Abstract
Vector-mediated delivery of short-hairpin RNA (shRNA) to regulate gene expression holds a great therapeutic promise. We hypothesize that gene expression can be autoregulated with RNA interference. We used inducible nitric oxide synthase (iNOS) as a gene model to test this hypothesis. Lipopolysaccharide dose-dependently increased iNOS in rat aortic smooth muscle cells and the nitrite production from these cells. These increases were attenuated in cells transfected with plasmids containing code for iNOS shRNA whose expression was controlled by an iNOS promoter. The production of shRNA was lipopolysaccharide dose-dependent. The lipopolysaccharide-induced iNOS expression in rat C6 glioma cells also was attenuated by transfection with plasmids containing the iNOS shRNA code. These results provide proof-of-concept evidence for using RNA interference technique to achieve autoregulation of gene expression.
Keywords: autoregulation, inducible nitric oxide synthase, RNA interference, short hairpin RNA
Introduction
RNA interference (RNAi) caused by endogenous microRNA is a naturally occurring molecular machinery to regulate protein expression [1]. To simulate this mechanism, small interfering RNA (siRNA) or short hairpin RNA (shRNA) can be introduced into cells to silence the expression of a selective protein. Direct delivery of siRNA to cells may have a limited application under in vivo conditions because of the difficulty for the siRNA to access target cells and the short life of these molecules. Vector-mediated delivery of shRNA holds great therapeutic potentials under in vivo conditions. Once the vector is delivered to cells, shRNA can be synthesized in these cells. One of popular methods for this purpose is to construct the vector in a way that the expression of shRNA is controlled by a polymerase (pol) III promoter [2]. Pol III promoters are usually short and well-defined, making them easy to be cloned into vectors and to be used to facilitate accurate transcription of shRNA.
Use of pol III promoters to control shRNA expression can have significant problems. Pol III promoters are constitutively active in many cell types. This feature makes them not suitable to provide spatial and temporal control of shRNA expression [3]. In addition, pol III promoters are usually very efficient. Robust expression of shRNA can saturate the endogenous microRNA processing machinery, which appears to induce cell toxicity in some cases [4]. To overcome these problems, various methods, such as introduction of an inducible element to the promoters and use of tissue specific pol II promoters, have been reported [5–7]. Although better spatial and temporal control can be achieved by these modifications, the expression of shRNA will be at its maximal level once the control mechanisms for the expression are activated.
Fine regulation of protein/gene expression may be desirable in most situations. For example, The damaging role of over-expression of inducible nitric oxide synthase (iNOS) after brain ischemia has been well established [8–10]. However, iNOS has many physiological functions, such as antitumoral and immunomodulatory effects [11,12] and is involved in neurogenesis after brain ischemia [13]. Thus, fine regulation of iNOS expression after brain ischemia may be necessary to reduce ischemic brain injury and to preserve neurogenesis. Such a fine regulation may be achieved via an autoregulation mechanism using RNAi technique. To provide proof-of-concept evidence, we constructed a vector in which the expression of iNOS shRNA was under the control of an iNOS promoter. This construction was designed to induce iNOS shRNA expression by the endogenous factors that could induce iNOS expression, allowing autoregulation of iNOS expression.
Materials and methods
DNA constructs
The backbone plasmid pcDNA6.2-GW/EmGFP-miR was from Block-iT PolII miR RNAi expression vector kit (Invitrogen, Carlsbad, CA). The plasmids pcDNA6.2-GW/EmGFP-miR-iNOS (pCMV-shRNA) containing shRNA sequences specifically targeting rat iNOS gene and pcDNA6.2-GW/EmGFP-miR-neg (pCMV-N) containing an unrelated insert were constructed using a method described in the manual for the Block-iT PolII miR RNAi expression vector kit. Six iNOS shRNA sequences were designed with the use of the website http://www.invitrogen.com/rnai and were shown as in table 1. The unrelated insert sequence was predicted not to target any known genes.
Table 1.
Sequences of selected iNOS shRNAs
| ShRNA1 | 5’-TGCTGTGTCCAGGGATTCTGGAACATGTTTTGGCCACTGACTGACATGTTCCAATCCCTGGACA-3’ |
| shRNA2 | 5’-TGCTGTGCATGTGCTTCATGAAGGACGTTTTGGCCACTGACTGACGTCCTTCAAAGCACATGCA-3’ |
| shRNA3 | 5’-TGCTGAATCGTTGTACTCTGAGGGCTGTTTTGGCCACTGACTGACAGCCCTCAGTACAACGATT-3’ |
| shRNA4 | 5’-TGCTGAGAAGTAATCCTCAACCTGCTGTTTTGGCCACTGACTGACAGCAGGTTGGATTACTTCT-3’ |
| shRNA5 | 5’-TGCTGTTCTGATGCAGTGCTACAGCTGTTTTGGCCACTGACTGACAGCTGTAGCTGCATCAGAA-3’ |
| shRNA6 | 5’-TGCTGTTTCAAAGACCTCTGGATCTTGTTTTGGCCACTGACTGACAAGATCCAGGTCTTTGAAA-3’ |
| Unrelated insert | 5’-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3’ |
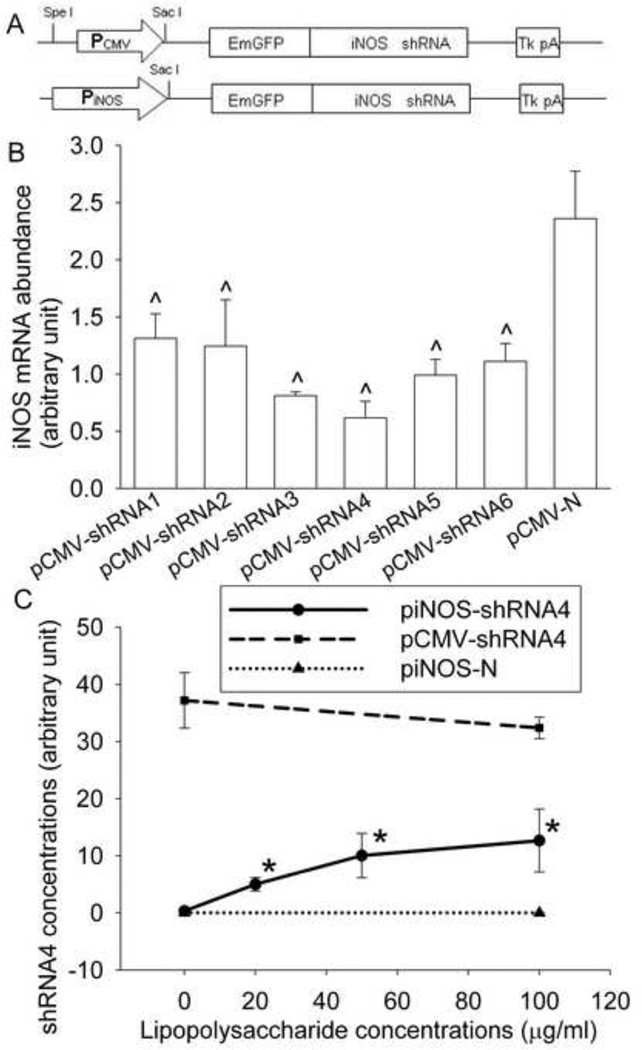
To replace the CMV promoter with the rat iNOS promoter in the above plasmids, we first cloned the iNOS promoter from rat C6 glioma cells (American Type Culture Collection, Manassas, VA) by PCR amplification using the forward primer 5’-AAAGTATTTGGGAGGAGGGGCTGAG-3’ and the reverse primer 5’-AGAGCTCACTCCCTGTAAAGCTGTGG-3’. This process introduced a Sac I site. The PCR product was blunted and then digested with Sac I. This process resulted in a 3.2 kb iNOS promoter that was found to have a full response to the regulation of various transcription factors for iNOS expression [14]. The plasmids containing CMV promoters were digested with Spe I first, blunted and digested with Sac I. The iNOS promoter then was sub-cloned into each plasmid to generate various piNOS-shRNAs and piNOS-N (Fig. 1).
Fig. 1. Effectiveness of the silencing effects of iNOS shRNAs and the induction of iNOS shRNA production.
A: A diagram showing the basic structure of the plasmids used in the study. B: Human 293FT cells were co-transfected with the plasmid pCMV-iNOS and various pCMV-shRNA plasmids indicated in the figure. RNA was prepared at 24 h after the transfection. Results are means ± S.D. (n = 3). ^ P < 0.05 compared with pCMV-N group. C: RASMCs were transfected with various plasmids indicated in the figure. Twenty four hours later, they were incubated with various concentrations of lipopolysaccharide for 12 h and then harvested for shRNA4 quantification. Results are means ± S.D. (n = 3). * P < 0.05 compared with piNOS-shRNA4 at 0 µg/ml lipopolysaccharide.
To construct the plasmid containing the code for rat iNOS, total RNA was extracted from rat aortic smooth muscle cells (RASMCs, Lonza, Walkersville, MD) simulated by lipopolysaccharide (LPS), an inducer of iNOS expression, and was reversely transcribed into cDNA. The cDNA then was amplified with the iNOS forward primer 5’-CCACCTTGGTGAGGGGACTGGA-3’ and the reverse primer 5’-AGGGCCAGATGCTGTAACTCTTCT-3’. The PCR product was cloned into the pTARGET vector (Promega, Madison, WI) to form pCMV-iNOS.
All plasmids were sequenced to confirm accuracy of their sequences and purified by using Qiagen Hispeed Plasmid Purification kit (Qiagen, Valencia, CA).
Cell culture and transfection
RASMCs were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium supplemented with 20% heat inactivated fetal bovine serum (FBS) at 37°C. Human 293FT cells were from Invitrogen and maintained in DMEM supplemented with 10% FBS, 0.1 mM MEM non-essential amino acids, 6 mM L-glutamine, 1 mM MEM sodium pyruvate and 500 µg/ml Geneticin. Rat C6 glioma cells were maintained in F-12K medium supplemented with 15% horse serum and 2.5% FBS at 37°C.
To determine the effectiveness of selected iNOS shRNAs in silencing the expression of rat iNOS, 1×105 293FT cells per well were plated in 12-well plates. Next day, the cells were transfected with 720 ng pCMV-iNOS and 240 ng pCMV-shRNA or pCMV-N using 1.5 µl FuGENE HD Transfection Reagent (Promega). RNA isolation from these cells was performed at 24 h after transfection.
To determine whether the production of iNOS shRNA and its silencing effect were LPS dose-dependent, 1×105 RASMCs per well were plated in 6-well plates. Cells were transfected with 2 µg piNOS-shRNA3, piNOS-shRNA4 or piNOS-N using 8 µl FuGENE HD Transfection Reagent. Since CMV promoter is much smaller than iNOS promoter, pCMV-shRNA was mixed with pCMV-N so that the same moles of pCMV-shRNA as contained in 2 µg piNOS-shRNA plasmids were used in the transfection. Twenty four hours after transfection, cells were washed, incubated in fresh medium with different concentrations of LPS for 12 h and then used for RNA preparation.
C6 cells were plated at 4×105 cells per well in 6-well plates. They were transfected with 2 µg plasmids per well using the GenJet transfection reagents (SignaGen laboratories, Rockville, MD).
Cell sorting
Twenty hours after transfection, C6 cells were trypsinized and suspended in ice-cold phosphate buffered saline. They were sorted based on the green fluorescent protein (GFP) tag, using an i-Cyte Reflection Cell Sorter (iCyt, Champaign, IL). Cells with GFP were collected and immediately plated onto 12-well plates at a density of 3×105 per well. Cells were cultured overnight, and LPS was added into the wells to make the final concentration of 1 µg/ml. Three hours later, cells were collected for RNA isolation.
Total RNA extraction and real-time PCR
Total RNA was extracted using RNeasy micro kit (Qiagen). Real-time PCR was performed as we described previously [15]. Primers for real-time PCR were designed based on reported sequence of rat iNOS gene using the Primer Express 3.0 software (Applied Biosystems, Carlsbad, CA) and selected to best fit the requirement of SYBR Green assays. The sequences of the primers are: forward, CACTGGGACTGCACAGAATGTT and reverse, CTCCATTGCCCCAGTTTTTG. These primers are not suitable to amplify human iNOS cDNA due to the sequence difference between the rat and human iNOS. Quantitative PCRs were carried out in triplicates using each cDNA sample that was equivalent to 50 ng of starting total RNA. Power SYBR Green Master Mix was used with the forward and reverse primers at the optimized concentrations in a total volume of 25 µl. Amplifying PCR and monitoring of the fluorescent emission in real-time were performed in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). To verify that only a single PCR product was amplified per transcript, dissociation curve data was analyzed through the 7900HT Sequence Detection Software. To account for possible differences in starting material, PCR of the housekeeping genes glyceraldehydes-3-phosphate dehydrogenase and actin also was carried out for each cDNA sample. The relative amount of iNOS mRNA in each sample was determined using the comparative threshold cycle method and then normalized to those of house keeping genes.
MicroRNA isolation and assay
MicroRNA was isolated using mirVana miRNA isolation kit (Ambion, Carlsbad, CA) and quantified by TaqMan microRNA assay kit (Applied Biosystems, Carlsbad, CA). Quantitative PCR was carried out in triplicates using U6 snRNA as control. Amplifying PCR and Monitoring of the fluorescent emission in real time were performed in the ABI Prism 7900HT Sequence Detection System.
Nitrite assay
Twenty-four hours after transfection, fresh medium with various concentrations of LPS was added to the cells. The culture medium was collected after a 24-h incubation. The nitrite concentrations were measured using the Griess Reagent kit (Invitrogen) as we described previously [16].
Data Analysis
The results are presented as means ± S.D. (n ≥ 3). Statistical analysis was performed by one way analysis of variance followed by the Tukey test after confirmation of normal distribution of the data or by Kruskal-Wallis analysis of variance on ranks followed by the Tukey test when the data are not normally distributed. A P < 0.05 was considered significant.
Results
The human 293FT cells, a cell line with a very high transfection rate, were co-transfected with the plasmids pCMV-shRNA and pCMV-iNOS to evaluate the effectiveness of the selected shRNAs to silence rat iNOS expression. This co-transfection system avoids interference of endogenous iNOS expression. As shown in Fig. 1, all six shRNAs significantly inhibited the rat iNOS expression.
Since the 3.2 kb iNOS promoter was found to be fully functional in RASMCs [14], we used these cells for the majority of our study. LPS induced a dose-dependent increase of shRNA4 in the RASMCs transfected with piNOS-shRNA4. A high level of shRNA4 was found in the RASMCs transfected with pCMV-shRNA4. However, the shRNA4 expression in these cells was not affected by LPS. Cells transfected with piNOS-N did not express reliably detectable shRNA4 by our method, no matter whether LPS was present (Fig. 1).
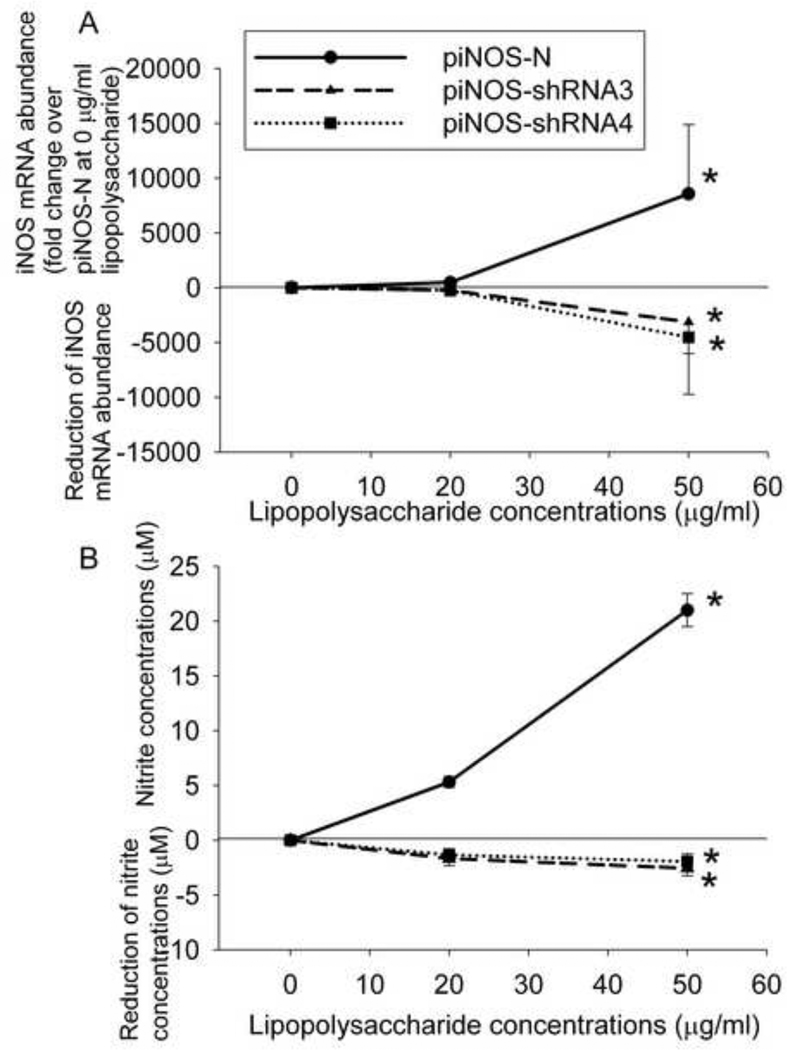
LPS caused a dose-dependent increase of iNOS mRNA in the RASMCs transfected with piNOS-N. This increase was attenuated in the RASMCs transfected with piNOS-shRNA3 or piNOS-shRNA4. The degree of this attenuation was LPS dose-dependent (Fig. 2). Similar change pattern was observed in the nitrite concentrations in the culture medium of the RASMCs (Fig. 2).
Fig. 2. Does-response of lipopolysaccharide to induce iNOS expression and nitrite production.
A: RASMCs were transfected with various plasmids indicated in the figure. Twenty four hours later, they were incubated with various concentrations of lipopolysaccharide for 12 h and then harvested for iNOS mRNA quantification. Results were normalized by the level of iNOS mRNA in cells transfected with piNOS-N at 0 µg/ml lipopolysaccharide. Reduction of iNOS mRNA abundance in cells transfected with piNOS-shRNA3 or piNOS-shRNA4 was calculated by subtracting mRNA levels in the cells transfected with piNOS-N from the mRNA levels in the cells transfected with those plasmids containing iNOS shRNA codes. Results are means ± S.D. (n = 3). * P < 0.05 compared with piNOS-N at 0 µg/ml lipopolysaccharide. B: The culture medium from the cells used in panel A was collected for nitrite measurement. Reduction of nitrite concentrations in cells transfected with piNOS-shRNA3 or piNOS-shRNA4 was calculated by subtracting nitrite concentrations of the cells transfected with piNOS-N from the concentration of the cells transfected with those plasmids containing iNOS shRNA codes. Results are means ± S.D. (n = 6). * P < 0.05 compared with piNOS-N at 0 µg/ml lipopolysaccharide.
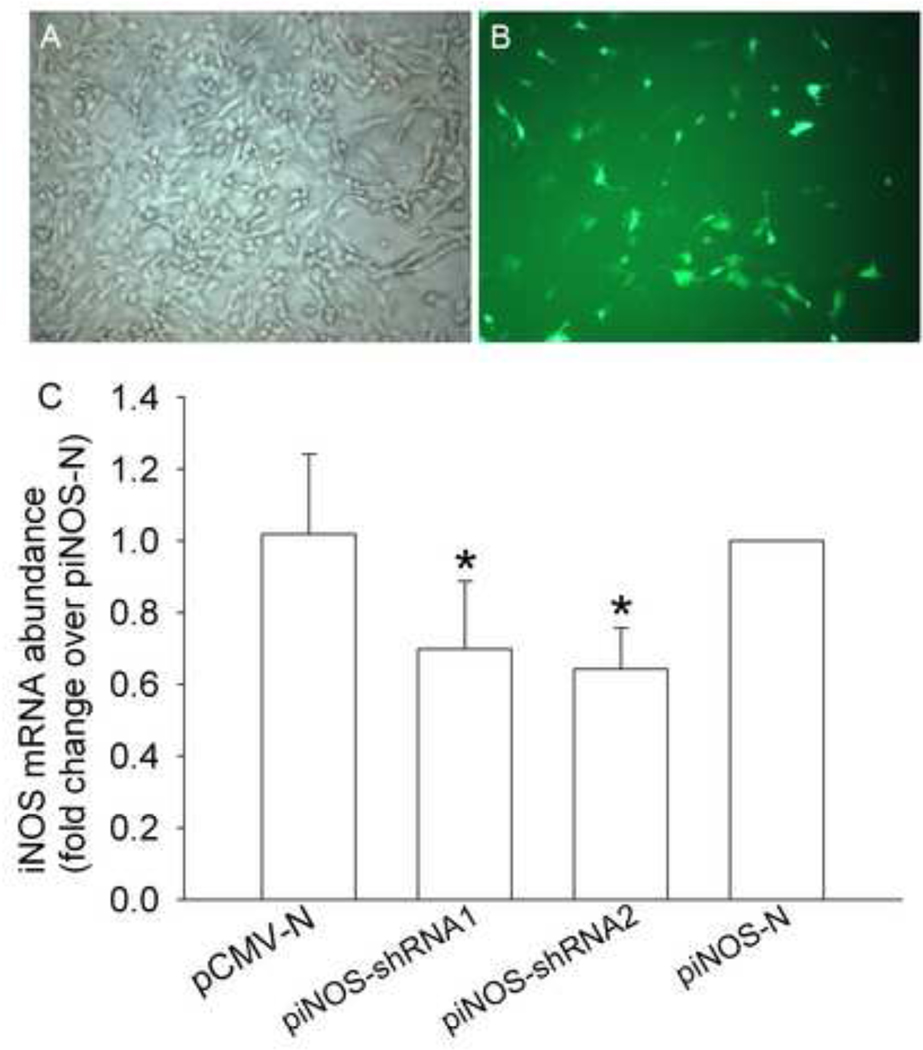
To determine whether the findings in the RASMCs were applicable to another type of cells, C6 cells were used. C6 cells transfected with piNOS-N expressed a significant amount of GFP (Fig. 3). Since the code for GFP was downstream of iNOS promoter in this plasmid, these results suggest that the iNOS promoter was activated in these cells. Since these cells had a much lower transfection rate (~25%) than human 293FT cells and RASMCs, we separated cells that were transfected successfully with piNOS-shRNA1, piNOS-shRNA2, pCMV-N or piNOS-N from those that were not by flow cytometry. Cells transfected with piNOS-shRNA1 or piNOS-shRNA2 expressed significantly less iNOS mRNA than cells transfected with piNOS-N after LPS stimulation (Fig. 3).
Fig. 3. Silencing effects of iNOS shRNAs in C6 cells.
A and B: Rat C6 cells were transfected with the plasmid piNOS-N that contained a code for green fluorescent protein. Photos were taken 24 h later. Panel A is a phase contrast picture to show all cells. Panel B is a fluorescent image to show cells transfected by the vector. C: C6 cells were transfected with or without plasmids indicated in the figure and were sorted based on their expression of green fluorescent protein. Cells that were transfected successfully by the corresponding plasmids were exposed to 1 µg/ml lipopolysaccharide for 3 h before they were harvested for real-time PCR for quantification of iNOS and actin mRNA. The iNOS results were normalized by actin data from the same sample. Results are means ± SD (n = 6). * P < 0.05 compared with iNOS-N group.
Discussion
We used iNOS as an example for a group of genes whose over-expression is harmful to cells. However, normal expression of these genes is necessary for various physiological functions. Thus, fine regulation of the expression of these genes is critical, especially in disease status. For example, robust iNOS expression is induced in microglia, astrocytes and neurons of ischemic brain [17–19]. Studies with iNOS inhibitors and iNOS knockout mice confirm that a decreased iNOS activity can reduce ischemic brain injury [9,20–26]. However, iNOS has been shown to participate in induction of ischemic tolerance [27,28] and neurogenesis after brain ischemia [13]. Thus, prevention of iNOS over-expression may block its damaging effects and preserve its beneficial effects after brain ischemia. To achieve this goal, we proposed to use an autoregulation mechanism in which the iNOS shRNA expression was controlled by an iNOS promoter. Thus, stimuli that induce iNOS expression also will cause the iNOS shRNA expression. Stronger stimuli for iNOS expression will induce a higher expression of iNOS shRNA, which then attenuates iNOS increase.
To test our theory, we first identified shRNAs that were effective in silencing rat iNOS expression. Among them, shRNA4 appeared to be the most effective. When the RASMCs were transfected with piNOS-shRNA4, LPS dose-dependently increased shRNA4 in these cells. The shRNA4 expression in the RASMCs transfected with pCMV-shRNA4 was not affected by LPS. These results suggest that our iNOS promoter is functional and that the expression of shRNA4 under the control of this promoter is inducible. These results, along with the results that a bigger decrease of iNOS mRNA abundance was seen when cells transfected with piNOS-shRNAs were stimulated by a higher LPS concentration, provide strong proof of concept evidence for using our technique to achieve autoregulation of gene expression. This gene autoregulation may have significantly affected the iNOS activity because the nitrite contents in the culture medium had a change pattern similar to that of iNOS mRNA expression. iNOS, once it is formed, produces nitric oxide, a short lived signaling molecule [29]. Nitrite is a stable oxidative product of nitric oxide and is often used to reflect nitric oxide levels [16].
In addition to the RASMCs, our piNOS-shRNAs were also effective to attenuate the LPS-stimulated iNOS expression in C6 cells. These results suggest that induction of autoregulation of iNOS expression is not cell type specific.
The novel concept, autoregulation of gene expression by RNAi, could have a broad application. For example, autoregulation of iNOS expression by RNAi may be used in brain ischemia to provide neuroprotection because iNOS contributes to ischemic brain injury [8–10]. The technique also can be used to autoregulate expression of other proteins, such as cytokines, and, thus, hinder many disease processes including inflammation in which iNOS and cytokines play an important role [30,31]. Use of suitable viral delivery system will facilitate the possible application of this technique under in vivo conditions.
Various modifications of our approach to achieve autoregulation of gene expression by RNAi can be performed. For example, it is possible to enhance or decrease shRNA expression by including or excluding certain regulatory elements in the promoter used to control shRNA expression. These fine tunes are necessary to achieve the intended goals under various laboratory and clinical conditions.
Research highlights.
Autoregulation of gene expression is achievable by RNA interference (RNAi)
Autoregulation of inducible nitric oxide synthase by RNAi is not cell type specific
This form of regulation of gene expression is a novel concept
Acknowledgement
This study was supported by a grant from the International Anesthesia Research Society (2007 Frontiers in Anesthesia Research Award to Z Zuo), Cleveland, OH, by a grant (R01 GM065211 Z Zuo) from the National Institutes of Health, Bethesda, MD, by a Grant-in-Aid from the American Heart Association Mid-Atlantic Affiliate (10GRNT3900019 to Z Zuo), Baltimore, MD, and the Robert M. Epstein Professorship endowment, University of Virginia, Charlottesville, VA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9–12. doi: 10.1016/s0168-9525(02)00005-7. [DOI] [PubMed] [Google Scholar]
- 3.Xia XG, Zhou H, Xu Z. Promises and challenges in developing RNAi as a research tool and therapy for neurodegenerative. Neurodegener. Dis. 2005;2:220–231. doi: 10.1159/000089629. [DOI] [PubMed] [Google Scholar]
- 4.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 5.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 6.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 7.Lin X, Yang J, Chen J, Gunasekera A, Fesik SW, Shen Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004;577:376–380. doi: 10.1016/j.febslet.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 8.Lipton P. Ischemic cell death in brain neurons. Physiol. Rev. 1999;79:1431–1568. doi: 10.1152/physrev.1999.79.4.1431. [DOI] [PubMed] [Google Scholar]
- 9.Danielisova V, Nemethova M, Burda J. The protective effect of aminoguanidine on cerebral ischemic damage in the rat brain. Physiol. Res. 2004;53:533–540. [PubMed] [Google Scholar]
- 10.Moro MA, Cardenas A, Hurtado O, Leza JC, Lizasoain I. Role of nitric oxide after brain ischaemia. Cell Calcium. 2004;36:265–275. doi: 10.1016/j.ceca.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 12.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 13.Luo CX, et al. Reduced neuronal nitric oxide synthase is involved in ischemia-induced hippocampal neurogenesis by up-regulating inducible nitric oxide synthase expression. J. Neurochem. 2007;103:1872–1882. doi: 10.1111/j.1471-4159.2007.04915.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Chen X, Teng X, Snead C, Catravas JD. Molecular cloning and analysis of the rat inducible nitric oxide synthase gene promoter in aortic smooth muscle cells. Biochem. Pharmacol. 1998;55:1873–1880. doi: 10.1016/s0006-2952(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 15.Zuo Z, Wang Y, Huang Y. Isoflurane preconditioning protects human neuroblastoma SH-SY5Y cells against in vitro simulated ischemia-reperfusion through the activation of extracellular signal-regulated kinases pathway. Eur. J. Pharmacol. 2006;542:84–91. doi: 10.1016/j.ejphar.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Zuo Z, Johns RA. Inhalational anesthetics up-regulate constitutive and lipopolysaccharide-induced inducible nitric oxide synthase expression and activity. Mol. Pharmacol. 1997;52:606–612. doi: 10.1124/mol.52.4.606. [DOI] [PubMed] [Google Scholar]
- 17.Han HS, Qiao Y, Karabiyikoglu M, Giffard RG, Yenari MA. Influence of mild hypothermia on inducible nitric oxide synthase expression and reactive nitrogen production in experimental stroke and inflammation. J. Neurosci. 2002;22:3921–3928. doi: 10.1523/JNEUROSCI.22-10-03921.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endoh M, Maiese K, Wagner J. Expression of the inducible form of nitric oxide synthase by reactive astrocytes after transient global ischemia. Brain. Res. 1994;651:92–100. doi: 10.1016/0006-8993(94)90683-1. [DOI] [PubMed] [Google Scholar]
- 19.Moro MA, et al. Neuronal expression of inducible nitric oxide synthase after oxygen and glucose deprivation in rat forebrain slices. Eur. J. Neurosci. 1998;10:445–456. doi: 10.1046/j.1460-9568.1998.00028.x. [DOI] [PubMed] [Google Scholar]
- 20.Niwa K, Takizawa S, Kawaguchi C, Kamiya U, Kuwahira I, Shinohara Y. Expression of inducible nitric oxide synthase immunoreactivity in rat brain following chronic hypoxia: effect of aminoguanidine. Neurosci. Lett. 1999;271:109–112. doi: 10.1016/s0304-3940(99)00534-0. [DOI] [PubMed] [Google Scholar]
- 21.Chang CP, Lee CC, Chen SH, Lin MT. Aminoguanidine protects against intracranial hypertension and cerebral ischemic injury in experimental heatstroke. J. Pharmacol. Sci. 2004;95:56–64. doi: 10.1254/jphs.95.56. [DOI] [PubMed] [Google Scholar]
- 22.Sugimoto K, Iadecola C. Effects of aminoguanidine on cerebral ischemia in mice: comparison between mice with and without inducible nitric oxide synthase gene. Neurosci. Lett. 2002;331:25–28. doi: 10.1016/s0304-3940(02)00834-0. [DOI] [PubMed] [Google Scholar]
- 23.Shirhan M, Moochhala SM, Siew Yang KL, Sng J, Ng KC, Mok P, Lu J. Preservation of neurological functions by nitric oxide synthase inhibitors in conscious rats following delayed hemorrhagic shock. Life Sci. 2004;76:661–670. doi: 10.1016/j.lfs.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Wada K, Chatzipanteli K, Kraydieh S, Busto R, Dietrich WD. Inducible nitric oxide synthase expression after traumatic brain injury and neuroprotection with aminoguanidine treatment in rats. Neurosurgery. 1998;43:1427–1436. doi: 10.1097/00006123-199812000-00096. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, et al. Neuroprotection by aminoguanidine after lateral fluid-percussive brain injury in rats: a combined magnetic resonance imaging, histopathologic and functional study. Neuropharmacology. 2003;44:253–263. doi: 10.1016/s0028-3908(02)00380-5. [DOI] [PubMed] [Google Scholar]
- 26.Chatzipanteli K, Garcia R, Marcillo AE, Loor KE, Kraydieh S, Dietrich WD. Temporal and segmental distribution of constitutive and inducible nitric oxide synthases after traumatic spinal cord injury: effect of aminoguanidine treatment. J. Neurotrauma. 2002;19:639–651. doi: 10.1089/089771502753754109. [DOI] [PubMed] [Google Scholar]
- 27.Zhao P, Zuo Z. Isoflurane preconditioning induces neuroprotection that is inducible nitric oxide synthase-dependent in the neonatal rats. Anesthesiology. 2004;101:695–702. doi: 10.1097/00000542-200409000-00018. [DOI] [PubMed] [Google Scholar]
- 28.Zhao P, Peng L, Li L, Xu X, Zuo Z. Isoflurane preconditioning improves long-term neurologic outcome after hypoxic-ischemic brain injury in neonatal rats. Anesthesiology. 2007;107:963–970. doi: 10.1097/01.anes.0000291447.21046.4d. [DOI] [PubMed] [Google Scholar]
- 29.Sessa WC. The nitric oxide synthase family of proteins. J. Vasc. Res. 1994;31:131–143. doi: 10.1159/000159039. [DOI] [PubMed] [Google Scholar]
- 30.Hierholzer C, Kalff JC, Billiar TR, Bauer AJ, Tweardy DJ, Harbrecht BG. Induced nitric oxide promotes intestinal inflammation following hemorrhagic shock. Am. J. Physiol. Gastrointest. Liver. Physiol. 2004;286:G225–G233. doi: 10.1152/ajpgi.00447.2002. [DOI] [PubMed] [Google Scholar]
- 31.Barone FC, Tuma RF, Legos JJ, Erhardt JA, Parsons AA. Brain inflammation, cytokines, and p38 MAP kinase signaling in stroke. In: Lin RCS, editor. New Concepts in Cerebral Ischemia. Boca Raton (Florida): CRC Press LLC; 2002. pp. 201–244. [Google Scholar]