and neuronal cell lines.
A, EAP1 represses transcription of all three KiSS1 promoter constructs in non-neuronal (GripTite™ 293 MSR) cells, n = 3, B, EAP1 represses the activity of two of the three constructs in the neuronal (GT1-7) cell line, n = 3. C, The repressive effect of EAP1 is abolished in both cell lines by deletion of the EAP1 RING finger domain. Bars are mean and vertical lines are SEM, n = 3; * = P< 0.05, and *** = P<0.001 compared to KiSS1 promoter alone.
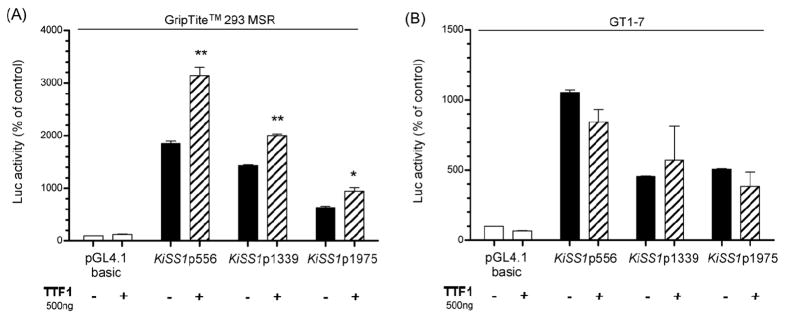
Luciferase reporter assays, using GripTite™ 293 MSR cells, showed that TTF1 consistently trans-activates the short KiSS1 promoter construct (Fig. 7A). However, this effect was not observed in GT 1–7 cells (Fig. 7B). In a previous study we showed that deletion of the TTF1 recognition site most proximal to the TSS (−109 to −100) or the simultaneous deletion of this site plus another site located at −1019 to −1010 in the KiSS1 promoter abolished the trans-activational effect of TTF1 on KiSS1 promoter activity (Mastronardi et al. 2006). Altogether, these results indicate that TTF1 is a trans-activator of KiSS1 transcription.
Figure 7. TTF1 enhances KiSS1 transcription in a non-neuronal cell line, but not in GT1-7 cells.
A, TTF1 trans-activates transcription of each KiSS1 construct tested in GripTite™ 293 MSR cells, n = 3, B, TTF1 does not modify KiSS1 promoter transcription in GT1-7 cells. Bars are mean and vertical lines are SEM, n = 3; * = P< 0.05, and ** = P< 0.01 compared to KiSS1 promoter alone.
3.6. CUX1, YY1, EAP1 and TTF1 are expressed in kisspeptin neurons
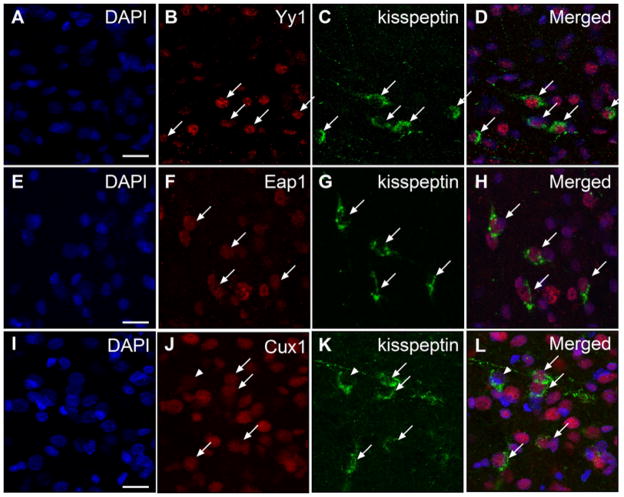
We previously demonstrated that the nuclei of kisspeptin neurons contain Ttf1 immunoreactive material (Mastronardi et al. 2006). To determine if they are also Yy1, Eap1 and/or Cux1 immunopositive, we performed double immunofluorescence studies using hypothalami from late juvenile female rats. The results indicated that each of these proteins is present in kisspeptin neurons of the medial basal hypothalamus (Fig. 8), and that as expected, they have a nuclear localization. However, not all kisspeptin neurons contain all three transcription factors, as shown by the absence of Cux1 in some kisspeptin neurons (Fig. 8; example denoted by arrowhead).
Figure 8. Kisspeptin neurons of the prepubertal female rat hypothalamus contain Eap1, Yy1 and Cux1 as detected by double immunohistofluorescence.
A–D, Yy1. E–H, Eap1. I–L, Cux1. Arrows point to examples of colocalization. Arrowhead in J–L points to a kisspeptin positive neuron lacking detectable Cux1 immunoreactivity. Notice the nuclear localization of Eap1, Yy1 and Cux1. Bar = 10 μm.
4. Discussion
Following the pioneer observations of two different groups (De Roux et al. 2003; Seminara et al. 2003) showing that mutations in the GPR54 gene result in hypogonadotropic hypogonadism, a substantial body of evidence has accumulated demonstrating that the kisspeptin-GPR54 receptor system is a key regulator of puberty and adult reproductive function [reviewed in (Oakley et al. 2009)]. This progress notwithstanding, the factors controlling KiSS1 expression at the transcriptional level have been only partially identified (Mitchell et al. 2006; Mitchell et al. 2007). In the present study, we have experimentally tested computational biology predictions implicating four TFs, previously postulated to be involved in the hypothalamic control of female puberty, as regulators of KiSS1 transcriptional activity (Roth et al. 2007). Not surprisingly, the picture that has emerged from this study is one of complexity, but also one which is consistent with the basic concept that ultimately KiSS1 transcription depends on a delicate balance between activators and repressors.
We found that the KiSS1 TSS is located in a region between −153 bp to −156 bp upstream of the ATG which is consistent with in silico predictions (NM_002256). The absence of other transcripts suggests that at least in placental tissue the KiSS1 gene utilizes a single TSS to generate a primary mRNA transcript. Analysis of the genomic region 5′ to this site revealed the presence of specific promoter elements such as a TATA-box, GC-rich regions, and binding sites for various members of the basic transcriptional machinery, like SP1 and AP1. To the best of our knowledge only one TSS (identical to the one we identified here) has been described for the human KiSS1 gene. An alternative upstream promoter expressed in placenta that includes sequences from the Golt1a has been reported to exist in the mouse (NM AY707856, AY707857, AY707859), but its biological significance has never been assessed experimentally. When examining in silico potential transcripts derived from this hypothetical TSS, we have not been able to obtain a translated kisspeptin sequence, suggesting that utilization of this upstream TSS may not generate a translatable transcript. Nevertheless, the possibility of an alternative TSS usage in nervous tissue is an issue that requires future consideration.
Promoter activity was highest when shorter 5′-flanking region fragments were used, suggesting that enhancer elements are preferentially located near the TSS, whereas repressor elements are located further upstream. Interestingly, the KiSS1 promoter activity of longer DNA fragments was dependent on the cell system used. It tended to be higher in the non-neuronal cell line GripTite™293 MSR than in neuronal GT1-7 cells. A potential explanation for the difference in basal transcriptional activity noted between these two cell lines is that they do not have the same complement of repressors/activators or the same abundance/availability of these proteins. Alternatively, the differences may have been simply due to different transfection efficiencies.
Because the transcriptional control of female puberty appears to be exerted by both TSGs and genes without tumor suppressor activity, we selected two members of each class for our study. EAP1 and TTF1 represent the latter category; both have been suggested to be upstream transcriptional regulators of mammalian puberty, based on physiological, cellular and genetic evidence (Heger et al. 2007; Mastronardi et al. 2006). Of the two, EAP1 has dual transcriptional activity. It trans-activates the GnRH promoter, but inhibits preproenkephalin gene transcription (Heger et al. 2007). Because RNAi-mediated knock-down of Eap1 in the preoptic region of prepubertal female rats results in delayed pubertal onset and disruption of estrous cyclicity (Heger et al. 2007), we expected that EAP1 would increase KiSS1 gene expression; instead, a repressive effect was observed. We do not understand well the physiological significance of such an inhibitory action, but speculate that EAP1 may behave differently in the presence of partners specifically expressed in kisspeptin neurons. It was recently shown that EAP1 physically associates with two other proteins to form a repressive complex that inhibits the expression of a downstream transcriptional repressor (Yeung et al. 2011). These findings suggest that in some cells EAP1 functions as a repressor of repressors. Within the hypothalamus, a function of EAP1 may be to repress genes involved in the inhibitory control of puberty/reproductive cyclicity, so that loss of EAP1 expression would result in pubertal delay and loss of reproductive cyclicity. This is, indeed, the case because knock-down of EAP1 expression in either the preoptic area of rats (Heger et al. 2007) or the arcuate nucleus of nonhuman primates (Dissen G, Lomniczi A, Heger S and Ojeda SR, in preparation) interrupts reproductive cyclicity. These findings and our own observation that EAP1 transactivates, instead of repressing, the GnRH promoter in GT1-7 cells (Heger et al. 2007), suggest that the absence/presence of appropriate partners or downstream targets in a given cell context may determine whether EAP1 will act as a transactivator or repressor. More work is obviously needed to clarify this issue.
In contrast to EAP1, our results show that TTF1 enhances KiSS1 expression. A previous study showed that this trans-activation requires a TTF1 binding site located near the TSS. TTF1 is expressed in kisspeptin neurons of the arcuate nucleus (Mastronardi et al. 2006). An earlier report showed that TTF1 expression increases in the hypothalamus during prepubertal development (Lee et al. 2001), preceding the increase in KiSS1 expression (Clarkson and Herbison 2006; Navarro et al. 2004; Takase et al. 2009). The possibility that this two events are causally related is supported by the present results showing that TTF1 is not only recruited to the KiSS1 promoter in vivo, but it also strongly trans-activates KiSS1 transcription in vitro. Unexpectedly, this trans-activational effect was observed in GripTiteMSR293 cells, but not in GT1-7 cells, suggesting that the ability of TTF1 to trans-activate the KiSS1 promoter is cell-dependent. Although GT1-7 cells are neuronal, they may not have the same complement of co-activators present in GripTiteMSR293 cells (and perhaps also in kisspeptin neurons themselves).
The two TSGs we examined also displayed contrasting activities. The activity of CUX1 was found to be isoform-dependent. Previously we showed that CUX1 mRNA abundance increases in the monkey hypothalamus at the time of puberty, and postulated that CUX1 acts as a central node of a TSG network that operates within the hypothalamus to control the onset of puberty (Roth et al. 2007; Ojeda et al. 2006). In general, CUX1 functions as a transcriptional repressor (Dufort and Nepveu 1994; Superti-Furga et al. 1989; Valarche et al. 1993; Coqueret et al. 1998). However, depending on the cellular context, CUX1 can also act as a transcriptional activator (Harada et al. 2008; Kim et al. 1997). In our experiments we used two different isoforms of CUX1 previously shown to have divergent transcriptional activities (Goulet et al. 2002; Sansregret and Nepveu 2008). Our results confirm this isoform-specific activity. While p200, the full- length form of CUX1, behaved as a trans-activator in non-neuronal cells and displayed limited repressive activity in GT1-7 cells, the proteolytically derived form p110 was consistently repressive. Whether p200 behaves as a repressor or activator in kisspeptin neurons themselves remains an open question. Our immunohistochemical studies show that the CUX1 protein is present in kisspeptin neurons. However, the lack of isoform-specific antibodies does not allow us to distinguish between the predominant form or to screen for a shift in relative abundance at the time of puberty. It may be speculated that if expression of the p200 and p110 forms decreases at puberty in kisspeptin neurons, a potentially repressive effect of CUX1 on KiSS1 transcription would be lifted. This shift would be expected to contribute to the pubertal increase in KiSS1 mRNA abundance (Clarkson and Herbison 2006; Navarro et al. 2004; Takase et al. 2009).
Like CUX1 and EAP1, YY1 can display both repressive and trans-activating activities (Shi et al. 1997; He and Casaccia-Bonnefil 2008; He et al. 2007). Our results show that YY1 is repressive in GripTiteMSR293 cells, but had no effect in GT1-7 cells. As in the case of TTF1, this difference may be due to the absence of appropriate partners in GT1-7 cells. It is not known if the repressive effect of YY1 we observed in GripTite™ 293 MSR cells occurs in kisspeptin neurons themselves. These cells may contain a different complement of trans-activators/repressors that may either reinforce the repressive effect of YY1 or switch it to activation, as shown in other systems (Gordon et al. 2006). Further studies are required to resolve this issue.
The repressive effect of YY1 on KiSS1 transcription is consistent with its well-established gene silencing activity (He et al. 2007; Wilkinson et al. 2006; Woo et al. 2010), and its contribution to the recruitment of PcG repressive proteins to gene promoters (Schwartz and Pirrotta 2007; Woo et al. 2010). Because of its known interaction with Sp1 (Lee et al. 1993), YY1 action was analyzed in combination with Sp1 and observed that, at least in GripTite™ 293 MSR cells, Sp1 does not alter the repressive activity of YY1. The increase in Kiss1 mRNA abundance observed in the rodent hypothalamus at puberty (Clarkson and Herbison 2006; Navarro et al. 2004; Takase et al. 2009) and the repression of KiSS1 transcription by YY1 we observed in GripTiteMSR293 cells, suggest that this repression is regulated developmentally. Should kisspeptin neurons expressed the appropriate partners for YY1 to exert its well-established repressive effects, one could envision a strong repression taking place during early juvenile days when KiSS1 expression is low, and loss of this inhibition near the time of puberty when KiSS1 expression increases.
Taken together, the present study provides experimental evidence supporting the in silico prediction that upstream regulators of the pubertal process control KiSS1 transcription by directly modifying KiSS1 promoter activity. The results also suggest that in vivo expression of the KiSS1 gene depends on a balance between repressors and inhibitors able to modify KiSS1 promoter activity. Although the above discussion focuses on the significance of these interactions for female puberty, it seems reasonable to infer that the KiSS1 promoter is similarly regulated in males.
Kisspeptin is a key component that controls puberty and reproductive development
Transcription of KiSS1 mRNA is initiated at a single transcription start site
KiSS1 promoter is activated by TTF1 and CUX1-p200
KiSS1 promoter is repressed by EAP1, YY1, and CUX1-p110
All four TFs are recruited in vivo to the KiSS1 promoter and are expressed in kisspeptin neurons
Acknowledgments
This work was supported by German Research Foundation Grant HE 3151/3-1, 3151/4-1 (SH), and NIH grants HD25123, the Eunice Kennedy Shriver NICHD/NIH through cooperative agreement [U54 HD18185] as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research, and RR000163 for the operation of the Oregon National Primate Research Center (SRO). We thank Ms. Maria Costa for performing the immunohistofluorescence and in situ studies and Dr. Anda Cornea (Director of the Imaging Core at the ONPRC) for her assistance with confocal microscopy imaging. We thank Prof. Dr. Sigurd Lenzen (Institute of Clinical Biochemistry, Hannover Medical School) for his excellent methodological support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Brann DW. Glutamate: A major excitatory transmitter in neuroendocrine regulation. Neuroendocrinology. 1995;61:213–225. doi: 10.1159/000126843. [DOI] [PubMed] [Google Scholar]
- Bruno MD, Bohinski RJ, Huelsman KM, Whitsett JA, Korfhagen TR. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. Journal of Biological Chemistry. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE. Development of GABA and glutamate signaling at the GnRH neuron in relation to puberty. Mol Cell Endocrinol. 2006;254–255:32–38. doi: 10.1016/j.mce.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Anglemont de Tassigny X, Fagg LA, Dixon JP, Day K, Leitch HG, Hendrick AG, Zahn D, Franceschini I, Caraty A, Carlton MB, Aparicio SA, Colledge WH. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci U S A. 2007;104:10714–10719. doi: 10.1073/pnas.0704114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufort D, Nepveu A. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- Goulet B, Watson P, Poirier M, Leduy L, Berube G, Meterissian S, Jolicoeur P, Nepveu A. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625–6633. [PubMed] [Google Scholar]
- Guazzi S, Price M, De Felice M, Damante G, Mattei MG, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO Journal. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada R, Berube G, Tamplin OJ, is-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada R, Vadnais C, Sansregret L, Leduy L, Berube G, Robert F, Nepveu A. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 2008;36:189–202. doi: 10.1093/nar/gkm970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Casaccia-Bonnefil P. The Yin and Yang of YY1 in the nervous system. J Neurochem. 2008;106:1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117:2145–2154. doi: 10.1172/JCI31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Shannon EM, Fritschy JM, Ojeda SR. Several GABAA receptor subunits are expressed in LHRH neurons of juvenile female rats. Brain Research. 1997;780:218–229. doi: 10.1016/s0006-8993(97)01152-9. [DOI] [PubMed] [Google Scholar]
- Kim EC, Lau JS, Rawlings S, Lee AS. Positive and negative regulation of the human thymidine kinase promoter mediated by CCAAT binding transcription factors NF-Y/CBF, dbpA, and CDP/cut. Cell Growth Differ. 1997;8:1329–1338. [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: Thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes and Development. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kordon C, Drouva SV, Martínez de la Escalera G, Weiner RI. Role of classic and peptide neuromediators in the neuroendocrine regulation of luteinizing hormone and prolactin. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2. Vol. 1. Raven Press; New York: 1994. pp. 1621–1681. [Google Scholar]
- Lapatto R, Pallais JC, Zhang D, Chan YM, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB. Kiss1−/− mice exhibit more variable hypogonadism than Gpr54−/− mice. Endocrinology. 2007;148:4927–4936. doi: 10.1210/en.2007-0078. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cho GJ, Norgren R, Junier MP, Hill DF, Tapia V, Costa ME, Ojeda SR. TTF-1, a homeodomain gene required for diencephalic morphogenesis, is postnatally expressed in the neuroendocrine brain in a developmentally regulated and cell-specific fashion. Molecular and Cellular Neurosciences. 2001;17:107–126. doi: 10.1006/mcne.2000.0933. [DOI] [PubMed] [Google Scholar]
- Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, Welch DR. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- Lee JS, Galvin KM, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci U S A. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X, Yu H, Wei X, Zhou Y, Wang W, Li P, Gan X, Wei D, Xiao J. GPR54 Polymorphisms in Chinese Girls with Central Precocious Puberty. Neuroendocrinology. 2007;86:77–83. doi: 10.1159/000107511. [DOI] [PubMed] [Google Scholar]
- Luong MX, van der Meijden CM, Xing DX, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Molecular and Cellular Biology. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi C, Smiley G, Kuswakabe T, Kawagushi A, Cabrera R, Mungenast A, Kimura S, Ojeda SR. Program No.143.2, 2004 Abstract Viewer. Washington, DC: Society for Neuroscience; 2004. Neuronal deletion of the T/EBP gene delays female puberty and causes premature reproductive senescence. 2004. Online. [Google Scholar]
- Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR. Deletion of the Ttf1 Gene in Differentiated Neurons Disrupts Female Reproduction without Impairing Basal Ganglia Function. Journal of Neuroscience. 2006;26:13167–13179. doi: 10.1523/JNEUROSCI.4238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DC, Stafford LJ, Li D, Bar-Eli M, Liu M. Transcriptional regulation of KiSS-1 gene expression in metastatic melanoma by specificity protein-1 and its coactivator DRIP-130. Oncogene. 2007;26:1739–1747. doi: 10.1038/sj.onc.1209963. [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Abdelrahim M, Weng J, Stafford LJ, Safe S, Bar-Eli M, Liu M. Regulation of KiSS-1 Metastasis Suppressor Gene Expression in Breast Cancer Cells by Direct Interaction of Transcription Factors Activator Protein-2{alpha} and Specificity Protein-1. Journal of Biological Chemistry. 2006;281:51–58. doi: 10.1074/jbc.M506245200. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Castellano JM, Fernández-Fernández R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology. 2004;145:4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O, Fujino M. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Lomniczi A, Mastronardi C, Heger S, Roth C, Parent AS, Matagne V, Mungenast AE. Minireview: The neuroendocrine regulation of puberty: Is the time ripe for a systems biology approach? Endocrinology. 2006;147:1166–1174. doi: 10.1210/en.2005-1136. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Prevot V, Heger S, Lomniczi A, Dziedzic B, Mungenast A. Glia-to neuron signaling and the neuroendocrine control of female puberty. Ann Med. 2003;35:244–255. doi: 10.1080/07853890310005164. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Skinner MK. Puberty in the rat. In: Neill JD, editor. The Physiology of Reproduction. 3. Academic Press/Elsevier; San Diego: 2006. pp. 2061–2126. [Google Scholar]
- Ojeda SR, Terasawa E. Neuroendocrine regulation of puberty. In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Moss R, Rubin R, editors. Hormones, Brain and Behavior. Vol. 4. Elsevier; New York: 2002. pp. 589–659. [Google Scholar]
- Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci U S A. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Kessel M. Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev Genes Evol. 1998;208:168–171. doi: 10.1007/s004270050170. [DOI] [PubMed] [Google Scholar]
- Plant TM, Witchel SF. Puberty in nonhuman primates and humans. In: Neill JD, editor. The Physiology of Reproduction. 3. Academic Press/Elsevier; San Diego: 2006. pp. 2177–2230. [Google Scholar]
- Price M, Lazzaro D, Pohl T, Mattei MG, Rüther U, Olivo JC, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8:241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Guerriero KA, Gibbs RB, Plant TM. Structural interactions between kisspeptin and GnRH neurons in the mediobasal hypothalamus of the male rhesus monkey (Macaca mulatta) as revealed by double immunofluorescence and confocal microscopy. Endocrinology. 2008;149:4387–4395. doi: 10.1210/en.2008-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth CL, Mastronardi C, Lomniczi A, Wright H, Cabrera R, Mungenast AE, Heger S, Jung H, Dubay C, Ojeda SR. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology. 2007;11:5147–5161. doi: 10.1210/en.2007-0634. [DOI] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008;Chapter 2(Unit) doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. New England Journal of Medicine. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- Semple RK, Achermann JC, Ellery J, Farooqi IS, Karet FE, Stanhope RG, O’Rahilly S, Aparicio SA. Two novel missense mutations in G protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. Journal of Clinical Endocrinology Metabolism. 2005;90:1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: An enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. Journal of Histochemistry and Cytochemistry. 1991a;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1…. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991b;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Research. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son YJ, Hur MK, Ryu BJ, Park SK, Damante G, D’Elia AV, Costa ME, Ojeda SR, Lee BJ. TTF-1, a homeodomain-containing transcription factor, participates in the control of body fluid homeostasis by regulating angiotensinogen gene transcription in the rat subfornical organ. Journal of Biological Chemistry. 2003;278:27043–27052. doi: 10.1074/jbc.M303157200. [DOI] [PubMed] [Google Scholar]
- Superti-Furga G, Barberis A, Schreiber E, Busslinger M. The protein CDP, but not CP1, footprints on the CCAAT region of the gamma-globin gene in unfractionated B-cell extracts. Biochim Biophys Acta. 1989;1007:237–242. doi: 10.1016/0167-4781(89)90046-8. [DOI] [PubMed] [Google Scholar]
- Takase K, Uenoyama Y, Inoue N, Matsui H, Yamada S, Shimizu M, Homma T, Tomikawa J, Kanda S, Matsumoto H, Oka Y, Tsukamura H, Maeda KI. Possible role of oestrogen in pubertal increase of Kiss1/kisspeptin expression in discrete hypothalamic areas of female rats. J Neuroendocrinol. 2009;21:527–537. doi: 10.1111/j.1365-2826.2009.01868.x. [DOI] [PubMed] [Google Scholar]
- Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum-Rakover Y, Commenges-Ducos M, Iovane A, Aumas C, Admoni O, De Roux N. Neuroendocrine phenotype analysis in five patients with isolated hypogonadotropic hypogonadism due to a L102P inactivating mutation of GPR54. J Clin Endocrinol Metab. 2007;92:1137–1144. doi: 10.1210/jc.2006-2147. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Fernandez DL. Neurobiological mechanisms of the onset of puberty in primates. Endocrine Reviews. 2001;22:111–151. doi: 10.1210/edrv.22.1.0418. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Valarche I, Tissier-Seta JP, Hirsch MR, Martinez S, Goridis C, Brunet JF. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. Journal of Biological Chemistry. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda S, Xu CF, Neubert TA, Samuels HH. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011:2011. doi: 10.1128/MCB.01381-10. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]