Abstract
Due to the fibrous shape and durability of multi-walled carbon nanotubes (MWCNT), concerns regarding their potential for producing environmental and human health risks, including carcinogenesis, have been raised. This study sought to investigate how previously identified lung cancer prognostic biomarkers and the related cancer signaling pathways are affected in the mouse lung following pharyngeal aspiration of well-dispersed MWCNT. A total of 63 identified lung cancer prognostic biomarker genes and major signaling biomarker genes were analyzed in mouse lungs (n = 80) exposed to 0, 10, 20, 40, or 80 μg of MWCNT by pharyngeal aspiration at 7 and 56 days post-exposure using quantitative PCR assays. At 7 and 56 days post-exposure, a set of 7 genes and a set of 11 genes, respectively, showed differential expression in the lungs of mice exposed to MWCNT vs. the control group. Additionally, these significant genes could separate the control group from the treated group over the time series in a hierarchical gene clustering analysis. Furthermore, 4 genes from these two sets of significant genes, coiled-coil domain containing-99 (Ccdc99), muscle segment homeobox gene-2 (Msx2), nitric oxide synthase-2 (Nos2), and wingless-type inhibitory factor-1 (Wif1), showed significant mRNA expression perturbations at both time points. It was also found that the expression changes of these 4 overlapping genes at 7 days post-exposure were attenuated at 56 days post-exposure. Ingenuity Pathway Analysis (IPA) found that several carcinogenic-related signaling pathways and carcinogenesis itself were associated with both the 7 and 11 gene signatures. Taken together, this study identifies that MWCNT exposure affects a subset of lung cancer biomarkers in mouse lungs.
Keywords: Nanoparticles, Lung cancer, Biomarker, Gene expression, Molecular network analysis
Introduction
Carbon nanotubes (CNT) represent an important class of engineered nanomaterials. They can be synthesized either as single-walled carbon nanotubes (SWCNT) or as multi-walled carbon nanotubes (MWCNT). SWCNT consist of a single sheet of graphite rolled-up in the form of a cylinder with a diameter of 1–2 nm and lengths ranging up to several micrometers. MWCNT consist of several stacked single wall carbon nanotubes with diameters up to 100 nm and lengths up to several micrometers (Pacurari et al., 2010). Due to their unique physicochemical properties, MWCNT have been widely used for various industrial applications, including, but not limited to, supercapacitors, batteries, automotive industry, aerospace industry, electronics, pharmaceutics, bio-engineering, medical devices, and biomedicine (Pacurari et al., 2010).
Concern over potential MWCNT-induced toxicity has emerged, particularly due to the structural similarity between asbestos and MWCNT. Asbestos is a group of naturally occurring hydrated silicate minerals with fibrous morphology (Pacurari et al., 2010). They consist of fibers of various lengths and diameters, with lengths ranging from 0.1 to greater than 200 μm. It has been well known that asbestos exposure leads to the development of pulmonary fibrosis (asbestosis), bronchogenic lung cancer, pleural plaques, and malignant mesothelioma (Pacurari et al., 2010). Numerous studies have demonstrated that fiber length, diameter, and durability are critical factors involved in the pathogenicity of asbestos (Pacurari et al., 2010). Although MWCNT have a tendency to bundle together, MWCNT aerosols may contain many long, thin, fiber-like nanotube structures similar to asbestos (Pacurari et al., 2010). In addition, the resistance of MWCNT to high temperature or acid treatment indicates that these engineered fibers are durable. It has been proposed that these unique asbestos-like properties of MWCNT may potentially induce pathogenic and carcinogenic effects (Service, 1999; Donaldson et al., 2006; Tian et al., 2006). Indeed, several studies reported that MWCNT induce inflammation, granuloma formation, and biopersistence in the rodent lung after 60 days post-exposure (Muller et al., 2005; Elgrabli et al., 2008; Porter et al., 2010). Other studies have shown that MWCNT exposure increases the genotoxic potential both in vivo and in vitro (Muller et al., 2008).
Recently, our group conducted an in vivo dose-response and time course study of MWCNT exposure in mice in order to investigate the ability of MWCNT to induce pulmonary inflammation, damage, and fibrosis (Porter et al., 2010). Mice were exposed to 0, 10, 20, 40, or 80 μg of MWCNT by pharyngeal aspiration. At 1, 7, 28, and 56 days post-exposure, MWCNT-induced pulmonary toxicity was evaluated. The results demonstrate that pulmonary inflammation and damage were dose-dependent, appeared 1 day post-exposure, and peaked 7 days post-exposure. In contrast, morphometric analysis of lung tissue from this study by Mercer et al. (Mercer et al., 2011) indicates that MWCNT-induced interstitial fibrosis increased at day 28 and progressed through 56 days post-exposure. These results indicate that MWCNT exposure rapidly produces significant pulmonary inflammation, damage, and fibrosis.
Several studies have indicated that fibrosis, a pulmonary fibrotic scarring, could be a precursor to lung cancer. A clinical study found that lung scarring was associated with elevated lung cancer risk, and furthermore, they found that pulmonary scarring and lung cancer occurred to the same lung and extended over time (Yu et al., 2008). In a review over a 21 year period of lung cancer patients, it was found that 45% of all peripheral lung cancers originated in a lung scar (Auerbach et al., 1979). A study with computed tomographic (CT) scans and pathologic specimen analysis found that 47 patients of 57 histologically proved lung cancers had pulmonary fibrosis, indicating a possible association between lung cancer and fibrosis (Sakai et al., 2003).
A casual relationship between lung fibrosis and lung cancer has been observed in crystalline silica exposed patients. Literature review indicated that an increased risk of lung cancer among patients with silicosis, a progressive lung fibrosis, might be an effect of the lung fibrosis rather than a direct effect of silica exposure (Peretz et al., 2006). Analysis of the relationship between tuberculosis-induced lung fibrosis and lung cancer indicated that there is an association between tuberculosis and increased lung cancer risk (Shiels et al., 2011), which implicates chronic pulmonary scarring in the etiology of lung cancer.
One study indicated that pulmonary fibrosis may not be a precursor for lung cancer, rather it may increase the susceptibility of fibrosis patients to develop a lung cancer over time. A comparative study of metaplastic epithelia in lungs of usual interstitial pneumonia (UIP), idiopathic pulmonary fibrosis, with or without lung cancer showed that 32 of 70 UIP autopsy cases had lung cancer, and quantitative assessment of the metaplastic epithelia in lungs demonstrated that squamous metaplasia occurred more frequently in UIP with lung cancer than in UIP without lung cancer (Hironaka and Fukayama, 1999).
Molecular biological analysis also supports that lung fibrosis and lung cancer could be associated. Immunohistochemical analysis found that TGF-beta expression is increased in asbestos-induced fibrosis (Jagirdar et al., 1997). TGF-beta is a ubiquitous and essential regulator of cellular proliferation, differentiation, migration, cell survival, and angiogenesis (Elliott and Blobe, 2005). Alterations in the TGF-beta have been associated with human cancers, including lung cancer. TGF-beta signaling promotes epithelial to mesenchymal transition, a process by which cells lose their epithelial characteristics and acquire more migratory and metastatic mesenchymal properties (Toonkel et al., 2010). An increase in TGF-beta expression in fibrosis may imply that fibrotic scarring may increase the risk of carcinogenesis. Several biological and pathological characteristics of lung fibrosis are similar to that of lung cancer, including genetic alterations, uncontrolled proliferation, and tissue invasion (MacKinnon et al., 2010; Vancheri et al., 2010).
Although more research is needed for the assessment of the clinical outcome of lung fibrosis, Vancheri et al. (Vancheri et al., 2010) suggested that the abnormal fibroblast proliferation observed in idiopathic pulmonary fibrosis may be associated with the development of lung cancer. The current study was designed to follow up our previous investigation of MWCNT-induced pulmonary inflammation, damage, and fibrosis in the mouse. The aim of this study was to successfully profile MWCNT-induced gene expression changes within a set of previously identified lung cancer biomarkers (Guo et al., 2006; Guo et al., 2008) using MWCNT-exposed mouse lungs from a previous study (Porter et al., 2010). Ultimately, the association between MWCNT-induced pulmonary inflammation, damage and fibrosis and genetic markers for lung carcinogenesis was investigated.
In our previous lung cancer-related genome-wide DNA microarray analysis, a 35-gene signature was identified from 86 lung adenocarcinoma patients in order to predict tumor recurrence (Guo et al., 2006). This 35-gene prognostic signature was further validated in 348 non-small cell lung cancer (NSCLC) patients using their transcriptional profiles generated from microarray studies (Guo et al., 2008). The gene expression of the 35 biomarkers was confirmed with real-time RT-PCR analysis of independent snap-frozen human lung cancer tumors (Guo et al., 2008). In the Director’s Challenge Study, a separate set of patient cohorts from multiple hospitals was analyzed, and a new 12-gene signature was identified from genome-wide DNA microarray analysis. This 12-gene signature provides accurate prognostic stratification of 442 lung adenocarcinoma patients including those with early stage tumors (Wan et al., 2010). We hypothesize that expression profiling of lung cancer biomarkers in MWCNT-exposed mouse lungs will identify a subset of biomarkers affected by the exposure, suggesting a possible association between MWCNT exposure and lung cancer risk. These biomarkers could be relevant for early detection or medical surveillance of lung cancer risk in occupationally-exposed workers. In the current study, in order to identify biomarkers relevant for risk prediction of lung carcinogenesis after MWCNT exposure, quantitative RT-PCR low density arrays (LDA) were designed to measure the gene expression profiles of the identified 47 lung cancer prognostic markers (Guo et al., 2006; Guo et al., 2008; Wan et al., 2010) as well as several major signaling transduction pathway genes in lung tissues from mice exposed to different doses of MWCNT at 7 and 56 days post-exposure (Table 1).
Table 1.
Information for the 63 genes analyzed in this study.
| Gene Symbol | Gene name | Assay ID | Function | Pathways | Remarks |
|---|---|---|---|---|---|
| Ahnak | Desmoyokin | Mm00613028_m1 | Protein:protein binding | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Akt1 | Protein kinase B subunit alpha | Mm01331624_m1 | Kinase activity | Signal transduction | Oncogen2 ((Sun et al., 2001) |
| Arhgap19 | Rho GTPase activating protein-19 | Mm01198653_m1 | GTPase activity | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Arhgdig | Rho-related GDP- dissociation inhibitor | Mm00433576_m1 | Cell signaling protein | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Atp5a1 | ATP synthase alpha- subunit-1 | Mm00431960_m1 | Atpase, hydrogen- transporting, ATP synthase activity | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) | |
| Atp6v0d1 | vacuolar (V−) ATPase proton pump subunit D1 | Mm00442694_m1 | Atpase H+-exporting | Organelles acidification | Prognostic maker for lung cancer (Wan et al., 2010) |
| Atp8a2 | P4-type ATPases or Phosphatidylserine Flippase | Mm00443740_m1 | Atpase, negative regulation of cell proliferation, aminophospholipid transport | Transport across membranes | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Atrx | Alpha thalassemia/mental retardation syndrome X- linked | Mm01225442_m1 | Transcriptional regulator, chromatin remodeling | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Bcl2 | B-cell leukemia/lymphoma-2 | Mm00477631_m1 | Protein binding, anti-apoptosis | Signal transduction | |
| Cav1 | Caveolin-1 | Mm00483057_m1 | Vasculogenesis, endocytosis | Tumor suppressor gene candidate | |
| Ccdc99 | Coiled-coil domain containing −99 | Mm00510371_m1 | Establishment of mitotic spindle orientation | Cell cycle | Prognostic marker for lung cancer (Wan et al., 2010) |
| Chd4 | Chromodomain helicase DNA binding protein-4 | Mm01190896_m1 | Transcription regulator | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Cox2 | Prostaglandin G/H synthase-2 | Mm01307329_m1 | Inflammation, mitogenesis | Signal transduction | |
| Creb3 | cAMP responsive element binding protein 3 | Mm00457268_m1 | Transcription factor | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Dhh | Deserted hedgehog | Mm01310203_m1 | Protein binding, peptidase activity, cell-cell signaling | Signal transduction | |
| Dlc1 | Deleted in liver cancer-1 | Mm00507957_m1 | GTPase activity | Signal transduction | A candidate tumor suppressor gene; Prognostic marker for lung cancer (Wan et al., 2010) |
| E2f4 | E2F transcription factor-4 | Mm00514160_m1 | Transcriptional factor, cell cycle apoptosis | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Egf | Epidermal growth factor | Mm01316968_m1 | Growth factor | Signal transduction | Oncogene; Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Erk1/Mapk 3 | Extracellular signal- regulated kinase-1 | Mm00662375_g1 | Serine/threonine kinase activity | Signal transduction | |
| Erk2/Mapk1 | Extracellular signal- regulated kinase-2 | Mm00442479_m1 | Serine/threonine kinase activity | Signal transduction | |
| Fam164A | Family with sequence similarity 164, member A | Mm00557410_m1 | Unknown | Unknown | Prognostic marker for lung cancer (Wan et al., 2010) |
| Fut7 | Fucosyltransferase-7 | Mm01330673_g1 | Protein glycosylation | Metabolism | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Gli1 | Glioma-associated oncogene homolog -1 | Mm00494645_m1 | DNA, protein binding | Transcription | |
| Gnb1 | Guanine nucleotide binding protein (G protein), beta polypeptide -1. | Mm00515002_m1 | GTPase activity | Signal transduction | |
| Gpx3 | Plasma gluthatione peroxidase-3 | Mm00492427_m1 | Response to oxidative stress | Antioxidant activity | |
| HnrnpA3 | Heterogeneous nuclear ribonucleoprotein-A3 | Mm00817148_g1 | mRNA binding, protein binding | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Igl-v1 | Immunoglobulin lambda variable 1–44 | Mm01611614_m1 | Signaling in immune system | Immunity | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Ihh | Indian hedgehog | Mm01259021_m1 | Development processes | Signal transduction | |
| Ilf3 | Interleukin enhancer- binding factor-3 | Mm00494960_m1 | DNA, protein binding | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Insr | Insulin receptor | Mm01211875_m1 | Growth factor receptor, insulin receptor | Signal transduction | Oncogene; Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Lmf1 | Lipase maturation factor-1 | Mm00512942_m1 | Maturation of specific proteins in ER, transport of lipoprotein lipase | Protein transport | Prognostic marker for lung cancer (Wan et al., 2010) |
| Mark2 | MAP/microtubule affinity-regulating kinase-2 | Mm00433039_m1 | Serine/threonin kinase activity | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Msx2 | Muscle segment homeobox gene-2 | Mm00442992_m1 | Transcription suppressor genes | Transcription | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Mt3 | Metallothionin-3 | Mm00496661_g1 | Cellular metal ion homeostasis, removal of superoxide radicals, binds to heavy metals | Antioxidant activity | Oncogene; Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Nos1 | Nitric oxide synthase-1 | Mm00435175_m1 | Nitric oxide synthesis, neurotransmiter | Signal transduction | |
| Nos2 | Nitric oxide synthase-2 | Mm00440488_m1 | Response to hypoxia; positive regulation of leukocyte mediated cytotoxicity, endothelial cell proliferation, blood vessel remodeling, nitric oxide synthesis | Signal transduction | Biomarker for bronchopulmonary dysplasia |
| Nos3 | Nitric oxide syntahse-3 | Mm01164908_m1 | Nitric oxide synthesis, diverse biological functions | Signal transduction | Biomarker for prostatic carcinoma |
| Nsfl | N-ethylmaleimide- sensitive factor | Mm01243864_g1 | Vesicle transport/target membrane fusion | Cellular transport | |
| Ogt | O-linked N- acetylglucosamine (GlcNAc) transferase | Mm00507317_m1 | Glycosilation | Metabolism | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Pdpk1 | Pyruvate dehydrogenase kinase, isozyme-1 | Mm00440707_m1 | Protein kinase activity | Signal transduction | Oncogene, Prognostic marker for lung cancer (Wan et al., 2010) |
| Pik3r1 | Phosphoinositide-3- kinase, regulatory subunit-1 (alpha) | Mm00803160_m1 | Protein localization in plasma membrane | Signal transduction | |
| Prmt1 | Protein arginine methyltransferase-1 | Mm00480133_m1 | Tranferease histone methyltransferase | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Ptch1 | Patched-1 | Mm00436014_m1 | Receptor activity | Signal transduction | Tumor suppressor gene |
| Rer1 | Retention in endoplasmic reticulum-1 | Mm00471276_m1 | Endoplasmic reticulum membrane proteins | Protein retention in ER | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Scly | Selenocysteine lyase | Mm00489563_m1 | Catalyzes the decomposition of L-selenocysteine | Metabolism | Prognostic marker (Wan et al., 2010) |
| Shh | Sonic hedgehog | Mm00436527_m1 | Proliferation, differentiation, development, specification, signaling, survival, migration, growth | Signal transduction | |
| Smo | Smoothened | Mm01162710_m1 | Signal transducer | Signal transduction | |
| Smpd1 | Sphingomyelin phosphodiesterase-1, acid lysosomal | Mm00488318_m1 | Converts sphingomyelin to ceramide | Metabolism | Prognostic marker for lung cancer (Wan et al., 2010) |
| Sod1 | Superoxide dismutase-1 | Mm01344231_g1 | Oxidative stress response | Antioxidant signaling | |
| Sod2 | Superoxide dismutase-2 | Mm00449726_m1 | Oxidative stress response | Antioxidant signaling | |
| Sod3 | Superoxide dismutase-3 | Mm00448831_s1 | Oxidative stress response, xenobiotic metabolism signaling | Antioxidant signaling | |
| Stk24 | Serine/threonine kinase- 24 | Mm00520930_m1 | Protein kinase activity | Signal transduction | Prognostic marker for lung cancer (Wan et al., 2010) |
| Tal2 | T-cell acute lymphocytic leukemia-2 | Mm00447439_s1 | DNA, and protein binding factor | Transcription regulation | T cell acute lymphoblastic leukemia.Oncogene; Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Tnfsf9 | Tumor necrosis factor (ligand) superfamily, member-9 | Mm00437154_m1 | Bidirectional signal transducer | Signal transduction | Oncogene; Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Tuba1a | Tubulin, alpha-1a | Mm00846967_g1 | Microtubule- based movement, protein polymerization | Structural activity | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Uba1 | Ubiquitin-like modifier activating enzyme-1 | Mm00493988_m1 | Ubiquitin- conjugation for protein degradation | Protein degradation | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Ube2i | Ubiquitin-conjugating enzyme E2I | Mm00495850_m1 | Ubiquitin- activating protein | Protein degradation | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Vac14 | Protein Vac14 homolog | Mm00461494_m1 | Regulates the level of PtdIns(3,5) P2 | Signal transduction | Prognostic maker for lung cancer (Guo et al., 2006; Guo et al., 2008) |
| Vegfa | Vascular endothelial growth factor A | Mm01281449_m1 | Angiogenesis | Signal transduction | |
| Wif1 | WNT inhibitory factor-1 | Mm00442355_m1 | Development, Wnt receptor signaling pathway | Signal transduction | Tumor suppressor gene |
| Wnt3a | Wingless-type MMTV integration site family, member-3A | Mm00437337_m1 | Protein binding, extracellular matrix structural constituent | Signal transduction | Oncogene |
| Xpo1 | Exportin-1 | Mm00506129_m1 | Protein transporter activity | Protein transport | Prognostic marker for lung cancer (Wan et al., 2010) |
| Zak | Leucine zipper- and sterile alpha motif- containing kinase | Mm00458198_m1 | Protein kinase activity, cell cycle control | Signal transduction | Biomarker for diagnosis of cervical cancer; Prognostic marker for lung cancer (Wan et al., 2010);. Oncogene |
| 18S | 18S ribosomal RNA | Protein synthesis |
Materials and Methods
MWCNT
MWCNT used in this study were a gift from Mitsui-&-Company (MWCNT-7, lot # 05072001K28). The characterization of MWCNT has been published (Porter et al., 2010). Briefly, the bulk MWCNT exhibit a distinctive crystalline structure with the number of walls ranging from 20 to 50 walls. Overall, MWCNT trace metal contamination was 0.78%, including sodium (0.41%) and iron (0.32%) with no other metals present above 0.02%. Transmission electron microscopy (TEM) micrographs of MWCNT dispersed in dispersion medium (DM) demonstrated that DM promotes significant dispersion of MWCNT. The quantitative analysis of TEM micrographs revealed that the median length of this MWCNT sample was 3.86 μm (GSD 1.94) and the count mean width was 49 ±13.4 (S.D.) nm. The zeta potential of the MWCNT in the DM was determined to be −11mV.
Animals
Male C57BL/6J mice (7 weeks old) were obtained from Jackson Laboratories (Bar Harbor, ME). Since male mice fight and injury each other if caged together, individual mice were housed one per cage in polycarbonate isolator ventilated cages, which were provided HEPA-filtered air, with fluorescent lighting from 0700 to 1900 hours. Autoclaved Alpha-Dri virgin cellulose chips and hardwood Beta-chips were used as bedding. Mice were monitored to be free of endogenous viral pathogens, parasites, mycoplasms, Helicobacter and CAR Bacillus. Mice were maintained on Harlan Teklad Rodent Diet 7913 (Indianapolis, IN), and tap water was provided ad libitum. Animals were allowed to acclimate for at least 5 days before use. All animals used in this study were housed in an AAALAC-accredited, specific pathogen-free, environmentally controlled facility.
MWCNT pharyngeal aspiration exposure
Suspensions of MWCNT were prepared in DM as previously described by our laboratory (Porter et al., 2008). Each treatment group consisted of 8 mice. Mice were anesthetized with isoflurane (Abbott Laboratories, North Chicago, IL). When fully anesthetized, the mouse was positioned with its back against a slant board and suspended by the incisor teeth using a rubber band. The mouth was opened, and the tongue gently pulled aside from the oral cavity. A 50μl aliquot of sample was pipetted at the base of the tongue, and the tongue was restrained until at least 2 deep breaths were completed (but for not longer than 15 seconds). Following release of the tongue, the mouse was gently lifted off the board, placed on its left side, and monitored for recovery from anaesthesia. Mice received either DM (vehicle control), 10, 20, 40 or 80 μg MWCNT.
RNA extraction
Total RNA was extracted from the frozen mouse lung tissue sample (−80 °C) in RNAlater using the RNeasy Fibrous Tissue Mini Kit according to the manufacturer’s protocol (Qiagen, USA). Total RNA was eluted in RNase-free water and stored at −80°C until further analysis. The quality and the concentration of each RNA sample was determined using a Nanodrop-1000 Spectrophotometer (NanoDrop Tech, Germany).
Quantitative real-time PCR (qPCR)
cDNA was generated using a TaqMan high capacity RT kit according to manufacturer’s protocol (Applied Biosystems, CA). Real-time qPCR was performed using TaqMan microfluidic low density arrays (LDA) with custom designed genes on an ABI 7900HT Fast RT-PCR instrument (Applied Biosystems). The reports were generated using SDS2.3 software (Applied Biosystems), and the expression level of each gene was given by the number of cycles required to reach threshold fluorescence (Ct). 18S was used as an endogenous control gene.
Statistical analysis
At every time point measured, each treatment and control group consisted of eight mice. To assess the gene expression fold change in each treated group versus the control group, the gene expression (Ct value) of three replicates was first averaged for each sample. Second, each gene expression value was normalized using the average expression of 18S for the entire plate, i.e. plate-wise normalization. Third, the fold change between treatment groups versus the control group was calculated using 2−ΔΔCt method (Livak and Schmittgen, 2001). Finally, the statistical significance was assessed with two-sample t-tests using software SAS 9.2. In order to assess post-exposure effects over the time series, a two-sample t-test was also performed on the fold change values between two time points for each gene.
Functional Pathway Analysis
Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) was used to derive curated molecular interactions reported in the literature. These interactions included both physical and functional interactions, as well as interactions representing pathway relevance. In this study, in order to delineate molecular networks of genes interacting with the 7-gene or 11-gene biomarker sets, a core analysis was employed to identify the most significant biological functions and processes from the derived interactions reported in mouse studies with IPA. The statistical significance of canonical signaling pathways and associated disease and disorder functions were assessed using Fisher’s exact tests adjusted with BH tests.
Hierarchical clustering analysis and heatmap
Hierarchical two-dimensional clustering analysis was performed using genes that exhibited significant differential expression after treatment at 7 and 56 days post-exposure, using the Heatplus function in the R package. Similarity metrics were manhattan distance, and the cluster method was complete linkage. Heatmaps were then generated in the R package.
Results
A 7-gene biomarker set identified at 7 days post-exposure
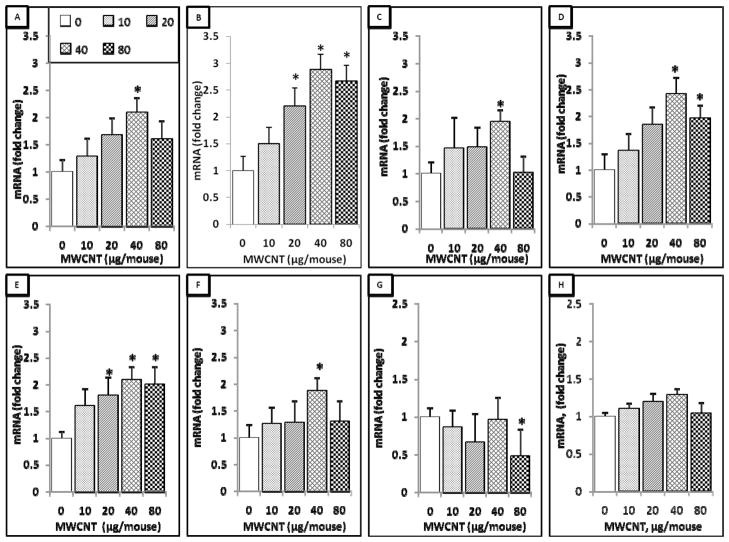
A total of 63 lung cancer prognostic and major signaling pathway biomarker genes were analyzed using qPCR assays in mouse lungs exposed by aspiration to 0, 10, 20, 40, or 80 μg of MWCNT at 7 days post-exposure. Overall, it was found that only 7 genes had different expression levels compared to the control group. These 7 genes were Rho GTPase activating protein-19 (Arhgap19), coiled-coil domain containing-99 (Ccdc99), muscle segment homeobox gene-2 (Msx2), methalothione-3 (Mt3), nitric oxide synthase-2 (Nos2), sonic hedgehog homolog (Shh), and wingless-type inhibitory factor-1 (Wif1). The expression levels of Arhgap19, Msx2, and Shh were significantly increased only at the exposure dose of 40 μg MWCNT. The expression level of Wif1 was found to be significantly decreased only at the dose of 80 μg MWCNT exposure. The expression levels of Ccdc99 and Nos2 were significantly increased at doses of 20, 40, and 80 μg of MWCNT, whereas the expression of Mt3 was significantly increased at doses of 40 and 80 μg of MWCNT (Figure 1). These results indicate that several lung cancer biomarker signature genes are affected during the earlier stage of MWCNT exposure.
Figure 1.
7-gene biomarker set in the mouse lung at 7 days post-exposure to MWCNT. Total RNA was extracted from the frozen lung tissue of mice that were exposed to different doses of MWCNT at 7 days post-exposure as indicated, followed by cDNA generation. Real-time PCR was performed to profile 63 lung cancer biomarker genes using LDA. Genes shown in the figure underwent significant changes in expression when compared to the negative control. A, Arhgap19. B, Ccdc99. C, Msx2. D, Mt3. E, Nos2. F, Shh. G, Wif1. H, 18S. Fold change was calculated after normalization to 18S and relative to control samples. Values given are mean ± SEM (p ≤ 0.05, n=8). * significantly different compared to control (unpaired t-tests).
An 11-gene biomarker set identified at 56 days post-exposure
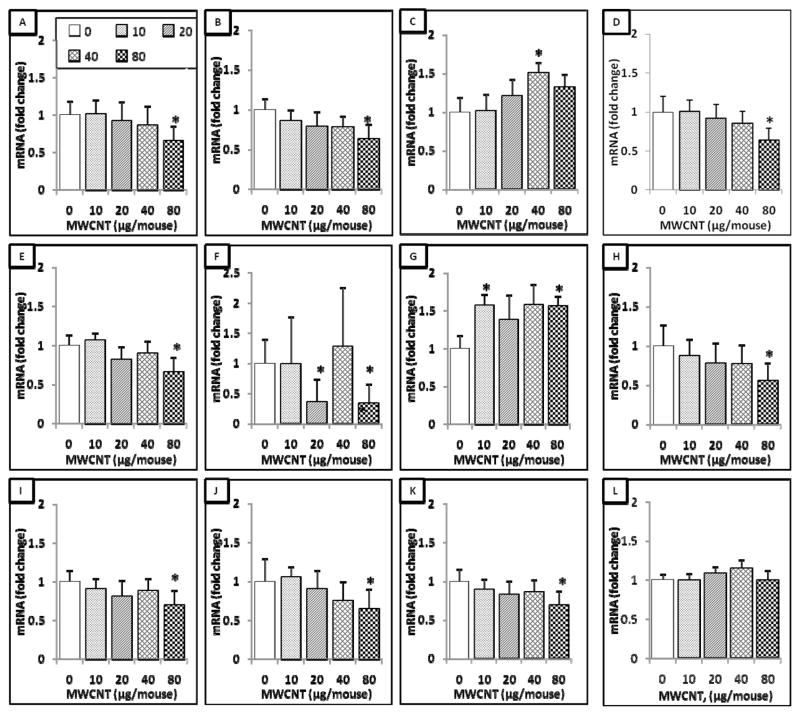
qRT-PCR analysis was applied to assess the expression changes of 63 lung cancer prognostic and major signaling biomarker genes in mouse lungs exposed by aspiration to 0, 10, 20, 40, or 80 μg of MWCNT at 56 days after a single exposure. At 56 days post-exposure, it was found overall that 11 genes exhibited differential expression levels compared to the control group (Figure 2). These 11 genes were B-cell leukemia/lymphoma-2 (Bcl2), caveolin-1 (Cav1), Ccdc9, desert hedgehog homolog (Dhh), gluthatione peroxidase-3 (Gpx3), Msx2, Nos2, phosphatidylinositol 3-kinase regulatory subunit-1 (Pi3kr1), patched-1 (Ptch1), Wif1, and leucine-zipper and sterile-alpha motif kinase (Zak) (Figure 2). Of these 11 genes, eight genes, Bcl2, Cav1, Dhh, Gpx3, Pi3kr1, Ptch1, Zak, and Wif1, had a significant decreased expression level at the highest tested dose (80 μg). The expression of Msx2 exhibited significant decreases at the doses of 20 and 80 μg. In contrast, Nos2 expression level was significantly increased at the doses of 10 and 80 μg. Ccdc99 expression was significantly increased at 40 μg of MWCNT (Figure 2). These results indicate that after 56 days post-exposure to MWCNT, the expression levels of several lung cancer biomarkers have differential expression levels compared to control samples.
Figure 2.
11-gene biomarker set in the mouse lung at 56 days post-exposure to MWCNT. Total RNA was extracted from the frozen stored lung tissue of mice that were exposed to different doses of MWCNT at 56 days post-exposure as indicated, followed by cDNA generation. Real-time PCR was performed to profile 63 lung cancer biomarker genes using LDA. Genes shown in the figure underwent significant changes in expression when compared to the negative control. A, Bcl2. B, Cav1. C, Ccdc99. D, Dhh. E, Gpx3. F, Msx2. G, Nos2. H, Pi3kr1. I, Ptch1. J, Wif1. K, Zak. L, 18S. Fold change was calculated after normalization to 18S and relative to control samples. Values given are mean ± SEM (p ≤ 0.05, n=8). * significantly different compared to control (unpaired t-tests).
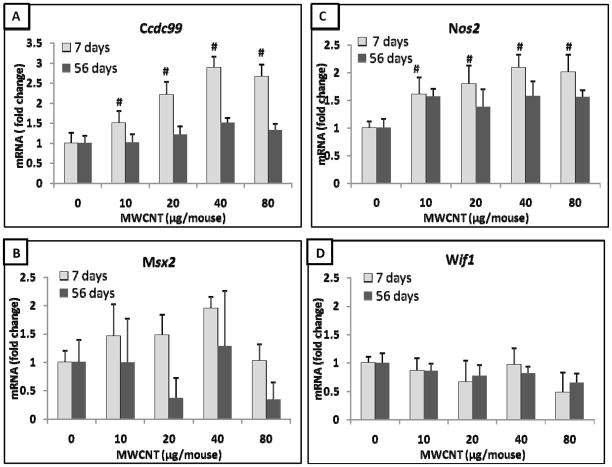
A common 4-gene biomarker set between 7 and 56 days post-exposure
Further analysis found that 4 genes appeared in the gene biomarker sets for both the 7-and 56 days post-exposure samples. These four overlapping genes were Ccdc99, Msx2, Nos2, and Wif1 (Figure 3). A comparison of the expression of these overlapping genes between 7 days (7-gene set) and 56 days (11-gene set) post-exposure samples revealed that the expression of three genes, Ccdc99, Nos2 and Msx2, decreased at 56 days post-exposure compared to their expression at 7 days post-exposure (Figure 3A-C). Statistical analysis demonstrated that the expression levels of Ccdc99 and Nos2 at 7 days were significantly higher compared with those at 56 days post-exposure (Figure 3A, 3C). These results further indicate that the expression of selected lung cancer biomarkers show differential expression levels during the more extended post-exposure interval.
Figure 3.
An overlapping 4-gene biomarker set in the mouse lung between 7- and 11-gene set at 7 and 56 days post-exposure to MWCNT, respectively. A, Ccdc99. B, Msx2. C, Nos2. D, Wif1. Fold change was calculated after normalization to 18S and relative to control samples. Values given are mean ± SEM (p ≤ 0.05, n=8). # significantly different compared to 56 days post-exposure.
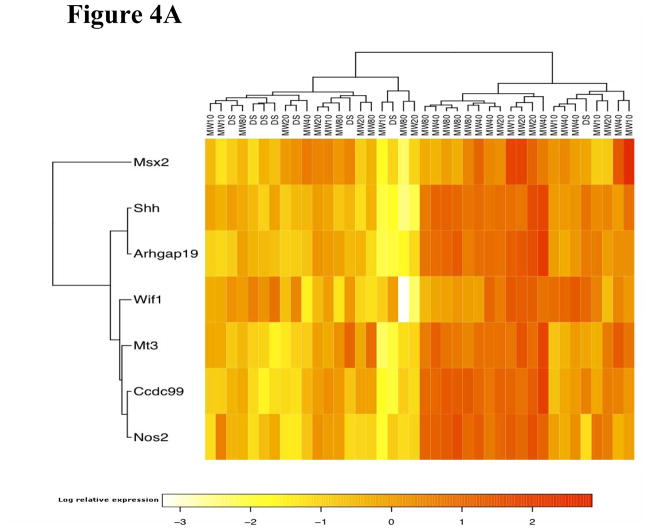
Gene clustering analysis
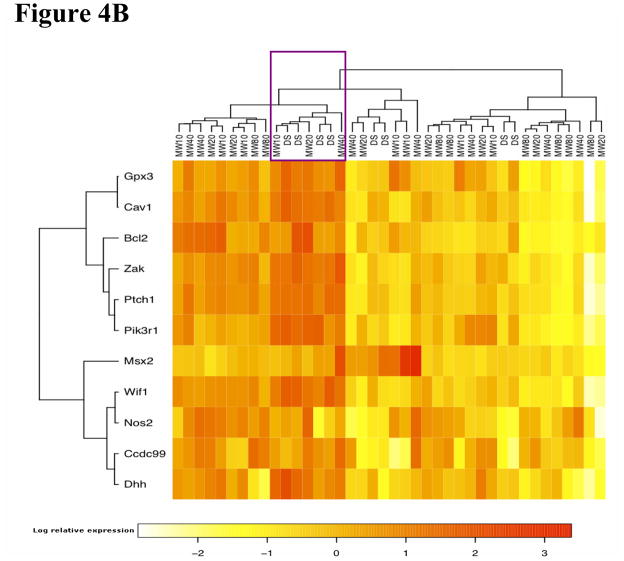
Hierarchical clustering analysis showed that the 7-gene biomarker set was able to separate MWCNT-treated animals from the control animals at 7 days post-exposure (Figure 4A). The 7-gene biomarker set showed elevated gene expression in the treated animals, except for Wif1 (Figure 4A and Figure 1). At 56 days post-exposure, the expression pattern of the 11-gene biomarker set was not as homogenous among the MWCNT-treated animals compared with that at 7 days post-exposure (Figure 4B). Most genes exhibited decreased expression in the majority of MWCNT-exposed animals compared with a middle cluster containing four vehicle control-exposed animals (marked in Figure 4B).
Figure 4.
Hierarchical clustering analysis. DS: control animal. The number after “MW” denotes the dosage used in the treatment. (A) Clustering analysis using the 7-gene biomarker set at 7 days post-exposure. (B) Clustering analysis using the 11-gene biomarker set at 56 days post-exposure. The marked cluster contains 4 control samples.
Molecular network analysis
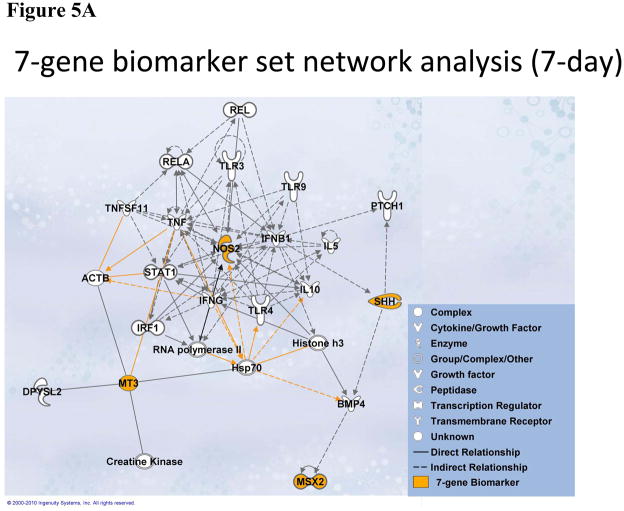
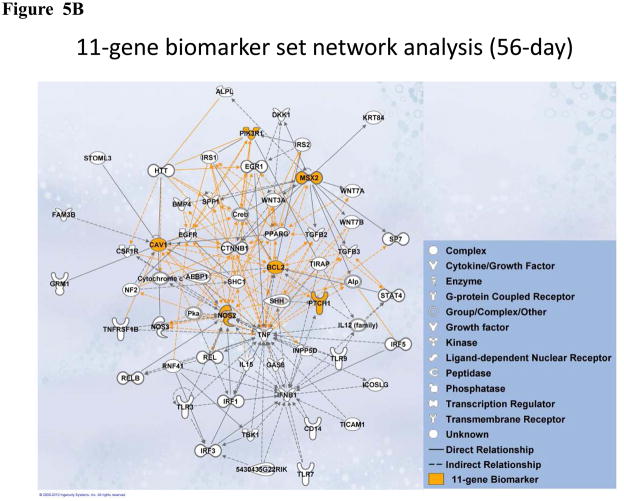
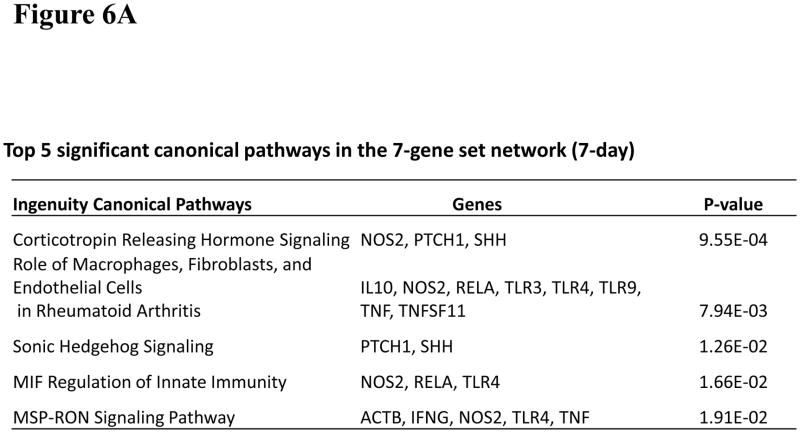
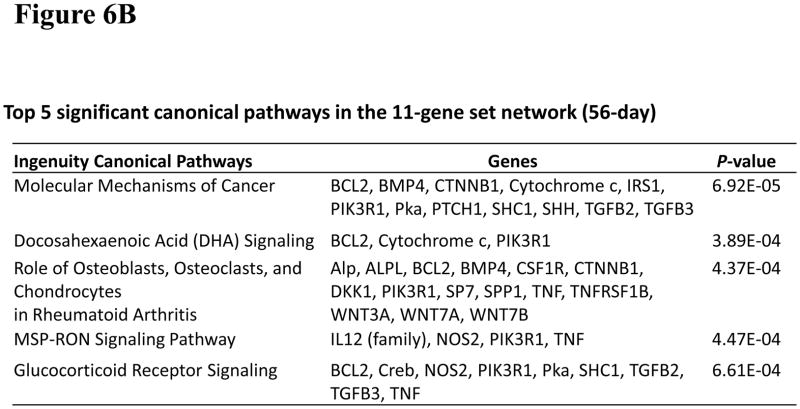
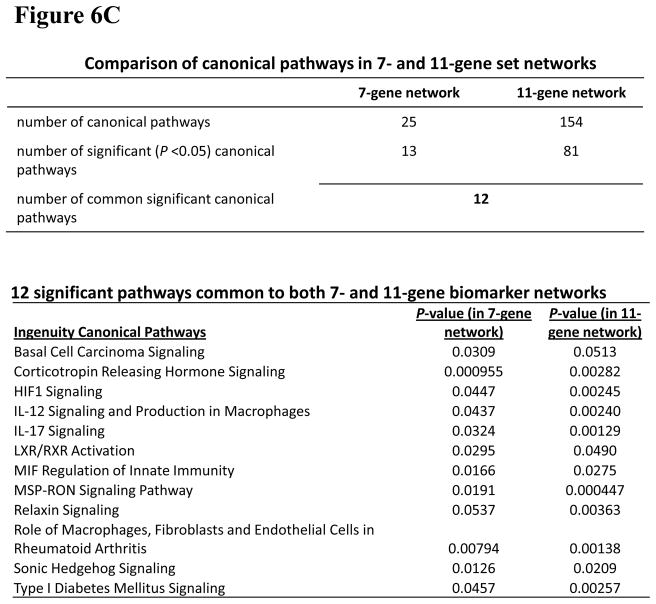
To investigate significant molecular network interactions and canonical signaling pathways associated with these differentially expressed genes, the Ingenuity Pathway Analysis (IPA) software was applied to analyze 7- and 11-gene biomarker sets, respectively. The results revealed that a total of 25 and 60 genes interacted with 7- and 11-gene biomarker sets, respectively (Figure 5). The analysis also demonstrated that 25 canonical pathways were associated with the 7-gene biomarker set. Among these 25 pathways, 13 were statistically significant (p<0.05). Ranking the pathways by their significance, the top five most significant were: corticotrophin releasing hormone signaling, the role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis, sonic hedgehog signaling, MIF regulation of innate immunity and MSP-RON signaling pathway (Figure 6A). For the 11-gene biomarker set, 154 associated canonical pathways were identified. Among these 154 pathways, 81 were statistically significant (p<0.05). Based on the ranking of the significance, the top five most significant pathways were: molecular mechanisms of cancer, docosahexaenoic acid (DHA) signaling, the role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis, MSP-RON signaling pathway and glucocorticoid receptor signaling (Figure 6B). Notably, the molecular mechanisms of cancer pathway was ranked as the most significant among the 11-gene associated canonical pathways at 56 days post-exposure.
Figure 5.
Molecular network analysis using IPA. (A). Network associated with the 7-gene biomarker set identified from animals treated with MWCNT at 7 days post-exposure. (B). Network associated with the 11-gene biomarker set identified from animals treated with MWCNT at 56 days post-exposure.
Figure 6.
Canonical pathways, diseases and functional disorders related to the networks generated from the 7- and 11-gene biomarker sets using IPA. (A). Canonical pathway analysis in the network related to the 7-gene biomarker set. (B). Canonical pathway analysis in the network related to the 11-gene biomarker set. (C). Comparison of canonical pathways in the 7- and 11-gene biomarker set network analysis. (D). Top 5 disease and disorder functions related to the 7-gene biomarker set network (at 7 day post-exposure) and the 11-gene biomarker set network (at 56 day post-exposure).
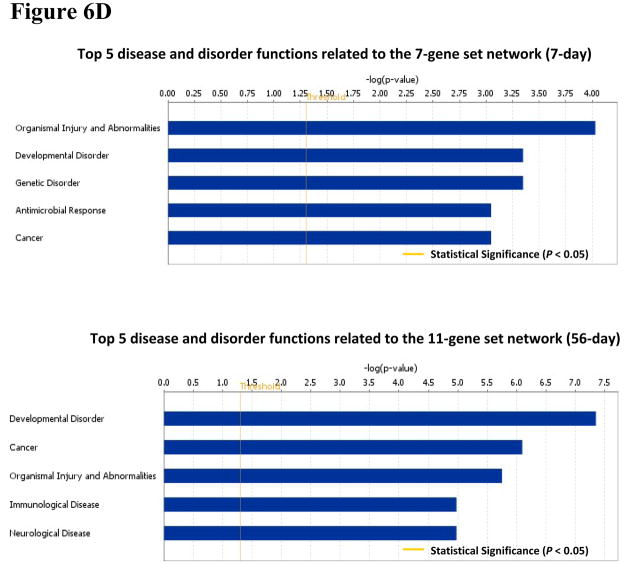
The IPA also identified 12 common canonical pathways between the 7- and 11-gene biomarker sets. Notably, basal cell carcinoma signaling pathway was ranked as the most significant among these 12 overlapping pathways (Figure 6C). An analysis of the 7- and 11-gene biomarker sets-associated potential diseases and biological function disorders showed that carcinogenesis was ranked fifth among the 7-gene set-associated diseases and function disorders (at 7 days post-exposure). For the 11-gene set network (at 56 days post exposure), carcinogenesis was ranked thesecond most significant disease function (Figure 6D). Taken together, these results indicate that exposure to MWCNT may be potentially carcinogenic in humans, which could be enhanced over a prolonged post-exposure period.
Discussion
Several studies have demonstrated that exposure to CNT substantially induces harmful effects on the lungs, including inflammatory granulomas and lung fibrosis in animal models (Lam et al., 2004; Muller et al., 2005; Shvedova et al., 2005; Porter et al., 2010). Furthermore, it has been proposed that CNT exposure can induce an asbestos-like pathogenicity and may pose a similar carcinogenic risk as exposure to asbestos fiber does (Takita et al., 1986; Donaldson et al., 2006; Poland et al., 2008). In fact, MWCNT are man-made fiber-shaped materials. The concern of their potential health hazards has been raised due to their unique physical properties, such as high aspect ratio (length/diameter), nanoscale diameters, micrometer length fiber like-shape and durability, which are analogous to asbestos fibers (Muller et al., 2005; Donaldson et al., 2006; Kobayashi et al., 2010). Particularly, the high aspect ratio and micrometer length are two important features that may contribute to the fiber toxicity (Stanton et al., 1981). Whether MWCNT fit the classical fiber ‘paradigm’ has not yet been fully established. Poland et al. (Poland et al., 2008) reported an asbestos-like acute inflammatory effect of long MWCNT when they were administered into abdominal cavity of mice. They found that the asbestos-like effects were fiber length-dependent since results showed that short fibers failed to induce abdominal inflammation or granulomas. Conversely, long fibers induced inflammation and granulomas at the peritoneal side of the diaphragm at 24 h and 7 days post-exposure. Another study by Takagi et al. (Takagi et al., 2008) reported that intra-abdominal exposure to MWCNT induced mesothelioma, a form of cancer of the lining of the body cavities, in p53+/− mice. In this study, the p53+/− mouse was chosen because it is sensitive to induction of tumors. Histopathological analysis of the peritoneal cavity of MWCNT-treated p53+/− mice found the presence of several asbestos-like pathological changes, such as fibrous scars, granulomas, and typical epithelial mesotheliomas. Particularly, the characteristics of MWCNT-induced mesotheliomas were hobnail appearance to large tumors along with high mitotic rate cells and central necrosis due to high grade of malignant mesothelioma. Furthermore, recent two studies show that MWCNT can reach the pleural tissue in mice, the site of mesothelioma, after pulmonary exposure to occupationally relevant burdens of MWCNT (Ryman-Rasmussen et al., 2009; Mercer et al., 2010) These studies conclude that the fibrous structure of MWCNT may pose a carcinogenic risk similar to that for asbestos fibers.
It has been well-established that the biopersistence and pulmonary penetration potential of asbestos are the critical factors associated with asbestos pathogenicity and carcinogenicity (Vallyathan et al., 1998; Shukla et al., 2003). Studies have shown that MWCNT exposure has a similar pattern of biopersistence and pulmonary penetration as asbestos (Elgrabli et al., 2008; Kim et al., 2010). Recent studies found that MWCNT were persistent in lungs up to 60 days post-exposure (Muller et al., 2005; Porter et al., 2010). In addition, histopathological analyses found that upon pulmonary exposure to MWCNT the particles penetrated through alveolar walls and were transported to the alveolar interstitium, subpleural tissue and subpleural lymphatics, finally reaching the intrapleural space (Porter et al., 2010).
Several in vitro studies have shown that MWCNT exposure induces genotoxicity. Zhu et al. (Zhu et al., 2007) found that MWCNT exposure induces DNA damage and increases mutation frequency in mouse embryonic stem cells. Karlsson et al.(Karlsson et al., 2008) demonstrated that MWCNT exposure significantly damages DNA in A549 type II epithelial cells. Sargent et al. (Sargent et al., 2011) found that MWCNT induce mitotic abnormality with one mitotic spindle pole, showing the evidence of the mechanism responsible for the disruption of cell division by MWCNT. Their results indicate that MWCNT are genotoxic, which may result in carcinogenic potential for MWCNT. These results are in agreement with our present results of MWCNT-induced lung cancer biomarker gene expression changes. Although whether MWCNT would induce tumors can’t be addressed in the present study, our data indicate an association between MWCNT exposure and lung cancer biomarker changes. Such an association has been used to justify a current project within NIOSH to evaluate lung tumor formation over the course of 1 year after a 2 week inhalation of MWCNT in a cancer susceptible mouse strain.
Taken together, the fibrous characteristics of MWCNT, their biopersistence, their ability to penetrate the lungs, and their genotoxicity have prompted the hypothesis that exposure to MWCNT may contribute to the initiation and progression of asbestos-like pathological responses, such as lung carcinogenesis (Donaldson et al., 2006; Pacurari et al., 2010).
In the present study, we sought to determine whether MWCNT exposure would affect lung cancer biomarker gene expression through profiling a set of lung cancer biomarker genes in mouse lungs following exposure to MWCNT. The design of this study was established based on our previous in vivo mouse model for pharyngeal aspiration of MWCNT (Porter et al., 2010). The mouse lung tissue specimens at both 7 and 56 days post-MWCNT exposure were taken from this previous investigation and were analyzed for the expression changes of a set of 63 lung cancer biomarker genes using RT-PCR analysis (Table 1). Of these 63 genes, 47 were previously shown to be prognostic markers of human lung cancer recurrence and metastasis (Guo et al., 2006; Guo et al., 2008; Wan et al., 2010), and the remaining 16 genes are well established hallmarks of cancer signaling pathways. The 47 lung cancer prognostic biomarkers were from a 35-gene (Guo et al., 2006) and a 12-gene signature (Guo et al., 2008), respectively. Both lung cancer gene signatures could identify more aggressive tumors from stage I non-small cell lung cancer tumors, which were at the early stage of cancer formation by the time of diagnosis. We have data showing that 45 out of 47 prognostic markers were also significantly differentially expressed in NSCLC tumors vs. normal lung tissues (currently unpublished results). Specifically, Msx2 and Ccdc99 were both over-expressed in lung cancer tumor tissues with a fold change of 2.3 and 1.8, respectively, which are concordant with the over-expression patterns observed in the MWCNT-treated mouse lungs in this study. The functions of these 63 genes and the associated signaling transduction pathways were extracted from the literatures and are listed in Table 1.
The results obtained from our study demonstrated that 7 genes out of these 63 lung cancer biomarker genes showed differential expression changes at 7 days post-exposure to MWCNT. These 7 genes are Arpgap19, Nos2, Shh, Wif1, Mt3, Ccdc99, and Msx2. The functional analysis of 7 genes showed that: 1) Arpgap19, Nos2, Shh, and Wif1 are involved in signaling transduction, 2) Mt3 is involved in metal detoxification, cell proliferation, and cell protection against damage, 3) Ccdc99 is involved in cell cycle regulation, and 4) Msx2 is a transcription factor and an oncogene (Table 1). Particularly, Shh, a tissue developmental and angiogenic factor, is involved in tumor angiogenesis in a variety of cancers, and Mt3, modulated by oxidative stress, plays a role in carcinogenesis (Pola et al., 2001; Garrett et al., 2002). The increase in expression of both genes upon MWCNT exposure in mouse lungs may indicate the potential molecular mechanisms related to MWCNT-induced potential carcinogenic effects. Such a relationship warrants further study.
At 56 days post-exposure to MWCNT, the gene profiling showed that 11 genes underwent different expression changes. These 11 genes are: Dhh, Nos2, Ptch1, Pik3r1, Zak, Wif1, Ccdc99, Cav1, Bcl2, Msx2, and Gpx3. The functional analysis of these 11 genes showed that: 1) Dhh, Nos2, Ptch1, Pik3r1, Zak, and Wif1 are involved in signaling transduction, 2) Ccdc99 is a cell cycle protein, 3) Cav1 is associated with plasma membrane, 4) Bcl2 is an anti-apoptotic gene, 5) Msx2 is a transcription factor and an oncogene, and 6) Gpx3 is involved in antioxidant activity (Table 1). Notably, Bcl2 is an anti-apoptotic gene and plays a major role in tumorigenesis (Coultas and Strasser, 2003). The decrease in Bcl-2 gene expression in this study indicates that MWCNT exposure may damage anti-oncogenic suppressor genes in mouse lungs.
Furthermore, these two sets of significant genes at 7 and 56 days post-exposure share 4 common genes between them. These genes are: Ccdc99, Msx2, Nos2, and Wif1 (Figure 3). The data analysis showed that the expression changes of these 4 overlapping genes at 7 days post-exposure were attenuated at 56 days post-exposure. Interestingly, our previous in vivo studies have shown that MWCNT-induced pulmonary inflammation and damage peaked at 7 days post-exposure of MWCNT (Porter et al., 2010); while interstitial fibrosis was significantly elevated at 28 days post-exposure and peaked at 56 days (Mercer et al., 2011). Therefore, the time-dependent changes in gene expression profiling correlate with the previously published results of histopathological measurements in mouse lungs, indicating a transient inflammatory/damage response and persistent fibrosis. These results demonstrate that gene profiling may well reveal alterations in the molecular processes at the level of the gene transcription and signaling transduction pathways upon MWCNT exposure in mouse lungs.
The notion of predicting chemical toxicity by gene expression profiling has been proposed and applied in genotoxicity for several decades (Aubrecht and Caba, 2005). Particularly, the recent development of high-throughput technologies has made it possible to identify key predictive biomarkers through global gene expression profiling. It has become increasingly important to use gene expression signatures identified with bioinformatics methods for toxicity predication (Shi et al., 2010), risk assessment, and screening (Afshari et al., 2011). A similar gene expression and copy number profiling study was conducted to identify important allelic imbalance in asbestos-related lung cancer (Wikman et al., 2007). Blood gene expression signatures have been used to predict exposure levels of APAP (Bushel et al., 2007). The unique advantage of these studies, as well our present study, is the ability to detect toxic injury at the molecular level and to identify the signaling pathways that lead to organ injury long before the clinical symptoms occur. In these biomarker and gene signature studies, the emphasis is on the prediction of potential risk or toxicity. In this study, the lung tissues were collected from the mice at 7 and 56 days post-exposure, which are not long enough for mice to develop lung cancer. In the case of asbestos exposure, the latency period between the first exposure and diagnosis of mesothelioma ranges from 20 to 40 years (Pacurari et al., 2010). An in vivo animal study demonstrated that asbestos exposure induces malignant mesothelioma in mouse lungs at 60 weeks post-exposure (Vaslet et al., 2002). Thus, neither asbestos nor MWCNT would be able to induce lung carcinogenesis in mice at 56 days post-exposure. In vivo animal model-generated gene profiling reveals information that approximates the complexity of the human body and its cellular, biochemical, and molecular systems that are involved in responses to chemical agents. Several studies have shown that animal model-based gene expression profiling can predict human target organ toxicities reliably for numerous human diseases, including cancer (Nuwaysir et al., 1999; Newton et al., 2004). The combined results from the gene expression profiling in this study and the histopathological changes observed in the previous study (Porter et al., 2010) indicate that exposure to MWCNT may potentially enhance the risk of pulmonary diseases, including lung carcinogenesis, in humans. Thus, results from the present study have been used to justify a further investigation by NIOSH to address whether the changes observed in gene expression in lung target cells are involved in the pathogenesis of lung cancer. Specifically, NIOSH is currently conducting a two week inhalation exposure study with histological evaluation through 1 year post-exposure in a cancer susceptible mouse strain.
Ingenuity Pathway Analysis (IPA) software analyzes the gene information from more than 200,000 published full text articles in order to create global gene networks, an interactome. At the same time, IPA relates specific biological functions and potential diseases and functional disorders to individual genes (Calvano et al., 2005; Long et al., 2008). The software is used to identify significant networks, biological functions, potential diseases and canonical signaling pathways associated with the differentially expressed gene profiles. The results obtained from IPA show a correlation between our gene expression profiling and an empirical classification of biological functions and potential diseases. Among the top five most significant canonical pathways associated with the 7-gene biomarker set, 4 pathways (corticotrophin releasing hormone signaling, Sonic hedgehog signaling, MIF regulation of innate immunity, and MSP-RON signaling pathway) are involved in carcinogenesis. Furthermore, the results of IPA showed that cancer is one of top 5 diseases and functional disorders associated with the 7-gene biomarker set. Similarly, 4 out of the 5 most significant canonical pathways associated with 11-gene biomarker set are involved in carcinogenesis and cancer is ranked as the second most significant of the potential diseases and functional disorders listed. Moreover, the comparison of results from analyses of both gene sets showed that there are 12 overlapping significant canonical signaling pathways and that basal cell carcinoma signaling is ranked as the most significant pathway.
Our IPA results also showed that MWCNT induced potentially carcinogenic alterations in the genes of mice and that these changes were time dependent. The Signaling Pathway of Molecular Mechanisms of Cancer is ranked as the most significant among the 11-gene biomarker set-associated canonical pathways at 56 days post-exposure. Notably, it is ranked much lower among the 7-gene biomarker set-associated canonical pathways at 7 days post-exposure. Furthermore, cancer is ranked as thesecond most significant among the 11-gene set-associated diseases and functional disorders whereas in the 7-gene set analysis, its significance falls to the fifth. These results are consistent with those of our histopathological measurements (Porter et al., 2010; Mercer et al., 2011). They demonstrate a shift from rapid/transient inflammation/damage to a persistent fibrotic response involving remodeling signals and growth factor damages, and as well as epithelial hyperplasia and hypertrophy (Porter et al., 2010). Such changes have been associated with initiation of carcinogenesis.
The IPA results were focused on the 7- and 11-gene sets identified in this study. As only cancer-related genes were initially analyzed in the experiments, the results would be biased toward cancer related pathways and functions, as opposed to those obtained from genome-wide expression studies. However, the identified biomarkers are involved in multiple diseases. Pathological disorders, such as organismal disorders and injuries (including fibrosis), were also retrieved from the IPA database as significant functions, indicating that the genes identified in this study were associated with the observed MWCNT-induced lung pathology.
Overall, the results obtained from this study have demonstrated that MWCNT exposure induces changes in lung cancer biomarker gene expression in mouse lungs, which may indicate a potential association between MWCNT exposure-induced lung inflammatory, damage and fibrotic responses and lung carcinogenesis. The results of the IPA also indicate that MWCNT exposure may induce alterations in several disease-related signaling transduction pathways. In light of this finding, long term inhalation studies of MWCNT to monitor possible induction of lung tumors and/or mesothelioma are justified. NIOSH is currently conducting a two week inhalation exposure to MWCNT in a cancer susceptible mouse strain with histological evaluation through 1 year post-exposure to address this issue. Additionally, the results obtained from this study indicate these gene signature alterations of MWCNT exposure and could conceivably be used for the medical surveillance of occupationally-exposed MWCNT workers.
Acknowledgments
We thank Rebecca Raese for her help in editing the manuscript. This study is supported by NIH/NLM R01LM009500 (PI: Guo) and NCRR P20RR16440 and Supplement (PD: Guo).
Footnotes
Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- Afshari CA, Hamadeh HK, Bushel PR. The Evolution of Bioinformatics in Toxicology: Advancing Toxicogenomics. Toxicological Sciences. 2011;120:S225–S237. doi: 10.1093/toxsci/kfq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrecht J, Caba E. Gene expression profile analysis: an emerging approach to investigate mechanisms of genotoxicity. Pharmacogenomics. 2005;6:419–428. doi: 10.1517/14622416.6.4.419. [DOI] [PubMed] [Google Scholar]
- Auerbach O, Garfinkel L, Parks VR. Scar cancer of the lung: increase over a 21 year period. Cancer. 1979;43:636–642. doi: 10.1002/1097-0142(197902)43:2<636::aid-cncr2820430234>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Coultas L, Strasser A. The role of the Bcl-2 protein family in cancer. Semin Cancer Biol. 2003;13:115–123. doi: 10.1016/s1044-579x(02)00129-3. [DOI] [PubMed] [Google Scholar]
- Donaldson K, Aitken R, Tran L, Stone V, Duffin R, Forrest G, Alexander A. Carbon nanotubes: a review of their properties in relation to pulmonary toxicology and workplace safety. Toxicological Sciences. 2006;92:5–22. doi: 10.1093/toxsci/kfj130. [DOI] [PubMed] [Google Scholar]
- Elgrabli D, Floriani M, Abella-Gallart S, Meunier L, Gamez C, Delalain P, Rogerieux F, Boczkowski J, Lacroix G. Biodistribution and clearance of instilled carbon nanotubes in rat lung. Part Fibre Toxicol. 2008;5:20. doi: 10.1186/1743-8977-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Garrett SH, Phillips V, Somji S, Sens MA, Dutta R, Park S, Kim D, Sens DA. Transient induction of metallothionein isoform 3 (MT-3), c-fos, c-jun and c-myc in human proximal tubule cells exposed to cadmium. Toxicol Lett. 2002;126:69–80. doi: 10.1016/s0378-4274(01)00448-9. [DOI] [PubMed] [Google Scholar]
- Guo L, Ma Y, Ward R, Castranova V, Shi X, Qian Y. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clinical Cancer Research. 2006;12:3344–3354. doi: 10.1158/1078-0432.CCR-05-2336. [DOI] [PubMed] [Google Scholar]
- Guo NL, Wan YW, Tosun K, Lin H, Msiska Z, Flynn DC, Remick SC, Vallyathan V, Dowlati A, Shi X, Castranova V, Beer DG, Qian Y. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clinical Cancer Research. 2008;14:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hironaka M, Fukayama M. Pulmonary fibrosis and lung carcinoma: a comparative study of metaplastic epithelia in honeycombed areas of usual interstitial pneumonia with or without lung carcinoma. Pathol Int. 1999;49:1060–1066. doi: 10.1046/j.1440-1827.1999.00989.x. [DOI] [PubMed] [Google Scholar]
- Jagirdar J, Lee TC, Reibman J, Gold LI, Aston C, Begin R, Rom WN. Immunohistochemical localization of transforming growth factor beta isoforms in asbestos-related diseases. Environ Health Perspect. 1997;105(Suppl 5):1197–1203. doi: 10.1289/ehp.97105s51197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson HL, Cronholm P, Gustafsson J, Moller L. Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem Res Toxicol. 2008;21:1726–1732. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- Kim JE, Lim HT, Minai-Tehrani A, Kwon JT, Shin JY, Woo CG, Choi M, Baek J, Jeong DH, Ha YC, Chae CH, Song KS, Ahn KH, Lee JH, Sung HJ, Yu IJ, Beck GR, Jr, Cho MH. Toxicity and clearance of intratracheally administered multiwalled carbon nanotubes from murine lung. J Toxicol Environ Health A. 2010;73:1530–1543. doi: 10.1080/15287394.2010.511578. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Naya M, Ema M, Endoh S, Maru J, Mizuno K, Nakanishi J. Biological response and morphological assessment of individually dispersed multi-wall carbon nanotubes in the lung after intratracheal instillation in rats. Toxicology. 2010 doi: 10.1016/j.tox.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Lam CW, James JT, McCluskey R, Hunter RL. Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intratracheal instillation. Toxicological Sciences. 2004;77:126–134. doi: 10.1093/toxsci/kfg243. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long JM, Bell CW, Fagg WSt, Kushman ME, Becker KG, McCubrey JA, Farwell MA. Microarray and pathway analysis reveals decreased CDC25A and increased CDC42 associated with slow growth of BCL2 overexpressing immortalized breast cell line. Cell Cycle. 2008;7:3062–3073. doi: 10.4161/cc.7.19.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon AC, Kopatz J, Sethi T. The molecular and cellular biology of lung cancer: identifying novel therapeutic strategies. Br Med Bull. 2010;95:47–61. doi: 10.1093/bmb/ldq023. [DOI] [PubMed] [Google Scholar]
- Mercer RR, Hubbs AF, Scabilloni JF, Wang L, Battelli LA, Schwegler-Berry D, Castranova V, Porter DW. Distribution and persistence of pleural penetrations by multi-walled carbon nanotubes. Part Fibre Toxicol. 2010;7:28. doi: 10.1186/1743-8977-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RR, Hubbs AF, Scabolloni JF, Wang L, Battelli LA, Friend S, Castranova V, Porter DW. Pulmonary fibrotic resposne to aspiration of multi-walled carbon nanotubes. Particle Fibre Toxicol. 2011 doi: 10.1186/1743-8977-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Decordier I, Hoet PH, Lombaert N, Thomassen L, Huaux F, Lison D, Kirsch-Volders M. Clastogenic and aneugenic effects of multi-wall carbon nanotubes in epithelial cells. Carcinogenesis. 2008;29:427–433. doi: 10.1093/carcin/bgm243. [DOI] [PubMed] [Google Scholar]
- Muller J, Huaux F, Moreau N, Misson P, Heilier JF, Delos M, Arras M, Fonseca A, Nagy JB, Lison D. Respiratory toxicity of multi-wall carbon nanotubes. Toxicol Appl Pharmacol. 2005;207:221–231. doi: 10.1016/j.taap.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Newton RK, Aardema M, Aubrecht J. The utility of DNA microarrays for characterizing genotoxicity. Environ Health Perspect. 2004;112:420–422. doi: 10.1289/ehp.6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol Carcinog. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Pacurari M, Castranova V, Vallyathan V. Single- and Multi-Wall Carbon Nanotubes Versus Asbestos: Are the Carbon Nanotubes a New Health Risk to Humans? Journal of Toxicology and Environmental Health-Part a-Current Issues. 2010;73:378–395. doi: 10.1080/15287390903486527. [DOI] [PubMed] [Google Scholar]
- Peretz A, Checkoway H, Kaufman JD, Trajber I, Lerman Y. Silica, silicosis, and lung cancer. Isr Med Assoc J. 2006;8:114–118. [PubMed] [Google Scholar]
- Pola R, Ling LE, Silver M, Corbley MJ, Kearney M, Blake Pepinsky R, Shapiro R, Taylor FR, Baker DP, Asahara T, Isner JM. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med. 2001;7:706–711. doi: 10.1038/89083. [DOI] [PubMed] [Google Scholar]
- Poland CA, Duffin R, Kinloch I, Maynard A, Wallace WAH, Seaton A, Stone V, Brown S, MacNee W, Donaldson K. Carbon nanotubes introduced into the abdominal cavity of mice show asbestos-like pathogenicity in a pilot study. Nature Nanotechnology. 2008;3:423–428. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- Porter D, Sriram K, Wolfarth M, Jefferson A, Schwegler-Berry D, Andrew M, Castranova V. A biocompatible medium for nanoparticle dispersion. Nanotoxicology. 2008;2:144–154. [Google Scholar]
- Porter DW, Hubbs AF, Mercer RR, Wu N, Wolfarth MG, Sriram K, Leonard S, Battelli L, Schwegler-Berry D, Friend S, Andrew M, Chen BT, Tsuruoka S, Endo M, Castranova V. Mouse pulmonary dose-and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology. 2010;269:136–147. doi: 10.1016/j.tox.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Ryman-Rasmussen JP, Cesta MF, Brody AR, Shipley-Phillips JK, Everitt JI, Tewksbury EW, Moss OR, Wong BA, Dodd DE, Andersen ME, Bonner JC. Inhaled carbon nanotubes reach the subpleural tissue in mice. Nat Nanotechnol. 2009;4:747–751. doi: 10.1038/nnano.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Ono M, Nishio T, Kawarada Y, Nagashima A, Toyoshima S. Lung cancer associated with diffuse pulmonary fibrosis: CT-pathologic correlation. J Thorac Imaging. 2003;18:67–71. doi: 10.1097/00005382-200304000-00002. [DOI] [PubMed] [Google Scholar]
- Sargent LM, Reynolds SH, Hubbs AF, Benkovic SA, Lowry DT, Kashon ML, Siergrist KT, Mustovich J, Sturgeon JL, Burker KL, Dinu CZ. Understanding carbon nanotubes genotoxicity. The Toxicologist. 2011;120:A59. [Google Scholar]
- Service RF. Meeting. American Chemical Society. Raising a glass to health and nanotubes. Science. 1999;285:2053–2055. doi: 10.1126/science.285.5436.2053. [DOI] [PubMed] [Google Scholar]
- Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JD, Jr, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels MS, Albanes D, Virtamo J, Engels EA. Increased risk of lung cancer in men with tuberculosis in the alpha-tocopherol, Beta-carotene cancer prevention study. Cancer Epidemiol Biomarkers Prev. 2011;20:672–678. doi: 10.1158/1055-9965.EPI-10-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Gulumian M, Hei TK, Kamp D, Rahman Q, Mossman BT. Multiple roles of oxidants in the pathogenesis of asbestos-induced diseases. Free Radic Biol Med. 2003;34:1117–1129. doi: 10.1016/s0891-5849(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Mercer R, Murray AR, Johnson VJ, Potapovich AI, Tyurina YY, Gorelik O, Arepalli S, Schwegler-Berry D, Hubbs AF, Antonini J, Evans DE, Ku BK, Ramsey D, Maynard A, Kagan VE, Castranova V, Baron P. Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice. Am J Physiol Lung Cell Mol Physiol. 2005;289:L698–708. doi: 10.1152/ajplung.00084.2005. [DOI] [PubMed] [Google Scholar]
- Stanton MF, Layard M, Tegeris A, Miller E, May M, Morgan E, Smith A. Relation of particle dimension to carcinogenicity in amphibole asbestoses and other fibrous minerals. J Natl Cancer Inst. 1981;67:965–975. [PubMed] [Google Scholar]
- Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A, Hirose A, Nishimura T, Fukumori N, Ogata A, Ohashi N, Kitajima S, Kanno J. Induction of mesothelioma in p53+/− mouse by intraperitoneal application of multi-wall carbon nanotube. Journal of Toxicological Sciences. 2008;33:105–116. doi: 10.2131/jts.33.105. [DOI] [PubMed] [Google Scholar]
- Takita M, Arisaka K, Kajita T, Kifune T, Koshiba M, Miyano K, Nakahata M, Oyama Y, Sato N, Suda T, Suzuki A, Takahashi K, Totsuka Y. Search for neutron-antineutron oscillation in 16O nuclei. Phys Rev D Part Fields. 1986;34:902–904. doi: 10.1103/physrevd.34.902. [DOI] [PubMed] [Google Scholar]
- Tian F, Cui D, Schwarz H, Estrada GG, Kobayashi H. Cytotoxicity of single-wall carbon nanotubes on human fibroblasts. Toxicol In Vitro. 2006;20:1202–1212. doi: 10.1016/j.tiv.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Toonkel RL, Borczuk AC, Powell CA. Tgf-beta signaling pathway in lung adenocarcinoma invasion. J Thorac Oncol. 2010;5:153–157. doi: 10.1097/JTO.0b013e3181c8cc0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallyathan V, Green F, Ducatman B, Schulte P. Roles of epidemiology, pathology, molecular biology, and biomarkers in the investigation of occupational lung cancer. J Toxicol Environ Health B Crit Rev. 1998;1:91–116. doi: 10.1080/10937409809524546. [DOI] [PubMed] [Google Scholar]
- Vancheri C, Failla M, Crimi N, Raghu G. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J. 2010;35:496–504. doi: 10.1183/09031936.00077309. [DOI] [PubMed] [Google Scholar]
- Vaslet CA, Messier NJ, Kane AB. Accelerated progression of asbestos-induced mesotheliomas in heterozygous p53+/− mice. Toxicol Sci. 2002;68:331–338. doi: 10.1093/toxsci/68.2.331. [DOI] [PubMed] [Google Scholar]
- Wan YW, Sabbagh E, Raese R, Qian Y, Luo D, Denvir J, Vallyathan V, Castranova V, Guo NL. Hybrid models identified a 12-gene signature for lung cancer prognosis and chemoresponse prediction. Plos One. 2010:5. doi: 10.1371/journal.pone.0012222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikman H, Ruosaari S, Nymark P, Sarhadi VK, Saharinen J, Vanhala E, Karjalainen A, Hollmen J, Knuutila S, Anttila S. Gene expression and copy number profiling suggests the importance of allelic imbalance in 19p in asbestos-associated lung cancer. Oncogene. 2007;26:4730–4737. doi: 10.1038/sj.onc.1210270. [DOI] [PubMed] [Google Scholar]
- Yu YY, Pinsky PF, Caporaso NE, Chatterjee N, Baumgarten M, Langenberg P, Furuno JP, Lan Q, Engels EA. Lung cancer risk following detection of pulmonary scarring by chest radiography in the prostate, lung, colorectal, and ovarian cancer screening trial. Arch Intern Med. 2008;168:2326–2332. doi: 10.1001/archinte.168.21.2326. discussion 2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chang DW, Dai L, Hong Y. DNA damage induced by multiwalled carbon nanotubes in mouse embryonic stem cells. Nano Lett. 2007;7:3592–3597. doi: 10.1021/nl071303v. [DOI] [PubMed] [Google Scholar]