Abstract
Sphingosine kinase 2 (SK2) catalyzes the conversion of sphingosine to the bioactive lipid sphingosine 1-phosphate (S1P). We report here, the stereospecific synthesis of an analogue of FTY720 called (R)-FTY720-OMe, which we show is a competitive inhibitor of SK2. (R)-FTY720-OMe failed to inhibit sphingosine kinase 1 activity, thereby demonstrating specificity for SK2. Prolonged treatment of HEK 293 cells with (R)-FTY720-OMe also induced a reduction in SK2 expression. In addition, (R)-FTY720-OMe inhibited DNA synthesis and prevented S1P-stimulated rearrangement of actin in MCF-7 breast cancer cells. These findings demonstrate that SK2 functions as a pro-survival protein and is involved in promoting actin rearrangement into membrane ruffles/lamellipodia in response to S1P in MCF-7 breast cancer cells.
INTRODUCTION
Sphingosine 1-phosphate (S1P) is a bioactive lipid that binds to specific G-protein coupled receptors (S1P1–5) (1). S1P is formed by the action of sphingosine kinase, which catalyses the phosphorylation of sphingosine. There are two isoforms termed SK1 and SK2. Degradation of S1P is catalysed by S1P lyase or dephosphorylated to sphingosine, catalysed by S1P phosphatase. S1P has been implicated in a number of diseases, including cancer (1) and therefore, there is a need to develop therapeutic agents that can interrupt S1P signalling in these diseases. Among the numberous agents that target S1P biology, the sphingosine analogue FTY720 has attracted considerable attention, and is now licensed (as Gilenya™) for oral treatment of multiple sclerosis (2, 3). FTY720 is a functional S1P1 antagonist that down-regulates S1P1 in T-lymphocytes (2). FTY720 (a synthetic sphingosine analogue) is phosphorylated to (S)-FTY720 phosphate by SK2 (4), and when released from cells binds to four of the five S1P receptors (S1P2 being the exception) (2) leading to proteasomal degradation of S1P1 in T-lymphocytes (5). S1P/S1P1 is required for the egress of T-lymphocyte from lymph nodes: FTY720 phosphate inhibits this process and therefore acts as an immunosuppressant (6).
The over-expression of SK2 causes suppression of cell growth and cell cycle arrest and requires nuclear localisation (7). SK2 can also induce apoptosis that is preceded by mitochondrial release of cytochrome c and activation of caspase-3 (8). Indeed, SK2 contains a BH3 domain that sequesters BCL-XL and abrogates its anti-apoptotic function (8). Thus, SK2 can be pro-apoptotic. However, other studies support a role for SK2 in promoting cancer cell survival. For instance, knock-down of SK2 in breast or colon cancer cells counters doxorubicin-induced expression of p21 (a cyclin-dependent kinase inhibitor) and G2/M arrest and increases doxorubicin-induced apoptosis (9) whereas knock-down of SK2 in glioblastoma cells inhibits proliferation more effectively than knock-down of SK1 (10).
Recently, histone deacetylase has been identified as an intracellular target for nuclear localised SK2-derived S1P (11). S1P binds to and inhibits histone deacetylases (HDAC1 and HDAC2) within repressor complexes that are enriched at the promoters of genes encoding p21 (a cyclin dependent kinase inhibitor) and c-fos (a transcriptional regulator). Thus, S1P promotes expression of p21 and c-fos by inhibiting HDACs and increasing histone acetylation. Therefore it appears appropriate to design and synthesise small molecule inhibitors that can specifically inhibit SK2 to reduce nuclear S1P levels, and thereby modulate the epigenetic effects of S1P that might be involved in diseases such as cancer.
In this regard, we have focussed on synthesising potential SK2 inhibitors. The studies described herein reveal that (R)-FTY720-OMe, a new analogue in which one of the prochiral hydroxyl groups of FTY720 has been replaced by a methoxy group, is a specific competitive inhibitor of SK2. The rationale for replacing one of the hydroxyl groups with a methylester was to block the site that is phosphorylated by SK2. The inhibition was enantioselective, since its enantiomer, (S)-FTY720-OMe, failed to inhibit SK2. (R)-FTY720-OMe also induced a reduction in the expression of SK2 and inhibited DNA synthesis in HEK 293 cells and stimulated actin focal adhesion assembly in MCF-7 cells. These findings indicate that (R)-FTY720-OMe exhibits novel properties that are favourable for anti-breast cancer activity.
MATERIALS AND METHODS
Materials
All general biochemicals and anti-actin antibody were from Sigma (Poole, UK). High glucose Dulbecco’s modified Eagle’s Medium (DMEM), Minimum Essential Medium (MEM), penicillin-streptomycin (10000 U/ml penicillin and 10 mg/ml streptomycin) and Lipofectamine 2000™ were from Invitrogen (Paisley, UK). MCF-7 parental and MCF-7 Neo cells were gifts from R. Schiff (Baylor College of Medicine, Houston, TX, USA). Anti-myc antibody was from Santa Cruz Biotechnology (USA). Anti-PARP antibody was from Cell Signalling Technology (supplied by New England Biolabs Ltd., Hitchin, UK). Sphingosine and S1P were from Avanti Polar Lipids (Alabaster, AL, USA). MG132 and purified SK2 were from Enzo Life Sciences (Exeter, UK). CA074Me was from Merck Biosciences (Nottingham, UK). 4′,6-Diamidino-2-phenylindole (DAPI) was from Vector Labs (UK).
Cell Culture
MCF-7 (parental, Neo) breast cancer cells were grown in a monolayer culture in high glucose DMEM with 10% European Fetal Calf Serum (EFCS) and 100 U/ml penicillin, 100 μg/ml streptomycin, 0.4% Geneticin (absent for parental cells), and 15 μg/ml insulin at 37°C with 5% CO2. HEK 293 cells were cultured in MEM supplemented with 10% EFCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% non-essential amino acids at 37°C in 5% CO2. HEK 293 cells were transfected with Lipofectamin 2000™ reagent and myc-tagged SK1 or SK2 plasmid constructs and grown for 48 h.
General Methods for Chemical Synthesis of (R)-FTY720-OMe
All reactions were carried out under a dry nitrogen atmosphere using oven-dried glassware and magnetic stirring. N,N-Dimethylformamide (DMF) was distilled over CaH2. Silica gel 60 F254 aluminum-backed TLC plates of 0.2-mm thickness were used to monitor the reactions. The spots were visualized with short wavelength ultraviolet light or by charring after spraying with 15% H2SO4. Flash chromatography was carried out with silica gel 60 (230–400 ASTM mesh). 1H and 13C NMR spectra were recorded at 400 and 100 MHz on a Bruker spectrometer, respectively. Chemical shifts (δ) were referenced on residual solvent peaks: CDCl3 (δ = 7.26 ppm for 1H NMR and 77.00 ppm for 13C NMR). Optical rotations were measured on a digital polarimeter at room temperature in a 1.0-dm cell. High-resolution mass spectra (HRMS) were acquired on an Agilent Technologies G6520A Q-TOF mass spectrometer using electrospray ionization (ESI).
Western Blotting
Cells were harvested in sample buffer (125 mM Tris, pH 6.7, 0.5 mM Na4P2O7, 1.25 mM EDTA, 0.5% w/v SDS containing 1.25% v/v glycerol, 0.06% w/v bromophenol blue and 50 mM dithiothreitol) and were subjected to SDS-PAGE and Western blotting as previously described (12).
Sphingosine Kinase Activity Assay
SK2 activity was optimised according to Liu et al. (13) where sphingosine was complexed with bovine serum albumin (final concentration 0.2 mg/ml) in reaction buffer 1 containing 20 mM Tris (pH 7.4), 1 mM EDTA, 1 mM Na3VO4, 40 mM β-glycerophosphate, 1 mM NaF, 0.007% (v/v) β-mercaptoethanol, 20% (v/v) glycerol, 10 μg/ml aprotinin, 10 μg/ml soybean trypsin inhibitor, 1 mM PMSF, and 0.5 mM 4-deoxypyridoxine and 400 mM KCl. Inhibition of SK2 activity was determined by incubating 15 ng of purified SK2 for 15–20 min at 30°C, in the presence of 0.5 to 20 μM sphingosine, 250 μM of [γ32P]ATP (4.4×104 cpm/nmol) in 10 mM MgCl2, and varying concentrations of inhibitors dissolved in DMSO or control (5% DMSO). SK1 activity was assayed as described previously (14). Sphingosine was solubilised in Triton X-100 (final concentration 0.063% w/v) and combined with buffer 1 without KCl. SK1 activity was determined by incubating 15 ng purified SK1 for 15–20 min at 30°C, in the presence of 10 μM sphingosine, 250 μM of [γ32P]ATP (4.4×104 cpm/nmol) in 10 mM MgCl2 with or without inhibitor dissolved in DMSO or control (5% DMSO). SK1 and SK2 reactions were terminated by the addition of 500 μ1 of 1-butanol and mixed with 1 ml of 2M KCl. The organic phase containing [32P]-S1P was then extracted by washing twice with 1 ml of 2 M KCl before quantification by Cerenkov counting. Kinetic parameters were calculated using the graph plotting and curve fitting programs Biograph (University of Strathclyde, Glasgow UK) and Prism 4.03 (GraphPad). Substrate kinetics were analysed according to the Michaelis-Menten equation and the inhibition constant (Kic) was determined using Dixon plots (15).
[3H]-Thymidine Incorporation Assay
MCF-7 cells were plated at 2.0–4.0 × 104 cells/well in 24-well plates and maintained overnight in complete medium. Subsequently, cells were treated with varying concentrations of the inhibitor dissolved in DMSO or vehicle control (0.1% DMSO). Cells were then incubated for another 24 h and pulsed with [3H]-thymidine (0.5 μCi/ml) for 5 h before termination by washing 3 times with 1 ml of ice-cold 10% (w/v) trichloroacetic acid. [3H]-Thymidine incorporated into DNA was isolated with 0.25 ml of 0.1% (w/v) SDS and 0.3 M NaOH and quantified by liquid scintillation counting with 2 ml of scintillation cocktail.
Fluorescence Microscopy
Cells were plated onto autoclaved 13-mm glass coverslips and grown to 60% confluence before serum-starvation for 48 h prior to stimulation. Cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min, and permeabilised with 0.1% Triton X-100 in PBS for 1 min before incubation in blocking solution (5% EFCS and 1% BSA in PBS) for 1 h at room temperature. Coverslips were then incubated with phalloidin red (1:100 dilution in blocking solution) for 1 h at room temperature. Coverslips were washed with PBS and mounted on glass slides using Vectashield® hard set mounting medium with DAPI. Actin rearrangement was assessed using phalloidin red staining. Fluorescence was visualised using a Nikon (Surrey, UK) E600 epifluorescence microscopy.
Chemical Synthesis
(2S)-2-(Methoxymethyl)-(2′-(trichloromethyl)-4′, 5′-dihydrooxazol-5-yl)-4-(4″-n-octylphenyl)-butan-1-ol [(S)-(+)-2] was prepared as follows. To a solution of (2S)-2-(2′-(trichloromethyl)-4′, 5′-dihydrooxazol-5-yl)-4-(4″-n-octylphenyl)-butan-1-ol ((S)-1, 20 mg, 0.046 mmol) in dry DMF (5 mL) was added NaH (3.7 mg, 60%, 0.092 mmol) portionwise at ice-bath temperature. The mixture was stirred at this temperature for 10 min before CH3I (5.7 μL, 0.092 mmol) was added. The reaction mixture was stirred overnight at room temperature, and then poured onto ice. The mixture was extracted with Et2O, and the combined organic extracts were dried (Na2SO4) and evaporated. The residue was purified by column chromatography (hexane/EtOAc 3:1) to give 17 mg (80%) of oxazoline methyl ether (S)-2 as a colorless oil: Rf 0.80 (hexane/EtOAc 3:1); [α]25D +25.1° (c 1.0, CHCl3). 1H NMR (CDCl3) δ 0.88 (t, 3H, J = 6.8 Hz), 1.26–1.29 (m, 10H), 1.56–1.59 (m, 2H), 1.91 (m, 1H), 2.04 (m, 1H), 2.56 (t, 2H, J = 7.6 Hz), 2.62 (m, 2H), 3.39 (s, 3H), 3.49 (s, 2H), 4.40 (d, 1H, J = 8.8 Hz), 4.61 (d, 1H, J = 8.8 Hz), 7.09 (s, 4H); 13C NMR (CDCl3) δ 14.1, 22.7, 29.1, 29.3, 29.4, 29.5, 31.6, 31.9, 35.6, 38.0, 59.6, 74.8, 86.6, 128.2, 128.5, 138.4, 140.7, 162.5. ESI-HRMS (M+H)+ m/z calcd for C22H33Cl3NO2+ 448.1571, found 448.1572.
(2R)-2-Amino-3-(O-methyl)-(2-(4′-n-octylphenyl)ethyl)propanol [(R)-FTY720-OMe] was prepared as follows. To a solution of 15 mg (0.033 mmol) of (S)-2 in 5 mL of tetrahydrofuran (THF) at room temperature was added 1 mL of 1 M HCl. After the reaction mixture was stirred overnight, the solvent was evaporated, and the residue was dissolved in EtOAc (20 mL) and then neutralized with saturated aqueous Na2CO3 solution. The organic phase was dried with MgSO4 and the solvents were removed. The residue was purified by chromatography (CHCl3/MeOH 4:1) to give 10 mg (93%) of (R)-FTY720-OMe as a pale yellow oil; Rf 0.33 (CHCl3/MeOH 4:1); [α]25D −2.8° (c 0.4, CHCl3). 1H NMR (CDCl3) δ 0.88 (t, 3H, J = 6.8 Hz), 1.26–1.33 (m, 10H), 1.58 (m, 2H), 1.72 (m, 2H), 2.05 (br s, 3H), 2.55 (t, 2H, J = 7.6 Hz), 2.60 (m, 4H), 3.32 (d, 1H, J = 9.2 Hz), 3.41 (d, 1H, J = 9.2 Hz), 3.44 (d, 1H, J = 10.8 Hz), 3.56 (d, 1H, J = 10.8 Hz), 7.09 (s, 4H); 13C NMR (CDCl3) δ 14.1, 22.7, 29.2, 29.3, 294, 29.5, 31.6, 31.9, 35.6, 37.7, 55.3, 59.4, 67.8, 78.5, 128.1, 128.5, 139.3, 140.5. ESI-HRMS (M+H)+ m/z calcd for C20H36NO2+ 322.2746, found 322.2743.
RESULTS AND DISCUSSION
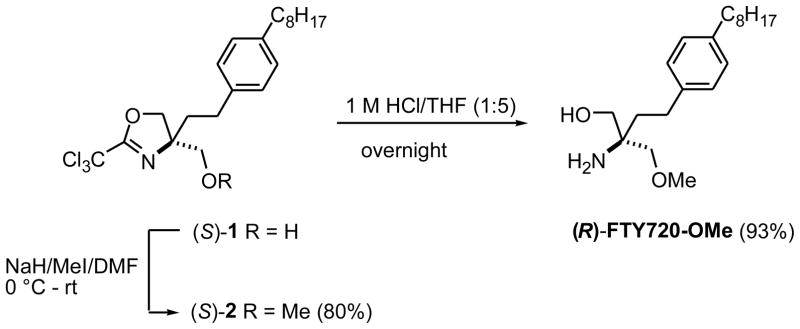
To prepare (R)-FTY720-OMe, one of the prochiral hydroxy groups of FTY720 was replaced by a methoxy group as depicted in Fig. 1. The starting oxazoline (S)-1 was prepared according to a reported procedure (16) and was converted to methylether (S)-2 in good yield by reaction with sodium hydride and methyl iodide in dry dimethylformamide (DMF). The hydroxy and amino groups were released by treatment with 1 M HCl, affording (R)-FTY720-OMe in 93% overall yield. (S)-FTY720-OMe was synthesized from (R)-1 in an analogous manner.
Fig. 1.
Chemical synthesis of (R)-FTY720-OMe.
(R)-FTY720-OMe is a Competitive Specific Inhibitor of SK2
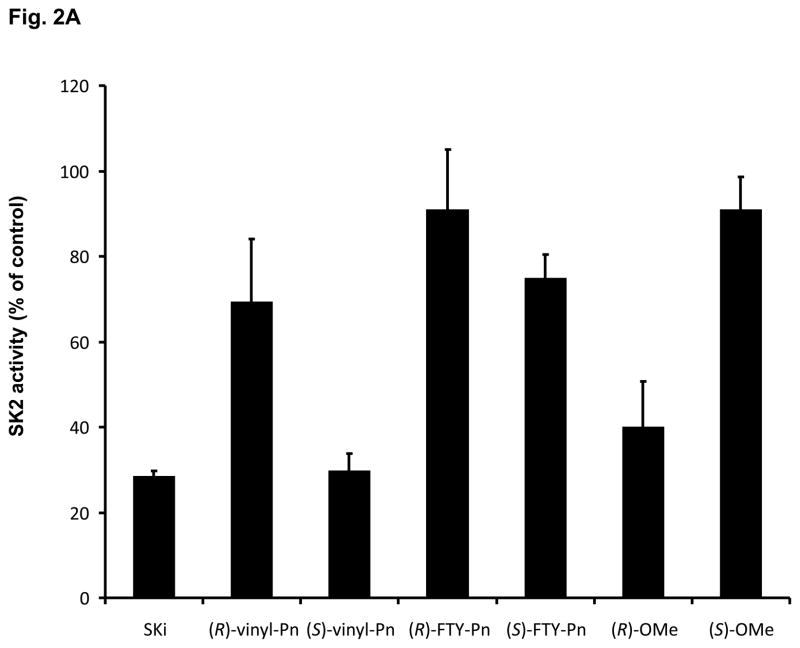
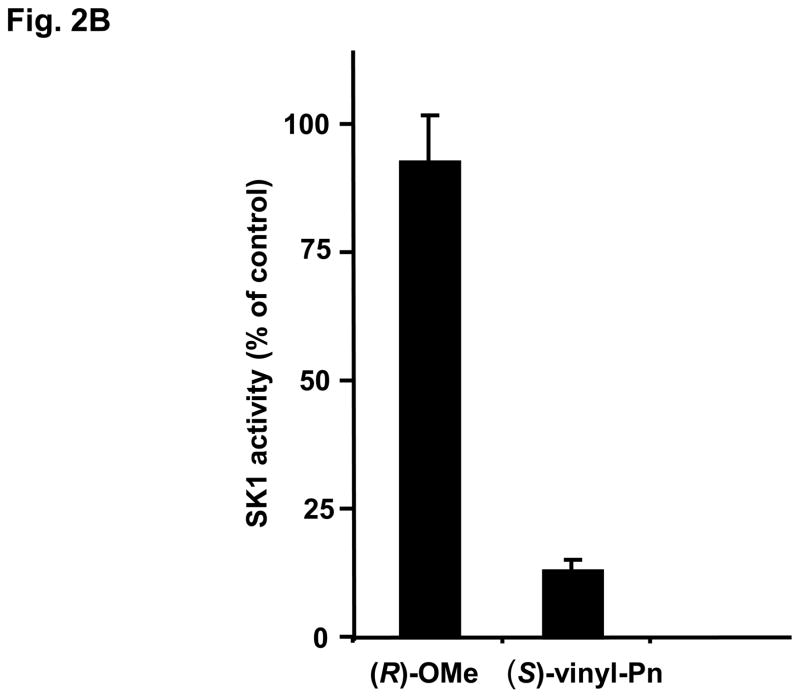
(R)-FTY720-OMe inhibited purified SK2 (Fig. 2A), but not SK1 activity (Fig. 2B). In contrast, (S)-FTY720-OMe did not inhibit purified SK2 activity (Fig. 2A), indicating that there is stereoselective inhibition with (R)-FTY720-OMe.
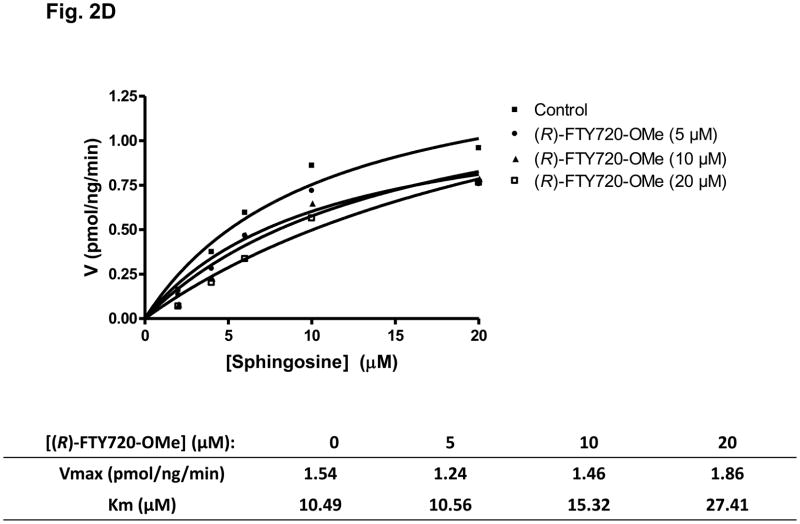
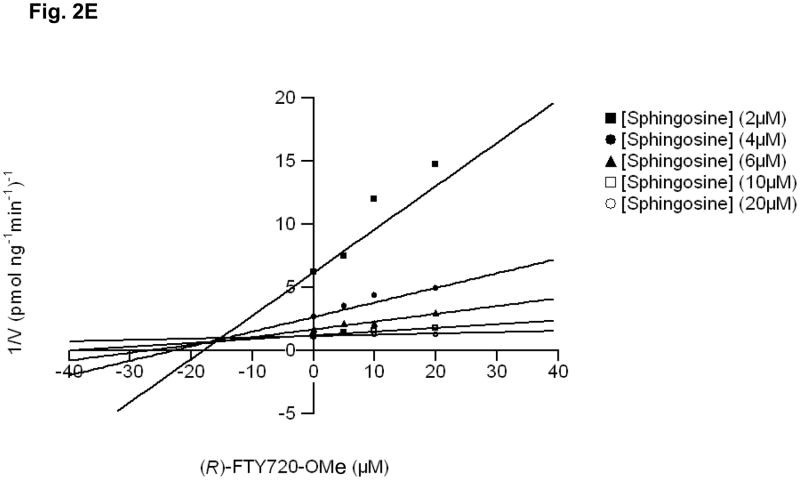
Fig. 2. Inhibitor kinetic analysis of SK2.
(A) Bar graph showing the effects of SKi, (S)- and (R)-FTY720 vinylphosphonate ((R) or (S)-vinyl-Pn) (S)- and (R)-FTY720 phosphonate (R)- or (S)-FTY-Pn, and (S)- and (R)-FTY720-OMe ((S)-OMe and (R)-OMe) (all at 50 μM) on purified SK2 activity (assayed with 10 μM sphingosine, n > 6). Results are expressed as % of SK2 activity in the absence of inhibitors (control = 100% SK2 activity). (B) Bar graph showing the effect of (S)-FTY720 vinylphosphonate (S)-vinyl-Pn or (R)-FTY720-OMe (R)-OMe (both at 50 μM) on purified SK1 activity (assayed with 10 μM sphingosine). Results are expressed as % of SK1 activity in the absence of (S)-FTY720 vinylphosphonate or (R)-FTY720-OMe (control = 100% SK1 activity). (C) Concentration dependence of inhibition of purified SK2 activity (assayed with 10 μM sphingosine) by (R)-FTY720-OMe. (D) V versus S non-linear regression analysis for purified SK2 with (R)-FTY720-OMe. (e) Dixon plot for Ki determinations for (R)-FTY720-OMe. Results are representative of three independent experiments.
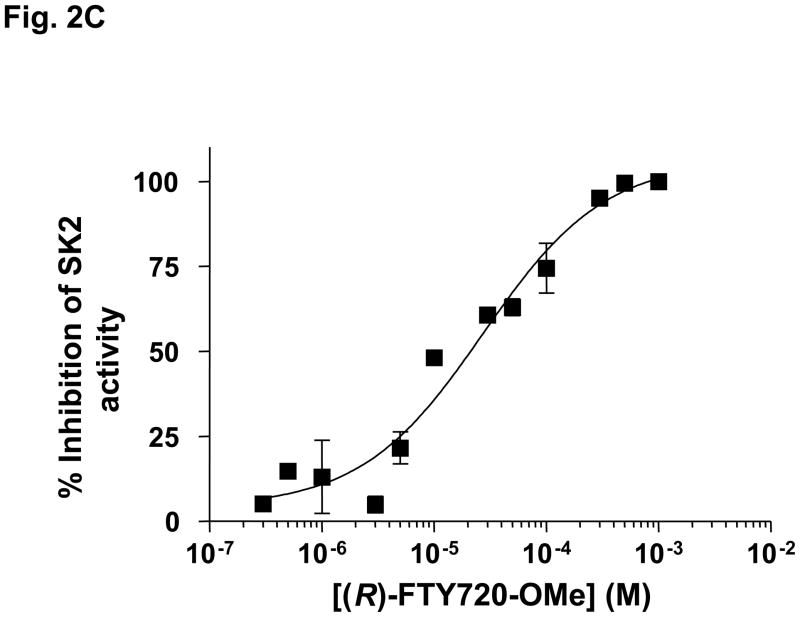
We have previously found that FTY720 inhibits purified SK1 activity (17). The (R) and (S) enantiomers of FTY720 vinylphosphonate (S > R) also inhibited purified SK1 activity, while their saturated counterpart, (R)- or (S)-FTY720 phosphonate, and (S)-FTY720 phosphate did not significantly inhibit the enzyme (17). 2-(p-Hydroxyanilino)-4-(p-chlorophenyl)thiazole, SKi (18) and (S)-FTY720 vinylphosphonate are also effective inhibitors of purified SK2 activity, while (R)-FTY720 vinylphosphonate ((R)-vinyl-Pn) and (S)-FTY720 phosphonate ((S)-FTY-Pn) were moderately active (20–30% inhibition) and (R)-FTY720 phosphonate ((R)-FTY-Pn) was inactive (Fig. 2A). (R)-FTY720-OMe is, therefore, the only compound tested that is an efficient specific inhibitor of only one isoform of SK, i.e. SK2. (R)-FTY720-OMe inhibited purified SK2 activity in a concentration-dependent manner with an IC50 = 27 +/− 1.3 μM and with a Hill coefficient of 0.8 +/− 0.2 (Fig. 2C).
Kinetic characterisation studies established that (R)-FTY720-OMe is a competitive inhibitor (with sphingosine) and exhibited a Kic = 16.5 +/− 1 μM (Fig. 2D, E). For instance, SK2 exhibits a Km = 10.5 μM for sphingosine (in the absence of (R)-FTY720-OMe), and this increases to 27.4 μM in the presence of 20 μM (R)-FTY720-OMe with no significant change in Vmax. (R)- FTY720-OMe is not a substrate for phosphorylation by SK2 (data not shown).
(R)-FTY720-OMe Induces a Reduction in SK2 Expression
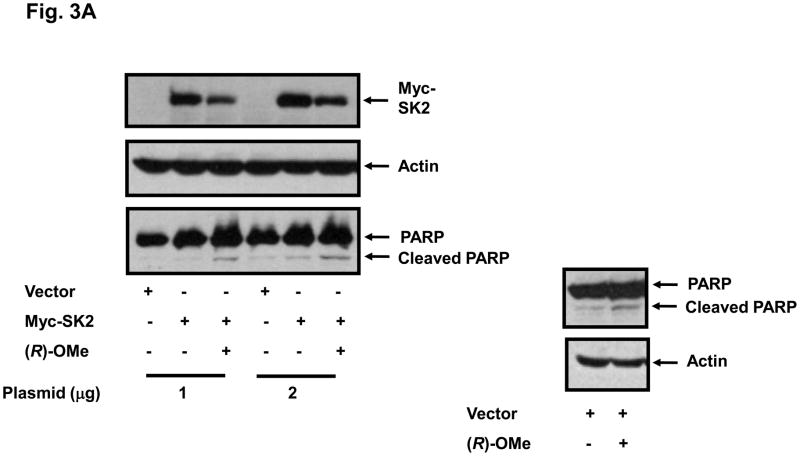
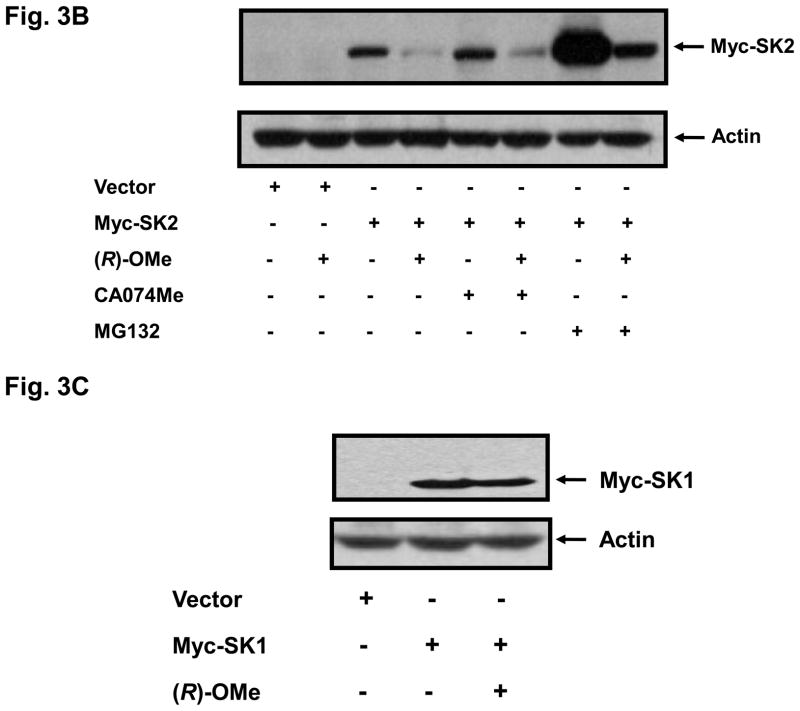
We have previously reported that SKi, FTY720 and (S)-FTY720 vinylphosphonate induce the MG132-sensitive proteasomal degradation of SK1 in breast and prostate cancer cells (17, 19). Therefore, we also investigated the effect of (R)-FTY720-OMe on SK2 expression. Indeed, we show for the first time that treatment of HEK 293 cells transiently over-expressing myc-tagged SK2 with (R)-FTY720-OMe for 24 h induced a reduction in the expression of SK2 (Fig. 3A,B), but not SK1 (Fig. 3C), and stimulated cleavage of the nuclear enzyme poly(ADP-ribose)polymerase (PARP) (Fig. 3A). The pre-treatment of HEK 293 cells with proteasomal inhibitor, MG132 alone substantially increased the expression of myc-tagged SK2 (Fig. 3B), suggesting that, in common with SK1, SK2 is regulated by the ubiquitin-proteasomal pathway under resting conditions. However, the effect (R)-FTY720-OMe on SK2 expression persisted in cells pre-treated with MG132 (Fig. 3B), thereby suggesting that (R)-FTY720-OMe does not enhance proteasomal degradation for SK2; the reduction in SK2 expression in response to (R)-FTY720-OMe in MG132-treated cells was similar to that observed in control cells not pretreated with MG132 (Fig. 3B). Moreover, the pre-treatment of cells with the cathepsin B inhibitor CA074Me (a lysosomal inhibitor) also had no effect on the (R)-FTY720-OMe-induced reduction in SK2 expression, thereby excluding autophagic/lysosomal processing of SK2 in response to (R)-FTY720-OMe.
Fig. 3. (R)-FTY720-OMe-induced reduction in SK2 expression.
(A) HEK 293 cells over-expressing myc-tagged SK2 (1 or 2 μg plasmid) or vector were treated with (R)-FTY720-OMe ((R)-OMe, 10 μM final concentration) for 24 h. Western blots probed with anti-myc antibody showing that (R)-FTY720-OMe reduces SK2 expression. PARP was detected with anti-PARP antibody. The western blot shows cleavage of PARP to form the p85 proteolytic fragment in response to (R)-FTY720-OMe. (B) Western blot probed with anti-myc antibody showing the lack of effect of MG132 or CA074Me (each at 10 μM) and used in a pre-treatment for 30 min prior to addition of (R)-FTY720-OMe ((R)-OMe) on the (R)-FTY720-OMe-induced reduction in SK2 expression. (C) Western blot probed with anti-myc antibody showing the lack of effect of (R)-FTY720-OMe ((R)-OMe) on Myc-tagged SK1 over-expressed in HEK 293 cells. Actin was detected with anti-actin antibody to show comparable protein loading between samples. Results are representative of three independent experiments.
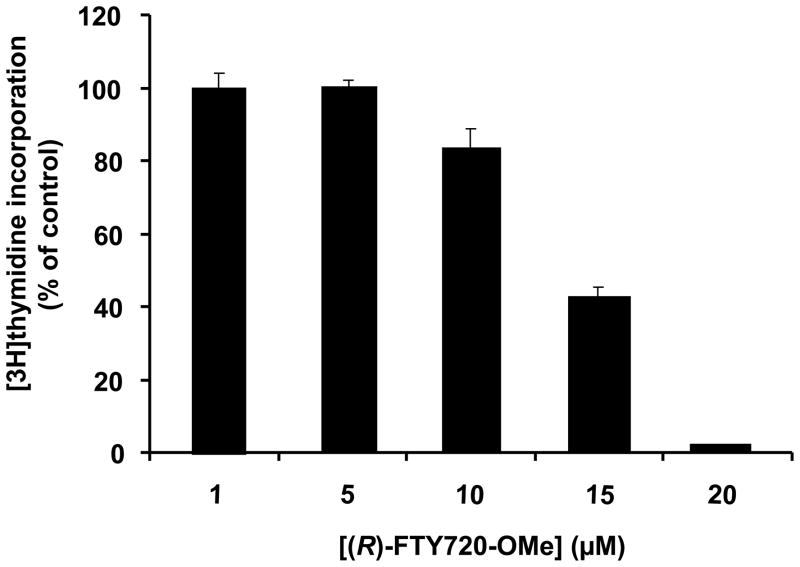
HEK 293 cells express endogenous SK2 (20), and indeed (R)-FTY720-OMe induced apoptosis of these cells, as assessed by increased cleavage of PARP (Fig. 3A). Therefore, our results suggest that SK2 functions as a pro-survival enzyme. Indeed, treatment of MCF-7 cells, which also express endogenous SK2 (9), with (R)-FTY720-OMe reduced basal [3H]-thymidine incorporation in a concentration-dependent manner (Fig. 4). Others have shown that knockdown of SK2 in glioblastoma cells inhibited proliferation, and this was more effective than knock-down of SK1 (10), suggesting a key role for SK2 in the survival of this type of cancer cell. This contrasts with the pro-apoptotic function of SK2 that has been proposed for this enzyme (7, 8), although this function might be dependent on the cell context specific sub-cellular localisation of SK2.
Fig. 4. DNA synthesis.
Bar graph showing the effect of (R)-FTY720-OMe on [3H]-thymidine incorporation into DNA in MCF-7 cells. Results are expressed as % of basal [3H]-thymidine incorporation in the absence of (R)-FTY720 (control = 100% basal [3H]-thymidine incorporation). Representative data of three independent experiments performed in triplicate (mean +/− SD).
Effects of (R)-FTY720-OMe on Actin Rearrangement in MCF-7 Neo cells
We also assessed the effect of (R)-FTY720-OMe on the actin cytoskeleton, since others have demonstrated that the migration of breast cancer cells in response to epidermal growth factor (EGF) requires ERK-1-catalysed phosphorylation of SK2 on Ser 351 and Thr578 (21), and migration involves dramatic rearrangement of the actin cytoskeleton. Indeed, we have previously shown that the siRNA knock down of SK1 induces actin focal adhesion assembly in MCF-7 Neo (transfected with a neomycin vector) (22). Therefore, we used these cells to allow direct comparison with the effects of inhibiting SK2 with (R)-FTY720-OMe.
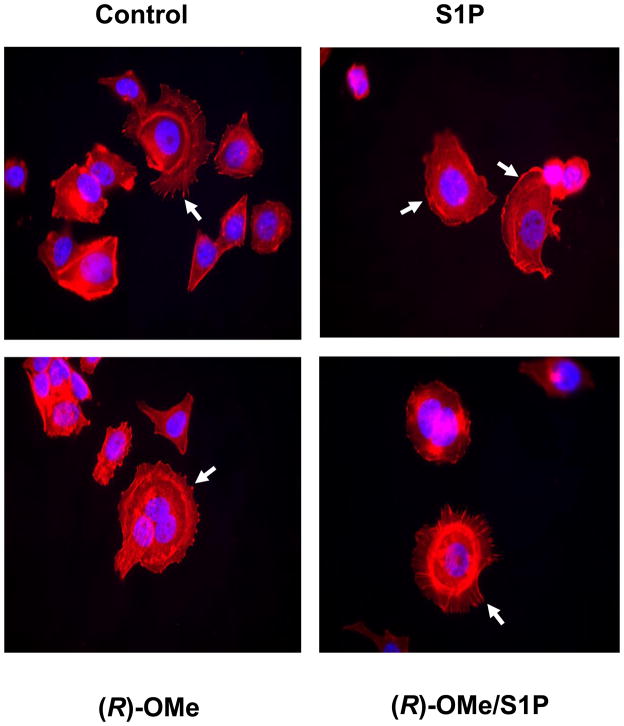
Actin is clustered into focal adhesions in MCF-7 Neo cells. Treatment of these cells with S1P stimulates the rearrangement of actin from focal adhesions into membrane ruffles/lamellipodia ((22), Fig. 5). Treatment of MCF-7 Neo cells with (R)-FTY720-OMe for 15 min reduced the S1P-stimulated rearrangement of actin into lamellipodia/membrane ruffles and increased actin focal adhesions (Fig. 5). These results suggest that as with SK1, SK2 might also regulate the redistribution of actin in response to exogenous S1P. Thus, SK2 appears to regulate the transition of MCF-7 cells from a stationary phenotype (as evidenced by focal adhesion assembly) to a migratory phenotype (as evidenced by accumulation of actin into membrane ruffles/lamellipodia).
Fig. 5. Actin rearrangement in MCF-7 cells.
MCF-7 Neo cells were treated with (R)-FTY720-OMe ((R)-OMe), (10 μM final concentration) for 15 min prior to stimulation with and without S1P (1 μM, 5 min). Actin was detected using phalloidin red staining. Arrows in the panel for S1P treatment identify actin localised to lamellipodia/membrane ruffles, while arrows in the other panels identify actin clustered into focal adhesions. Nuclei were stained with DAPI. Results are representative of three independent experiments.
There is only one other SK2-specific inhibitor reported in the literature (23). ABC294640 (3-(4-chlorophenyl)-adamantane-1-carboxylic acid (pyridin-4-ylmethyl)amide) was very recently found to be a competitive inhibitor with sphingosine with a Kic of 10 μM, which is very similar to that found with (R)-FTY720-OMe (Kic=16.5 +/− 1 μM). However, an important advance here is that the potency of (R)-FTY720-OMe is significantly augmented by efficacy, as evidenced by the substantial reduction of SK2 expression in cells with prolonged treatment.
In conclusion, (R)-FTY720-OMe is an effective specific SK2 inhibitor that also substantially reduces expression of SK2 in cells. In addition, (R)-FTY720-OMe exhibits properties that are conducive to anti-cancer activity. Using this inhibitor we have established that SK2 plays an important role in regulating the survival of MCF-7 breast cancer cells.
Acknowledgments
This work was supported by a Strathclyde University Scholarships to K. G Lim and NIH grant HL083187 (RB).
ABBREVIATIONS
- ERK
extracellular signal regulated kinase-1/2
- PARP
poly ADP ribose polymerase
- (R)-FTY720-OMe
(2R)-2-amino-3-(O-methyl)-(2-(4′-n-octylphenyl)ethyl)propanol
- Pn
phosphonate
- S1P
sphingosine 1-phosphate
- SK2
sphingosine kinase 2
- SKi
2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole
- (S)-vinyl-Pn
(S) FTY720 vinylphosphonate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pyne NJ, Pyne S. Nature Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P, Foster CA, Zollinger M, Lynch KR. J Biol Chem. 2002;277:21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, Aradhye S, Burtin P. Nat Rev Drug Discov. 2010;9:883–897. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez T, Estrada-Hernandez T, Paik JH, Wu MT, Venkataraman K, Brinkmann V, Claffey K, Hla T. J Biol Chem. 2003;278:47281–47290. doi: 10.1074/jbc.M306896200. [DOI] [PubMed] [Google Scholar]
- 5.Gräler MH, Goetzl EJ. FASEB J. 2004;18:551–553. doi: 10.1096/fj.03-0910fje. [DOI] [PubMed] [Google Scholar]
- 6.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei GJ, Card D, Koehane C, Rosenbach M, Hale J, Lynch CL, Rupprecht K, Parsons W, Rosen H. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 7.Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura S. J Biol Chem. 2003;278:46832–46839. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Toman RE, Goparaju SK, Maceyka M, Nava VE, Sankala H, Payne SG, Bektas M, Ishii I, Chun J, Milstien S, Spiegel S. J Biol Chem. 2003;278:40330–40336. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- 9.Sankala HM, Hait NC, Paugh SW, Shida D, Lépine S, Elmore LW, Dent P, Milstien S, Spiegel S. Cancer Res. 2007;67:10466–10474. doi: 10.1158/0008-5472.CAN-07-2090. [DOI] [PubMed] [Google Scholar]
- 10.Van Brocklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, Prior TW. J Neuropathol Exp Neurol. 2005;64:695–705. doi: 10.1097/01.jnen.0000175329.59092.2c. [DOI] [PubMed] [Google Scholar]
- 11.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alderton F, Rakhit S, Kong KC, Palmer T, Sambi S, Pyne S, Pyne NJ. J Biol Chem. 2001;276:28578–28585. doi: 10.1074/jbc.M102771200. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poilton S, Milstien S, Kohama T, Spiegel S. J Biol Chem. 2000;275:19513–19520. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 14.Delon C, Manifava M, Wood E, Thompson D, Krugmann S, Pyne S, Ktistakis NT. J Biol Chem. 2004;279:44763–44774. doi: 10.1074/jbc.M405771200. [DOI] [PubMed] [Google Scholar]
- 15.Cortes A, Cascante M, Cardenas ML, Cornish-Bowden A. Biochem J. 2001;357:263–268. doi: 10.1042/0264-6021:3570263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Sun C, Valentine WJES, Liu J, Tigyi G, Bittman R. J Org Chem. 2009;74:3192–3195. doi: 10.1021/jo900023u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonelli F, Lim K, Loveridge C, Long J, Pitson SM, Tigyi G, Bittman R, Pyne S, Pyne NJ. Cell Signal. 2010;22:1536–1542. doi: 10.1016/j.cellsig.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French KJ, Schrecengost RS, Lee BD, Zhuang Y, Smith SN, Eberly JLM, Yun JK, Smith CD. Cancer Res. 2003;63:5962–5969. [PubMed] [Google Scholar]
- 19.Loveridge C, Tonelli F, Leclecq T, Lim KG, Long S, Berdyshev E, Tate RJ, Natarajan V, Pitson Pyne NJ, Pyne S. J Biol Chem. 2010;285:38841–38852. doi: 10.1074/jbc.M110.127993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hait NC, Sarkar S, Le Stunff H, Mikami A, Maceyka M, Milstien S, Spiegel S. J Biol Chem. 2005;280:29462–29469. doi: 10.1074/jbc.M502922200. [DOI] [PubMed] [Google Scholar]
- 21.Hait NC, Bellamy A, Milstien S, Kordula T, Spiegel S. J Biol Chem. 2007;282:12058–12065. doi: 10.1074/jbc.M609559200. [DOI] [PubMed] [Google Scholar]
- 22.Long JS, Edwards J, Watson C, Tovey S, Mair K, Schiff R, Natarajan V, Pyne NJ, Pyne S. Mol Cell Biol. 2010;30:3827–3841. doi: 10.1128/MCB.01133-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.French KJ, Zhuang Y, Maines LW, Gao P, Wang W, Beljanski V, Upson JJ, Green CL, Keller SN, Smith CD. J Pharmacol Exp Ther. 2010;333:129–139. doi: 10.1124/jpet.109.163444. [DOI] [PMC free article] [PubMed] [Google Scholar]